Abstract
Physical interactions between genetic elements located throughout the genome play important roles in gene regulation and can be identified with the Chromosome Conformation Capture (3C) methodology. 3C converts physical chromatin interactions into specific ligation products, which are quantified individually by PCR. Here we present a high-throughput 3C approach, 3C-Carbon Copy (5C), that employs microarrays or quantitative DNA sequencing using 454-technology as detection methods. We applied 5C to analyze a 400-kb region containing the human β-globin locus and a 100-kb conserved gene desert region. We validated 5C by detection of several previously identified looping interactions in the β-globin locus. We also identified a new looping interaction in K562 cells between the β-globin Locus Control Region and the γ–β-globin intergenic region. Interestingly, this region has been implicated in the control of developmental globin gene switching. 5C should be widely applicable for large-scale mapping of cis- and trans- interaction networks of genomic elements and for the study of higher-order chromosome structure.
Intense efforts are under way to map genes and regulatory elements throughout the human genome (ENCODE Project Consortium 2004). These studies are expected to identify many different types of elements, including those involved in gene regulation, DNA replication, and genome organization in general. Analysis of only 1% of the human genome has already revealed that genes are surrounded by a surprisingly large number of putative regulatory elements (data available at http://genome.cse.ucsc.edu/encode/).
In order to fully annotate the human genome and to understand its regulation, it is important to map all genes and functional elements and also to determine all relationships between them. For instance, all regulatory elements of each gene must be identified. This endeavor is complicated by the fact that the genomic positions of genes and elements do not provide direct information about functional relationships between them. A well-known example is provided by enhancers that can regulate multiple target genes that are located at large genomic distances or even on different chromosomes without affecting genes immediately next to them (Spilianakis et al. 2005; West and Fraser 2005).
Recent evidence indicates that regulatory elements can act over large genomic distances by engaging in direct physical interactions with their target genes or with other elements (Dekker 2003; de Laat and Grosveld 2003; Chambeyron and Bickmore 2004; West and Fraser 2005). These observations indicate that the genome may be organized as a complex three-dimensional network that is determined by physical interactions between genes and elements. Therefore, we hypothesize that functional relationships between genes and regulatory elements can be determined by analyzing this network through mapping of chromatin interactions.
Physical interactions between elements can be detected with the Chromosome Conformation Capture (3C) method (Dekker et al. 2002; Dekker 2003; Splinter et al. 2004; Miele et al. 2006). 3C uses formaldehyde cross-linking to covalently trap interacting chromatin segments throughout the genome. Interacting elements are then restriction-enzyme-digested and intramolecularly ligated (Fig. 1A). The frequency with which two restriction fragments become ligated is a measure of the frequency by which they interact in the nucleus (Dekker et al. 2002).
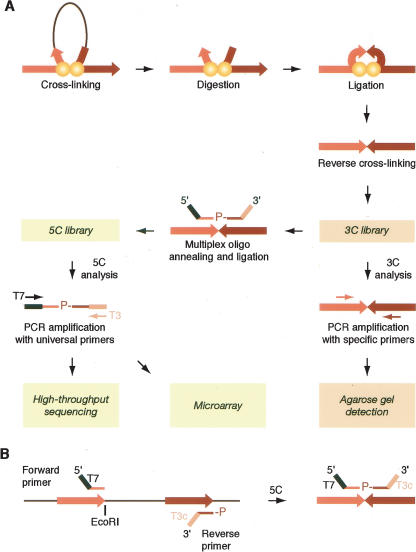
Figure 1.
Schematic representation of 5C. (A) A 3C library is generated by conventional 3C and then converted into a 5C library by annealing and ligating 5C oligonucleotides in a multiplex setting. 5C libraries are then analyzed on a microarray or by quantitative sequencing. (B) 5C primer design. Forward 5C primers anneal to the sense strand of the 3′-end of restriction fragments and include half of the selected restriction site. All forward primers feature a common 5′-end tail containing the T7 promoter sequence. Reverse 5C primers anneal to the antisense strand of the 3′-end of restriction fragments, including half of the restriction site. All reverse primers contain a common 3′-end tail featuring the complementary T3 sequence (T3c) and are phosphorylated at the 5′-end. 5C forward and reverse primers anneal to the same strand of head-to-head ligation products present in the 3C library.
3C was initially used to study the spatial organization of yeast chromosome III (Dekker et al. 2002) and has since been applied to the analysis of several mammalian loci such as the β-globin locus (Tolhuis et al. 2002; Palstra et al. 2003; Vakoc et al. 2005), the T-helper type 2 cytokine locus (Spilianakis and Flavell 2004), the immunoglobulin κ locus (Liu and Garrard 2005), and the Igf2 imprinted locus (Murrell et al. 2004). These studies revealed direct interactions between enhancers and promoters of target genes, with the linking DNA looping outward. 3C was also used to detect trans interactions between yeast chromosomes (Dekker et al. 2002) and between functionally related elements located on different mouse chromosomes (Spilianakis et al. 2005; Ling et al. 2006; Xu et al. 2006). Together, these studies suggest that long-range cis and trans interactions play widespread roles in the regulation of the genome and that 3C is a convenient approach to map this network of interactions.
3C uses PCR to detect individual chromatin interactions, which is particularly suited for relatively small-scale studies focused on the analysis of interactions between a set of candidate elements. However, PCR detection is not conducive to ab initio and large-scale mapping of chromatin interactions. To overcome this problem, 3C libraries need to be analyzed using a high-throughput detection method such as microarrays or DNA sequencing. The extreme complexity of the 3C library and the low relative abundance of each specific ligation product make direct large-scale analysis difficult. Here we present a novel 3C-based methodology for large-scale parallel detection of chromatin interactions. We refer to this method as 3C-Carbon Copy, or “5C.” 5C uses highly multiplexed ligation-mediated amplification (LMA) to first “copy” and then amplify parts of the 3C library followed by detection on microarrays or by quantitative DNA sequencing.
5C was developed and validated by analyzing the human β-globin locus and a conserved gene desert region located on human chromosome 16. We find that 5C quantitatively detects several known DNA looping interactions. Interestingly, 5C analysis also identified a looping interaction between the β-globin Locus Control Region (LCR) and the γ–δ intergenic region. Previously, several lines of evidence have suggested that this region plays a role in regulating the developmentally controlled switching from γ-globin expression in fetal cells to β-globin expression in adult cells (Calzolari et al. 1999; Gribnau et al. 2000).
5C should be widely applicable to determine the cis and trans connectivity of regulatory elements throughout large genomic regions. In addition, 5C experiments can be designed so that complete interaction maps can be generated for any large genomic region of interest, which can reveal locations of novel gene regulatory elements and may also provide detailed insights into higher-order chromosome folding.
Results
The 3C method has been described in detail (Dekker et al. 2002; Splinter et al. 2004; Vakoc et al. 2005; Dekker 2006; Miele et al. 2006) and is illustrated in Figure 1A. A 3C experiment generates a complex library of ligation products that reflects all chromatin interactions that occur throughout the genome. The abundance of each specific ligation product in the library is a measure of the frequency of interaction of the two corresponding loci.
In a typical 3C analysis, individual interaction frequencies are determined by quantifying the formation of predicted “headto-head” ligation products using semiquantitative PCR (Fig. 1A). As a PCR control, a library is used that contains all ligation products in equimolar amounts. The control library is generated by mixing equimolar amounts of minimally overlapping BAC clones covering the genomic region of interest (Palstra et al. 2003; Dekker 2006). This mixture is then digested and randomly ligated. Interaction frequencies are determined by calculating the ratio of PCR product obtained with the 3C library and the amount obtained with the control library.
Outline of the 5C technology
We have developed 5C to detect ligation products in 3C libraries by multiplex LMA (Fig. 1A). LMA is widely used to detect and amplify specific target sequences using primer pairs that anneal next to each other on the same DNA strand (Fig. 1; Landegren et al. 1988; Li et al. 2005; Peck et al. 2006). Only primers annealed next to each other can be ligated. Inclusion of universal tails at the ends of 5C primers allows subsequent amplification of ligated primers. LMA-based approaches are quantitative and can be performed at high levels of multiplexing using thousands of primers in a single reaction (Fan et al. 2004; Bibikova et al. 2005; Hardenbol et al. 2005; Wang et al. 2005).
To analyze chromatin interactions by 5C, a 3C library is first generated using the conventional 3C method. A mixture of 5C primers is then annealed onto the 3C library and ligated. Two types of 5C primers are used: 5C forward and 5C reverse primers. These primers are designed so that forward and reverse primers anneal across ligated junctions of head-to-head ligation products present in the 3C library (Fig. 1A,B). 5C primers that are annealed next to each other are then ligated with Taq ligase. This step generates a 5C library, which is amplified with universal PCR primers that anneal to the tails of the 5C primers. Forward and reverse 5C primers are only ligated when both are annealed to a specific 3C ligation product. Therefore, the 3C library determines which 5C ligation products are generated and how frequently. As a result, the 5C library is a quantitative “carbon copy” of a part of the 3C library, as determined by the collection of 5C primers.
Forward and reverse 5C primers are designed to contain a unique sequence corresponding to the sense and antisense strand of the 3′-ends of restriction fragments, respectively (Fig. 1B; for more details, see the Supplemental material). The primers also contain universal tails for amplification (T7 at the 5′-ends of forward primers and T3c at the 3′-ends of reverse primers) (Fig. 1B). To analyze interactions between many restriction fragments, multiple forward and reverse primers are mixed together in the same multiplex 5C reaction. Since predicted forward and reverse primers of each restriction fragment are complementary, only one primer per fragment, either a forward or a reverse, can be used in a given 5C experiment. To facilitate ligation, all reverse primers are phosphorylated at their 5′-ends. This 5C primer design allows simultaneous amplification of all potential interactions between all restriction fragments recognized by a forward primer and all those recognized by a reverse primer.
Chromatin looping in the human β-globin locus
We developed and optimized the 5C approach by analyzing the human β-globin locus. This locus was selected because several looping interactions have previously been detected by 3C as well as by a second method, RNA-TRAP (Carter et al. 2002; Tolhuis et al. 2002), and therefore detection of these looping interactions using 5C can be used to validate the new method.
The human β-globin locus consists of five developmentally regulated β-globin-like genes (ε, HBE; Aγ and Gγ, HBG1 and HBG2; δ, HBD; and β, HBB), one pseudogene (HBpsi), and a Locus Control Region (LCR) located upstream of the gene cluster (Fig. 2A). The LCR is characterized by five DNaseI hypersensitive sites (HS1–5) and is required for tissue-specific and position-independent expression of downstream β-globin genes (Li et al. 2002; Stamatoyannopoulos 2005). Previous 3C analysis of the murine β-globin locus revealed transcription-factor-mediated looping interactions between the LCR and transcribed globin genes (Drissen et al. 2004; Vakoc et al. 2005). The LCR was also found to interact with HS elements located upstream (HS-62.5/ HS-60) and downstream (3′-HS1) from the locus (Tolhuis et al. 2002).
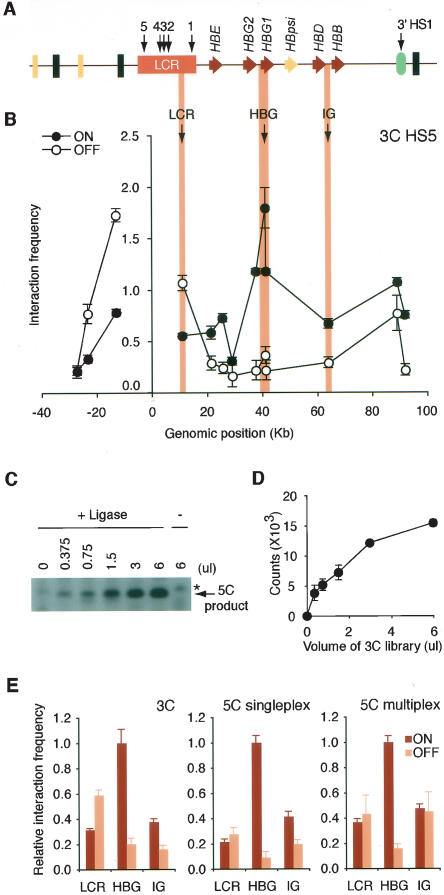
Figure 2.
Analysis of the human β-globin locus and development of 5C. (A) Schematic representation of the human β-globin locus. The β-globin genes and pseudogene are illustrated by red and yellow arrows, respectively. The positions of HSs are indicated with black arrows. Olfactory genes and pseudogenes are represented by black and yellow rectangles, respectively. The location of a gene of unknown function (EST BU661736) is indicated by a green oval. (B) 3C analysis of interactions between the LCR (HS5) and the rest of the β-globin locus. Interaction frequencies were measured by semiquantitative PCR in K562 (locus ON) and GM06990 (locus OFF) cells. The y-axis indicates normalized interaction frequencies; the x-axis shows the genomic position relative to the LCR. Each data point is the average of at least three PCR reactions; bars represent the standard error of the mean (S.E.M.). (C) Representative 3C library titration in singleplex LMA with 5C primers. Increasing volumes of 3C library from K562 cells were analyzed with gene desert 5C primers, and the amplified 5C ligation products were analyzed on an agarose gel. An asterisk indicates a nonspecific band. (D) Quantification of titration shown in C. Each data point corresponds to the average of three PCR reactions; bars represent the S.E.M. (E) 3C, singleplex, and sixplex LMA detection of looping interactions between HS5 and the Aγ-globin gene HBG1. Interaction frequencies from both cell lines were expressed relative to the K562 LCR–Aγ-globin interaction (HBG1), which was set at 1. Each histogram value represents the average of at least three PCRs; bars represent the S.E.M.
We first verified the presence of chromatin loops in the human β-globin locus using the conventional 3C method. The locus was analyzed in the erythroleukemia cell line K562 and in the EBV-transformed lymphoblastoid cell line GM06990. K562 cells express high levels of ε- and γ-globin, whereas GM06990 cells do not express the β-globin locus (Supplemental Fig. 1). 3C libraries were generated from both cell lines and a control library, which was generated using a series of minimally overlapping BAC clones (see Methods). Interaction frequencies between the EcoRI fragment overlapping the HS5 element of the LCR and EcoRI restriction fragments throughout the β-globin locus were determined by PCR. To allow direct quantitative comparison of interaction frequencies determined in K562 cells and GM06990 cells, interaction frequencies were normalized using a set of 12 interaction frequencies detected in a control region, a conserved gene desert region on chromosome 16 (ENCODE region ENr313) (ENCODE Project Consortium 2004).
The normalized results are presented in Figure 2B. In both cell lines, we find that HS5 interacts frequently with adjacent DNA fragments. These interactions likely reflect nonfunctional random collisions resulting from the intrinsic close proximity of neighboring restriction fragments (Dekker et al. 2002; Dekker 2006). The frequent random interactions between adjacent genomic fragments are likely dependent on local physical properties of the chromatin fiber and limit the ability to detect specific looping interactions between elements separated by small genomic distances (2–5 kb) (Dekker 2006; Gheldof et al. 2006). Importantly, random collisions are predicted to decrease progressively for sites separated by increasingly large genomic distances. In K562 cells, high interaction frequencies were observed specifically between the LCR and a restriction fragment located ∼40 kb downstream and overlapping the Aγ-globin gene (HBG1), indicating the presence of a strong looping interaction. We also detected a frequent interaction between the LCR and the 3′-HS1 element. Interestingly, this interaction was also present in GM06990 cells. Previous studies of the murine locus have shown that the analogous interaction also occurs in nonexpressing erythroid precursor cells (Palstra et al. 2003).
Finally, our analysis revealed less frequent random collisions between neighboring restriction fragments around the LCR in K562 cells as compared to GM06990 cells. We have observed similar differences in random collisions around the active and inactive FMR1 promoter (Gheldof et al. 2006) and have proposed that these differences may reflect transcription-dependent modulation of chromatin expansion or changes in subnuclear localization.
Based on this analysis, we conclude that the conformation of the human β-globin locus is comparable to the murine locus with looping interactions between the LCR and 3′-HS1 in both expressing and nonexpressing blood-derived cells. The interaction between the LCR and the active Aγ-globin gene is only observed in globin-expressing K562 cells.
LMA detection of 3C ligation products
We used detection of chromatin loops in the β-globin locus to develop and optimize the 5C technology. LMA was first performed with a single pair of 5C forward and reverse primers to verify that this method can quantitatively detect a ligation product in the context of a 3C library. We designed a 5C primer pair that recognizes a ligation product that is formed by two adjacent restriction fragments located in the gene desert control region. LMA was performed with this primer pair in the presence of increasing amounts of 3C library (generated from K562 cells), and the formation of ligated forward and reverse primers was quantified by PCR amplification with the pair of universal T7 and T3 primers. We find that ligation of 5C primers is not observed when nonspecific DNA is present, is dependent on the amount of the 3C library, and requires Taq ligase (Fig. 2C,D). We conclude that LMA can be used to quantitatively detect ligation products present in the 3C library.
LMA detection of looping between the LCR and the Aγ-globin gene
We determined whether singleplex LMA can be used to quantitatively detect chromatin looping interactions in the β-globin locus. We focused on interactions involving the LCR and three diagnostic fragments in the β-globin locus (indicated as bars in Fig. 2A): a restriction fragment located just downstream from the LCR, a restriction fragment that overlaps the Aγ-globin gene HBG1, and a restriction fragment located in between the δ- and β-globin genes. We designed a reverse 5C primer for the restriction fragment overlapping HS5 of the LCR and forward 5C primers for the three other fragments. The linear range of 5C detection was determined with individual pairs of 5C primers in the presence of increasing amounts of 3C libraries (from K562 and GM06990 cells) or control library (Supplemental Fig. 2).
Interaction frequencies between HS5 and the three other sites in the β-globin locus were determined by calculating the amount of ligated 5C primers obtained with the 3C library and the amount obtained with the control library. The interaction frequency between the two adjacent restriction fragments located in the gene desert control region was used for normalization. Normalized interaction frequencies are shown in Figure 2E, middle panel. We find that the data obtained with LMA closely reproduced the 3C data, including the looping interaction between the LCR and the Aγ-globin gene.
We then tested whether the four interaction frequencies studied here (three in the β-globin locus and one in the control region) can be detected and quantified in a multiplex LMA reaction. We performed LMA with a mix of six 5C primers and used PCR with specific primers to quantify the frequency with which specific pairs of 5C primers were ligated. We then calculated normalized interaction frequencies as described above. We again obtained similar results as with conventional 3C (Fig. 2E, right panel). Together, these experiments demonstrate that LMA can be used to quantitatively detect chromatin interactions.
Generation of complex 5C libraries using highly multiplexed LMA
Comprehensive 5C analysis of chromatin interactions throughout large genomic regions would require high levels of multiplexing in combination with a high-throughput method for analysis of 5C libraries. Therefore, we tested LMA at higher levels of multiplexing and explored two high-throughput detection methods to analyze 5C libraries: microarrays and quantitative DNA sequencing.
We designed 5C reverse primers for each of the three EcoRI restriction fragments that overlap the LCR and 5C forward primers for 55 restriction fragments throughout a 400-kb region around the LCR. This primer design allows detection of looping interactions between each of the three sections of the LCR and the surrounding chromatin in parallel in a single experiment (see below). We also designed 10 5C forward and 10 5C reverse primers throughout a 100-kb region in the gene desert control region. Forward and reverse primers were designed to recognize alternating restriction fragments. This primer design scheme allows the detection of a matrix of interactions throughout the control region (see below).
We performed LMA with a mixture of all 78 5C primers using 3C libraries from K562 and GM06990 and the control library as templates. Each 5C library contained up to 845 different 5C ligation products (the products of 13 reverse primers and 65 forward primers). These products included 165 possible interactions within the β-globin locus, 100 interactions throughout the gene desert, and 590 interactions between the two genomic regions. We verified that the 5C libraries represented quantitative copies of the selected fraction of the 3C libraries. To do this, we again analyzed the same set of four interaction frequencies as in Figure 2E. We used specific PCR primers to quantify the abundance of specific 5C ligation products in the 5C libraries and determined normalized interaction frequencies between the LCR and the three positions of the β-globin locus as described above. We found that the 5C data closely reproduced the 3C interaction profile in both cell lines (Supplemental Fig. 3) with strong looping interactions between the LCR and the Aγ-globin gene HBG1 in K562 cells.
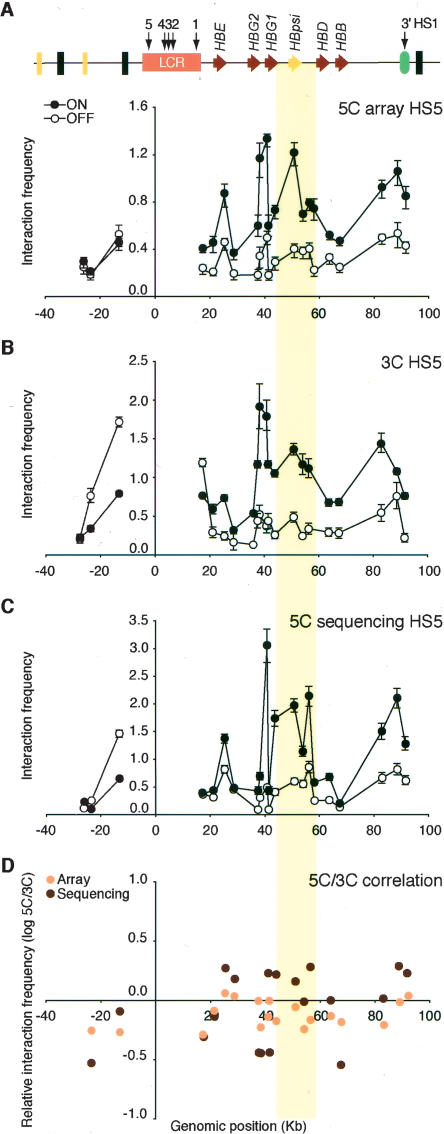
Figure 3.
Microarray and DNA sequencing analysis of 5C libraries recapitulates 3C interaction profiles. (A) 5C analysis of human β-globin locus HS5 chromatin interactions in K562 (ON) and GM06990 (OFF) cells detected by microarray. The y-axis indicates normalized interaction frequencies. The x-axis indicates the genomic position relative to the LCR. Each data point is the average of six hybridization signals obtained with probes of 38–48 bases, and error bars represent the S.E.M. A schematic representation of the entire locus is presented at the top. (B) Conventional 3C analysis of human β-globin HS5 chromatin interactions. (C) 5C analysis of human β-globin HS5 interactions as detected by quantitative DNA sequencing. Error bars indicate the standard deviation. We assumed that count data were drawn for a Poisson distribution and, thus, that the standard deviation equals the square root of the count number. Standard deviations were propagated using the delta method. (D) Correlation between 3C and 5C human β-globin locus profiles from K562 cells. Microarray and DNA sequencing 5C results were compared to corresponding 3C data by calculating the fold difference (log ratio 5C/3C) for each pair of interacting fragments (y-axis). The x-axis indicates the genomic position relative to the LCR. The colored vertical bar across all panels highlights the interaction between the LCR and the region between the γ- and δ-globin genes that contains the β-globin pseudogene.
Analysis of 5C libraries on microarrays and by quantitative sequencing
We tested whether microarray detection and quantitative sequencing can be used for comprehensive analysis of the composition of 5C libraries. First, to facilitate microarray detection, we amplified the 5C libraries described above with Cy3-labeled universal primers. The labeled 5C libraries were then hybridized to a custom-designed microarray that can detect specific 5C ligation products. Since each 5C product is composed of two half-sites, each corresponding to a 5C primer, cross-hybridization of non-specific 5C products can occur to probes that share one half-site. To assess half-site cross-hybridization, the microarray also contained probes that recognize only one of the 78 5C primers present in the library. To determine the optimal length of the microarray probes that allows the least cross-hybridization, each probe was spotted with 18 different lengths of half-sites ranging from 15 to 32 bases (total probe length ranging from 30 to 64 bases). 5C libraries were hybridized to the array, and specific and half-site hybridization was quantified. We found that probes that are composed of two half-sites with a length ranging from 19 to 24 bases displayed the lowest relative level of cross-hybridization of half-sites (see Supplemental Fig. 4). Data obtained with these six feature lengths were averaged, and interaction frequencies were calculated by dividing the hybridization signal obtained with a 5C library by the signal obtained with the control library (see below; Supplemental Table 7).
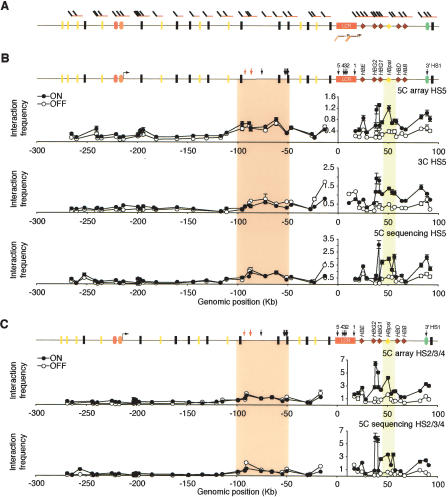
Figure 4.
Large-scale 5C analysis of the human β-globin locus. (A) Positions of forward (top) and reverse (bottom) 5C primers within the β-globin locus. (B) Chromatin interaction profile of HS5 with a 400-kb region surrounding the LCR. Physical interactions in K562 (ON) and GM06990 (OFF) cells were measured by 5C and microarrays (top), 3C (middle), and 5C and quantitative sequencing (bottom) analysis. The β-globin locus illustrated above the profiles includes features described in Figure 2A. The forward-facing arrow indicates the location of an alternative ε-globin transcription start site. Light and dark orange ovals represent the Ubiquilin 3 (UBQLN3) and MGC20470 hypothetical genes, respectively. Arrows indicate HSs identified in K562 cells. Red arrows indicate HSs identified in K562 cells that have been proposed to be orthologous to HS-62.5/-62.0 in the murine locus (Farrell et al. 2002; Bulger et al. 2003). (C) Chromatin interaction profile of HS2/3/4 of the LCR with the 400-kb region around the LCR as determined by microarray (top) and quantitative sequencing (bottom). Colored vertical bars highlight the interactions between the LCR and upstream HSs and the γ–δ intergenic region.
Second, we analyzed the composition of 5C libraries by quantitative sequencing. 5C libraries are composed of linear DNA molecules that all are ∼100 bp long, which makes them ideally suited for high-throughput single-molecule pyrosequencing. We generated similar 5C libraries as used for microarray detection, except that five of the 65 forward primers were left out. We analyzed 5C and control libraries using the GS20 platform developed by 454 Life Sciences Corp. (Margulies et al. 2005). For each library, we obtained at least 160,000 sequence reads (Supplemental Table 4). For each sequence, we determined which pair of ligated 5C primers it represented and the number of times each specific 5C ligation product was sequenced was counted (see Methods; Supplemental Table 5). As each ligation product was sequenced many times (the median count for intrachromosomal interactions was 133 for K562, 53 for GM06990, and 134 for the control library), a quantitative determination of interaction frequencies was obtained. Interaction frequencies were calculated by dividing the number of times a 5C product was sequenced in a 5C library by the number of times it was sequenced in the control library (see Supplemental Table 6).
Large-scale 5C analysis of the β-globin locus
We first analyzed the interaction profiles of HS5 throughout the 100-kb β-globin locus as detected on the microarray (Fig. 3A) and by quantitative sequencing (Fig. 3C). For comparison, the same interaction profile was also determined by conventional 3C, and data were normalized using interaction frequencies determined within the control region (Fig. 3B). Both microarray detection and quantitative sequencing reproduced the overall 3C interaction profile of the β-globin locus in K562 and GM06990 cells. In all three data sets, we found that the LCR specifically and strongly interacted with the γ-globin genes in K562 cells. In both cell lines, we detected the looping interaction between the LCR and the 3′-HS1 element.
Interestingly, 3C and 5C analyses also revealed strong interactions between the LCR and a region located between the γ- and δ-globin genes in K562 cells. This region contains the β-globin pseudogene, which is weakly expressed in K562 cells but is silent in GM06990, and the initiation site for an intergenic transcript (Gribnau et al. 2000).
We compared the 5C and 3C data sets directly by calculating for each pair of interacting fragments the fold difference between their interaction frequencies as determined by 5C and 3C. We find that the difference in 5C data obtained by microarray detection and conventional 3C is generally less than twofold (Fig. 3D). However, larger differences were observed when 5C data obtained by quantitative sequencing were compared to 3C data (Fig. 3D). This may be due to the fact that the dynamic range of quantitative sequencing appears to be higher than that of semiquantitative PCR or microarrays, which results in higher peaks and lower valleys in the profile obtained by sequencing as compared to semiquantitative PCR. We also determined the correlation between 5C and 3C data directly by plotting the data in a scatter plot (Supplemental Fig. 5). We found a high degree of correlation (r 2 = 0.73 for the data set obtained by sequencing and r 2 = 0.75 for the data set obtained on the microarray).
Figure 5.
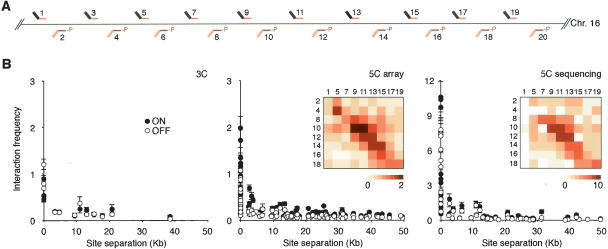
Analysis of the conformation of the gene desert control region. (A) Positions of alternating 5C forward (top) and reverse (bottom) primers throughout the gene desert control region. (B) Chromatin interaction frequencies of the gene desert region as determined by conventional 3C (left panel), by 5C and microarrays (middle panel), and by 5C and quantitative sequencing (right panel). Interaction frequencies are plotted vs. genomic distance between the interacting fragments. Bars represent the S.E.M. Interaction frequencies detected in K562 cells are also presented as two-dimensional heat maps in which the color of each square is a measure of the interaction frequency. Note: Primers 3, 6, and 20 recognize repetitive sequences. These primers were included to generate 5C libraries, but data obtained with these primers are not included in B. Interaction frequencies obtained with these primers are presented in Supplemental Table 7.
Taken together, these results provide clear proof of principle that 5C in conjunction with microarray detection or quantitative sequencing is a powerful methodology to quantitatively detect chromatin interactions in a high-throughput setting.
Interactions between HS5 and upstream elements
The mouse LCR interacts with HS elements (HS-62.5/HS-60) located up to 40 kb upstream of the LCR (Tolhuis et al. 2002; Palstra et al. 2003). It is not known whether functionally equivalent interacting elements are present in this region of the human genome. It has been noted that olfactory receptor genes located ∼90 kb upstream of the LCR are orthologous to ones located 40 kb upstream of the murine locus (Bulger et al. 2000), indicating that these regions are related. In addition, the murine HS-62.5/ HS-60 element is embedded in a sequence that is similar to a sequence located ∼90 kb upstream of the human LCR (Bulger et al. 2003). These observations indicate that the region located ∼90 kb upstream of the human LCR is orthologous to the region located ∼40 kb upstream of the murine locus, suggesting that this region may also interact with the LCR in human cells. To assess in an unbiased fashion whether the LCR interacts with any upstream elements in the human locus, the 5C experiment described above was designed to include analysis of a large region located upstream of the LCR.
We analyzed the interaction profiles obtained by microarray detection and quantitative sequencing of HS5 with a region up to 280 kb upstream of the LCR. Both data sets showed that interactions throughout this region are generally much lower than those observed between the LCR and the β-globin locus. However, in both cell lines, we did detect elevated interaction frequencies throughout a large domain located 50–100 kb upstream of the LCR (Fig. 4A). This result was confirmed by conventional 3C analysis (Fig. 4A, cf. top, middle, and bottom panels). This region contains three olfactory receptor genes and multiple HS sites (Bulger et al. 2003).
These results suggest that the region located 50–100 kb upstream of the LCR in the human genome is in relatively close proximity to the LCR and therefore may be functionally equivalent to the genomic region located 40 kb upstream of the LCR in the murine locus. In addition, these results illustrate that large-scale mapping of interactions using 5C can greatly facilitate the discovery of the locations of novel putative regulatory elements.
Parallel analysis of multiple interaction profiles
A major advantage of 5C is the fact that interactions between multiple elements of interest and other genomic elements can be analyzed in parallel in a single experiment. The 5C experiment described here was designed to illustrate this aspect of the methodology. As described above, 5C forward and reverse primers were designed to allow simultaneous detection of interaction profiles of each of the three subsections of the LCR with the 400 kb of surrounding chromatin. Data obtained by microarray analysis and quantitative sequencing of 5C libraries showed that the interaction profile of the restriction fragment overlapping HS2/3/4 of the LCR fragment is very similar to that of HS5 (Fig. 4C). Interestingly, we found that in K562 cells, HS2/3/4 interacted more frequently with sites throughout the β-globin locus than HS5, suggesting that these HSs may contribute most to the formation of the chromatin loops with the LCR. 5C analysis of the LCR 3′-end, which contains HS1, did not yield sufficient levels of ligation products to obtain a significant number of sequence reads. Analysis of microarray hybridization signals confirmed the low levels of ligation products formed with the 5C primer for HS1, but the general patterns of interaction frequencies for K562 and GM06990 cells were consistent with the patterns obtained for HS5 and HS2/3/4 (Supplemental Table 7).
Large-scale 5C analysis of the gene desert control region
The 5C analysis of the β-globin locus was focused on the mapping of interactions between a fixed regulatory element, the LCR, and the surrounding chromatin. 5C experiments can also be designed so that a more global data set is obtained, which is particularly useful when the positions of regulatory elements are poorly defined. Here we provide an example of an alternative 5C primer design scheme that provides insights into the general spatial conformation of a genomic region.
The 5C analysis described above included 10 forward and 10 reverse 5C primers for restriction fragments located in the gene desert control region (Fig. 5A). Importantly, forward and reverse primers were designed for alternating restriction fragments (Fig. 5A). Combined, these primers detected chromatin interactions throughout the region. Interaction frequencies determined by microarray analysis and quantitative sequencing were plotted against the genomic distance between the interacting restriction fragments. In both cell lines, interaction frequencies were found to decrease with increasing genomic distance (Fig. 5B). Similar results were obtained by conventional 3C analysis.
The graphs in Figure 5 do not reveal where along the chromosome particular interaction frequencies were measured. To better illustrate that a global interaction map is obtained, we also presented interaction frequencies between forward and reverse 5C primers as two-dimensional heat maps in which the color of each square is an indication of the interaction frequency between restriction fragments. Interaction frequencies displayed along the diagonal reflect interactions between fragments located closely together along the chromosome. The overall pattern of interactions observed in this gene desert region is very different from that observed in the β-globin locus and is consistent with random-coil behavior of the chromatin fiber without long-range looping interactions (Rippe 2001; Dekker et al. 2002; Dekker 2006).
Discussion
The development of 3C has greatly facilitated the detection and study of cis and trans interactions between genes and regulatory elements. Here we developed 5C technology, a novel extension of 3C that should significantly expand the range of 3C applications by allowing comprehensive and large-scale mapping of chromatin interactions. Large-scale application of 5C will provide information about relationships between genes and regulatory elements and can be used to identify novel regulatory elements and to reveal higher-order chromosome structural features.
Validation of 5C through analysis of the β-globin locus
We have validated 5C by detection of chromatin looping interactions in the human β-globin locus. The most prominent interaction was observed between the LCR and the expressed γ-globin genes, which was specifically observed in γ-globin-expressing K562 cells. In addition, in both K562 and GM06990 cells, the LCR also interacted with the 3′-HS1 element and a large domain located 50– 100 kb upstream. The latter region corresponds to the region around HS-62.5/ HS-60 in the murine locus (Farrell et al. 2002; Bulger et al. 2003). Similar long-range interactions between HSs were observed in the mouse. The clustering of these HSs has been proposed to create a chromatin hub, or a specialized nuclear compartment dedicated to the transcription of the β-globin genes (Tolhuis et al. 2002; de Laat and Grosveld 2003). Several of the HSs in the β-globin locus bind the insulator binding protein CTCF (Farrell et al. 2002; Bulger et al. 2003) (and for human HS5, see Supplemental Fig. 6), and this protein has been proposed to mediate their interactions and formation of the chromatin hub (Patrinos et al. 2004). However, the functional significance of some of these interactions is not well understood as deletion of some of the elements does not directly affect β-globin expression (Bender et al. 2006). Our observation that interactions between several of the HSs also occur in GM06990 cells, which will never express β-globin, suggests that they may not directly regulate β-globin expression but are involved in some aspect of higher-order chromosome architecture.
The interaction profiles of HS5 and HS2/3/4 are very similar, except that HS2/3/4 interacted more frequently with the β-globin locus specifically in K562 cells. This result is in agreement with observations that HS2 and HS3 have the strongest enhancer activity (Fraser et al. 1993; Peterson et al. 1996). In addition, RNA-TRAP showed that the expressed globin gene interacted most strongly with HS2 (Carter et al. 2002).
We identified a new chromatin looping interaction between the LCR and the region between the γ- and δ-globin genes. This result is intriguing given that this region has been implicated in developmental control of the β-globin locus (O'Neill et al. 1999; Chakalova et al. 2005). This region contains a β-globin pseudogene that is activated in K562 cells (Supplemental Fig. 1), suggesting that the looping interaction may be involved in transcription activation. This region also contains a promoter for a large intergenic transcript whose expression may be related to activation of the adult β-globin gene (Gribnau et al. 2000). Certain patients who suffer from hereditary persistence of fetal hemoglobin carry deletions in this region and display defects in β-globin expression (Chakalova et al. 2005). In contrast to some other looping interactions in the locus, CTCF may not play a role in the interaction between the LCR and the γ–δ intergenic regions, as we did not detect CTCF binding to several sites within the intergenic region, despite the presence of several weak putative CTCF binding sites (Supplemental Fig. 6).
Microarray detection and quantitative sequencing
Results obtained with microarray detection, quantitative sequencing and semiquantitative PCR are generally very comparable. However, we also observed several differences. First, the dynamic range of microarray detection was smaller than that of quantitative sequencing, as has been observed before (Yuen et al. 2002). Second, small quantitative differences were observed between the data sets obtained by microarray analysis, quantitative sequencing, and semiquantitative PCR, e.g., in the γ–δ intergenic region (Figs. 3, 4). These differences may reflect intrinsic biases in the detection methods, but may also be due to experimental variation between independently generated 5C libraries.
Both detection methods have advantages. DNA sequencing displays a larger dynamic range and obviates the need to design a specific array for each genomic region of interest. On the other hand, microarray analysis is currently more cost-effective, particularly when a given genomic region needs to be analyzed under a large number of different conditions.
5C data obtained by DNA sequencing allowed an estimate of the background of the LMA-based approach. We quantified 451 interactions between the β-globin locus and the control gene desert region, which are located on different chromosomes. These interactions are detected by forward primers located on one chromosome and reverse primers located on the other, and vice versa. There is no biological indication that the β-globin locus and the gene desert region should preferentially interact. Therefore, we reasoned that these interchromosomal interaction frequencies would correspond to background signals. Generally, we detected very low background interaction frequencies between the two regions (average interaction frequency 0.08 [standard error of the mean, or S.E.M. = 0.02] for K562 and 0.08 [S.E.M. = 0.01] for GM06990) (Supplemental Table 6), which is 75-fold lower than the interaction frequency between HS2/3/4 and the β-globin gene in K562 cells. However, we also detected a few higher interaction frequencies. We do not know whether these represent true interchromosomal contacts, false positives due to primer design, or experimental noise. Future large-scale 5C analyses will provide more detailed insights into these issues.
5C applications
5C relies on multiplexed ligation-mediated amplification and thus is potentially limited by the number of 5C primers that can be used in a single reaction. Other assays based on LMA have successfully used many thousands of primers in a single reaction. For example, the methylation status of 1534 CpG sites was assessed using a mixture of ∼6000 primers (Bibikova et al. 2005). Another example is the use of highly multiplexed LMA with up to 20,000 Molecular Inversion Probes in a single reaction to detect single nucleotide polymorphisms (SNPs) (Hardenbol et al. 2005; Wang et al. 2005). When 5C is performed at a similar level of multiplexing, e.g., using 10,000 5C primers in a single experiment, up to 25 million distinct chromatin interactions can be detected in parallel involving up to 40 Mb (10,000 4-kb restriction fragments) of DNA.
For highly multiplexed 5C analyses, it is important to carefully design 5C primers. Nine 5C primers that were used to generate the 5C libraries analyzed here perfectly recognized abundant interspersed repeats, and these primers were found to produce excessively large numbers of ligation products (see Supplemental Table 5B). Thus, repeated sequences must be avoided.
We anticipate two types of 5C applications that are distinguished by 5C primer design schemes. First, 5C can be used for large-scale mapping of chromatin looping interactions between specific genomic elements of interest, similar to the analysis of the β-globin locus described here. Such studies are focused on mapping interactions between a “fixed” element (e.g., the LCR) and other restriction fragments located in cis or in trans, in order to identify elements with which it interacts. 5C allows simultaneous quantification of interaction profiles of many such “fixed” elements in parallel in a single reaction followed by analysis on a custom-designed microarray or by direct quantitative sequencing. To do this, reverse 5C primers are designed for each fixed fragment of interest, and forward 5C primers are designed for all other restriction fragments, as shown in Figure 4A. This type of analysis will allow rapid detection of networks of interactions among multiple genes and regulatory elements throughout large segments of the genome.
Second, 5C analysis can be used to generate dense interaction maps that cover most or all potential interactions between all fragments of any genomic region. Dense interaction maps can provide a global overview of the conformation of a given genomic region. For example, when 5C forward and reverse primers are designed for alternating restriction fragments, as performed here for the gene desert control region (Fig. 5), a relatively dense matrix of interaction frequencies can quickly be obtained throughout a genomic region. Although powerful, such a 5C analysis will not provide a complete interaction map for a given genomic region, as interactions between two fragments that are both recognized by forward primers or by reverse primers cannot be detected. This limitation can be overcome by performing several 5C analyses, each with a permuted 5C primer design scheme to obtain partially overlapping interaction matrices. When combined, these maps can yield complete interaction maps containing interaction frequencies of all pairs of restriction fragments throughout a region of interest. Each row and column of such matrices would correspond to a “fixed” element experiment as described above. Generation of complete interaction matrices can be used as a discovery tool for unbiased detection of chromatin looping interactions between previously unannotated elements. Analysis of a matrix of interaction frequencies can also provide global information regarding the general spatial conformation of a genomic region (Dekker et al. 2002; Gheldof et al. 2006).
Methods
BAC selection and control library preparation
A control library for the human β-globin locus and gene desert regions (ENCODE regions ENm009 and ENr313, respectively) was generated as described (Dekker 2006; Miele et al. 2006). Briefly, an array of bacterial artificial chromosomes (BACs) covering both genomic regions was mixed, digested with EcoRI, and randomly ligated. In this study, the BAC arrays from the β-globin locus and gene desert regions were mixed in a 4:1 ratio to obtain strong signals for the β-globin locus. Interaction frequencies were adjusted accordingly. The following seven BAC clones were used for the β-globin region: CTC-775N13, RP11-715G8, CTD-3048C22, CTD3055E11, CTD-2643I7, CTD-3234J1, and RP11-589G14. A set of four BAC clones was selected to cover the 0.5-Mb gene desert region, and include RP11-197K24, RP11-609A13, RP11-454G21, and CTD-2133M23. BAC clones were obtained from Invitrogen and the Children's Hospital Oakland Research Institute (CHORI).
Cell culture and 3C analysis
The GM06990 cell line was derived from EBV-transformed B-lymphocytes and was obtained from Coriell Cell Repositories (CCR). This cell line was cultured in Roswell Park Memorial Institute medium 1640 (RPMI 1640) supplemented with 2 mM L-glutamine and 15% fetal bovine serum (FBS). The K562 cell line was obtained from the American Type Culture Collection (ATCC) and cultured in RPMI 1640 supplemented with 2 mM L-glutamine and 10% FBS. Both cell lines were grown at 37°Cin5% CO2 in the presence of 1% penicillin-streptomycin. 3C analysis was performed with log-phase GM06990 and K562 cells using EcoRI as previously described (Dekker et al. 2002; Vakoc et al. 2005; Miele et al. 2006). The primer sequences are presented in Supplemental Table 2.
Real-time PCR quantification
Total RNA from log-phase cells was isolated with the RNeasy Mini Kit as described by the manufacturer (Qiagen). cDNA was synthesized with oligo(dT)20 (Invitrogen) using the Omniscript Reverse Transcription Kit (Qiagen). β-Globin transcripts were quantified by real-time PCR in the presence of SYBR Green I stain (Molecular Probes). The specific human β-globin primers used in this analysis are summarized in Supplemental Table 1.
5C primer design
Forward and reverse primers were designed to recognize the top or bottom strand of the 3′-end of EcoRI restriction fragments. Primer homology lengths varied from 24 to 40 nucleotides, and melting temperatures were centered at 72°C. The genomic uniqueness of all primers was verified with the SSAHA algorithm (Ning et al. 2001). Forward 5C primers were designed to include a 5′-end tail that includes (5′–3′) CTG followed by one MmeI restriction site (TCCAAC) and a modified T7 Universal primer sequence (TAATACGACTCACTATAGCC). Reverse 5C primers were designed to include a 3′-end tail that includes (5′–3′) a modified complementary T3 Universal sequence (TCCCTTTAGT GAGGGTTAATA) and one MmeI restriction site (GTCGGA), followed by CTC. 5C forward and reverse primers each contained half of the EcoRI restriction site, and only the reverse primers were phosphorylated at the 5′-end. All 5C primers are presented in Supplemental Table 3.
5C library preparation
The 3C library (representing ∼150,000 genome copies) or control library (5 ng) was mixed with salmon testis DNA (Sigma) to a total DNA mass of 1.5 μg, and with 1.7 fmol of each 5C primer in a final volume of 10 μL of annealing buffer (20 mM Tris-acetate at pH 7.9, 50 mM potassium acetate, 10 mM magnesium acetate, and 1 mM DTT). Samples were denatured for 5 min at 95°C and annealed for 16 h at 48°C. Annealed primers were ligated for 1 h at 48°C by adding 20 μL of ligation buffer (25 mM Tris-HCl at pH 7.6, 31.25 mM potassium acetate, 12.5 mM magnesium acetate, 1.25 mM NAD, 12.5 mM DTT, 0.125% Triton X-100) containing 10 units of Taq DNA ligase (NEB). Reactions were terminated by incubating samples at 65°C for 10 min. 5C ligation products were amplified by PCR using forward (T7modif, CTGTCCAACTAA TACGACTCACTATAGCC) and reverse (T3modif, GAGTCCGAC TATTAACCCTCACTAAAGGGA) primers. Six microliters of ligation reaction were amplified with 10 pmol of each primer in 25-μL PCR reactions (32 cycles of 30 sec of denaturing at 95°C, 30 sec of annealing at 60°C, and 30 sec of extension at 72°C). 5C libraries were purified with MinElute Reaction Cleanup Kit (Qiagen) to remove unincorporated primers and other contaminants as recommended by the manufacturer.
Singleplex and sixplex 5C analysis
5C libraries from K562 and GM06990 (each representing ∼150,000 genomes) or control libraries (5 ng) were incubated with individual 5C primer pairs and processed as described above, except that ligation reactions were amplified by 35 PCR cycles of 30 sec of denaturing at 95°C, 30 sec of annealing at 60°C, and 30 sec of extension at 72°C. Amplified 5C ligation products were resolved on 2% agarose gels and visualized with ethidium bromide (0.5 μg/mL). Sixplex 5C analysis was performed by mixing six distinct 5C primers with 3C or control libraries. Individual 5C ligation products of sixplex samples were detected by PCR with specific internal PCR primers and measured on agarose gels as described above. Linear-range PCR detection of 5C products was verified by twofold serial dilution titrations of multiplex samples. The sequences of internal primers are available on request.
5C library microarray analysis
5C libraries were prepared by performing multiplex LMA with 78 5C primers, and amplified with a 5′-Cy3-labeled reverse PCR primer complementary to the common 3′-end tail sequence of reverse 5C primers (Cy3-T3modif). Maskless array synthesis and hybridization were carried out with 100 ng of amplified 5C libraries at NimbleGen Systems Inc. as previously described (Singh-Gasson et al. 1999; Nuwaysir et al. 2002; Kim et al. 2005; Selzer et al. 2005). Each array featured the sense strand of predicted 5C ligation products. The arrays also contained several inter-region negative controls. The arrays contained 18 replicates of increasing feature lengths ranging from 30 to 64 nt, which were used to identify optimal array probe lengths (Supplemental Fig. 4). A full description of the array design is available on request. Arrays were scanned using a GenePix4000B scanner (Axon Instruments, Molecular Devices Corp.) at 5-μm resolution. Data from scanned images were extracted using NimbleScan 2.0 extraction software (NimbleGen Systems, Inc.).
5C library high-throughput DNA sequencing analysis
5C libraries were generated with 73 5C primers, as described in the Results section. Each library was amplified with 5′-end phosphorylated PCR primers and processed for single-molecule pyrosequencing as previously described (Margulies et al. 2005). Using the GS20 platform developed by 454 Life Sciences Corp., 550,189 sequence reads totaling 60 million bases were obtained. The mean read length was 108 bases (mode, 112 bases). Each read was BLASTed against all forward and reverse primers. For each sample, the number of reads that matched each of the 682 possible primer pairs (62 forward × 11 reverse) was counted. These combinations include 159 possible interactions in the β-globin locus, 72 interactions in the gene desert region, and 451 interregion interactions. The data are summarized in Supplemental Tables 4 and 5.
ACKNOWLEDGMENTS
We thank the members of our laboratories for stimulating and helpful discussions, and Drs. J. Perry, J. Teodoro, and M. Walhout for critical reading of the manuscript. This work was supported by grants from the National Institutes of Health to J.D. (NHGRI-ENCODE HG003143), to R.D.G. (NHGRI-ENCODE HG003129), and to A.K. (CA109597). R.A.A. was supported through the NSF/ NIH Joint Program in Mathematical Biology (NIH grant R01GM078986) and Jeffrey Epstein's sponsorship of the Program for Evolutionary Dynamics (Harvard University).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5571506.
References
- Bender M.A., Byron R., Ragoczy T., Telling A., Bulger M., Groudine M., Byron R., Ragoczy T., Telling A., Bulger M., Groudine M., Ragoczy T., Telling A., Bulger M., Groudine M., Telling A., Bulger M., Groudine M., Bulger M., Groudine M., Groudine M. Flanking HS-62.5 and 3′HS1, and regions upstream of the LCR, are not required for β-globin transcription. Blood. 2006;108:1395–1401. doi: 10.1182/blood-2006-04-014431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M., Lin Z., Zhou L., Chudin E., Garcia E.W., Wu B., Doucet D., Thomas N.J., Wang Y., Vollmer E., Lin Z., Zhou L., Chudin E., Garcia E.W., Wu B., Doucet D., Thomas N.J., Wang Y., Vollmer E., Zhou L., Chudin E., Garcia E.W., Wu B., Doucet D., Thomas N.J., Wang Y., Vollmer E., Chudin E., Garcia E.W., Wu B., Doucet D., Thomas N.J., Wang Y., Vollmer E., Garcia E.W., Wu B., Doucet D., Thomas N.J., Wang Y., Vollmer E., Wu B., Doucet D., Thomas N.J., Wang Y., Vollmer E., Doucet D., Thomas N.J., Wang Y., Vollmer E., Thomas N.J., Wang Y., Vollmer E., Wang Y., Vollmer E., Vollmer E., et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2005;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M., Bender M.A., van Doorninck J.H., Wertman B., Farrell C.M., Felsenfeld G., Groudine M., Hardison R., Bender M.A., van Doorninck J.H., Wertman B., Farrell C.M., Felsenfeld G., Groudine M., Hardison R., van Doorninck J.H., Wertman B., Farrell C.M., Felsenfeld G., Groudine M., Hardison R., Wertman B., Farrell C.M., Felsenfeld G., Groudine M., Hardison R., Farrell C.M., Felsenfeld G., Groudine M., Hardison R., Felsenfeld G., Groudine M., Hardison R., Groudine M., Hardison R., Hardison R. Comparative structural and functional analysis of the olfactory receptor genes flanking the human and mouse β-globin gene clusters. Proc. Natl. Acad. Sci. 2000;97:14560–14565. doi: 10.1073/pnas.97.26.14560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M., Schubeler D., Bender M.A., Hamilton J., Farrell C.M., Hardison R.C., Groudine M., Schubeler D., Bender M.A., Hamilton J., Farrell C.M., Hardison R.C., Groudine M., Bender M.A., Hamilton J., Farrell C.M., Hardison R.C., Groudine M., Hamilton J., Farrell C.M., Hardison R.C., Groudine M., Farrell C.M., Hardison R.C., Groudine M., Hardison R.C., Groudine M., Groudine M. A complex chromatin landscape revealed by patterns of nuclease sensitiv ity and histone modification within the mouse β-globin locus. Mol. Cell. Biol. 2003;23:5234–5244. doi: 10.1128/MCB.23.15.5234-5244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzolari R., McMorrow T., Yannoutsos N., Langeveld A., Grosveld F., McMorrow T., Yannoutsos N., Langeveld A., Grosveld F., Yannoutsos N., Langeveld A., Grosveld F., Langeveld A., Grosveld F., Grosveld F. Deletion of a region that is a candidate for the difference between the deletion forms of hereditary persistence of fetal hemoglobin and δβ-thalassemia affects β- but not γ-globin gene expression. EMBO J. 1999;18:949–958. doi: 10.1093/emboj/18.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter D., Chakalova L., Osborne C.S., Dai Y.-F., Fraser P., Chakalova L., Osborne C.S., Dai Y.-F., Fraser P., Osborne C.S., Dai Y.-F., Fraser P., Dai Y.-F., Fraser P., Fraser P. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- Chakalova L., Osborne C.S., Dai Y.F., Goyenechea B., Metaxotou-Mavromati A., Kattamis A., Kattamis C., Fraser P., Osborne C.S., Dai Y.F., Goyenechea B., Metaxotou-Mavromati A., Kattamis A., Kattamis C., Fraser P., Dai Y.F., Goyenechea B., Metaxotou-Mavromati A., Kattamis A., Kattamis C., Fraser P., Goyenechea B., Metaxotou-Mavromati A., Kattamis A., Kattamis C., Fraser P., Metaxotou-Mavromati A., Kattamis A., Kattamis C., Fraser P., Kattamis A., Kattamis C., Fraser P., Kattamis C., Fraser P., Fraser P. The Corfu δβ thalassemia deletion disrupts γ-globin gene silencing and reveals post-transcriptional regulation of HbF expression. Blood. 2005;105:2154–2160. doi: 10.1182/blood-2003-11-4069. [DOI] [PubMed] [Google Scholar]
- Chambeyron S., Bickmore W.A., Bickmore W.A. Does looping and clustering in the nucleus regulate gene expression? Curr. Opin. Cell Biol. 2004;16:256–262. doi: 10.1016/j.ceb.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Dekker J. A closer look at long-range chromosomal interactions. Trends Biochem. Sci. 2003;28:277–280. doi: 10.1016/S0968-0004(03)00089-6. [DOI] [PubMed] [Google Scholar]
- Dekker J. The 3 C's of chromosome conformation capture: Controls, controls, controls. Nat. Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- Dekker J., Rippe K., Dekker M., Kleckner N., Rippe K., Dekker M., Kleckner N., Dekker M., Kleckner N., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- de Laat W., Grosveld F., Grosveld F. Spatial organization of gene expression: The active chromatin hub. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- Drissen R., Palstra R.J., Gillemans N., Splinter E., Grosveld F., Philipsen S., de Laat W., Palstra R.J., Gillemans N., Splinter E., Grosveld F., Philipsen S., de Laat W., Gillemans N., Splinter E., Grosveld F., Philipsen S., de Laat W., Splinter E., Grosveld F., Philipsen S., de Laat W., Grosveld F., Philipsen S., de Laat W., Philipsen S., de Laat W., de Laat W. The active spatial organization of the β-globin locus requires the transcription factor EKLF. Genes & Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- Fan J.B., Yeakley J.M., Bibikova M., Chudin E., Wickham E., Chen J., Doucet D., Rigault P., Zhang B., Shen R., Yeakley J.M., Bibikova M., Chudin E., Wickham E., Chen J., Doucet D., Rigault P., Zhang B., Shen R., Bibikova M., Chudin E., Wickham E., Chen J., Doucet D., Rigault P., Zhang B., Shen R., Chudin E., Wickham E., Chen J., Doucet D., Rigault P., Zhang B., Shen R., Wickham E., Chen J., Doucet D., Rigault P., Zhang B., Shen R., Chen J., Doucet D., Rigault P., Zhang B., Shen R., Doucet D., Rigault P., Zhang B., Shen R., Rigault P., Zhang B., Shen R., Zhang B., Shen R., Shen R., et al. A versatile assay for high-throughput gene expression profiling on universal array matrices. Genome Res. 2004;14:878–885. doi: 10.1101/gr.2167504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell C.M., West A.G., Felsenfeld G., West A.G., Felsenfeld G., Felsenfeld G. Conserved CTCF insulator elements flank the mouse and human β-globin loci. Mol. Cell. Biol. 2002;22:3820–3831. doi: 10.1128/MCB.22.11.3820-3831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P., Pruzina S., Antoniou M., Grosveld F., Pruzina S., Antoniou M., Grosveld F., Antoniou M., Grosveld F., Grosveld F. Each hypersensitive site of the human β-globin locus control region confers a different developmental pattern of expression on the globin genes. Genes & Dev. 1993;7:106–113. doi: 10.1101/gad.7.1.106. [DOI] [PubMed] [Google Scholar]
- Gheldof N., Tabuchi T.M., Dekker J., Tabuchi T.M., Dekker J., Dekker J. The active FMR1 promoter is associated with a large domain of altered chromatin conformation with embedded local histone modifications. Proc. Natl. Acad. Sci. 2006;103:12463–12468. doi: 10.1073/pnas.0605343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribnau J., Diderich K., Pruzina S., Calzolari R., Fraser P., Diderich K., Pruzina S., Calzolari R., Fraser P., Pruzina S., Calzolari R., Fraser P., Calzolari R., Fraser P., Fraser P. Intergenic transcription and developmental remodeling of chromatin subdomains in the human β-globin locus. Mol. Cell. 2000;5:377–386. doi: 10.1016/s1097-2765(00)80432-3. [DOI] [PubMed] [Google Scholar]
- Hardenbol P., Yu F., Belmont J., Mackenzie J., Bruckner C., Brundage T., Boudreau A., Chow S., Eberle J., Erbilgin A., Yu F., Belmont J., Mackenzie J., Bruckner C., Brundage T., Boudreau A., Chow S., Eberle J., Erbilgin A., Belmont J., Mackenzie J., Bruckner C., Brundage T., Boudreau A., Chow S., Eberle J., Erbilgin A., Mackenzie J., Bruckner C., Brundage T., Boudreau A., Chow S., Eberle J., Erbilgin A., Bruckner C., Brundage T., Boudreau A., Chow S., Eberle J., Erbilgin A., Brundage T., Boudreau A., Chow S., Eberle J., Erbilgin A., Boudreau A., Chow S., Eberle J., Erbilgin A., Chow S., Eberle J., Erbilgin A., Eberle J., Erbilgin A., Erbilgin A., et al. Highly multiplexed molecular inversion probe genotyping: Over 10,000 targeted SNPs genotyped in a single tube assay. Genome Res. 2005;15:269–275. doi: 10.1101/gr.3185605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Richmond T.A., Wu Y., Green R.D., Ren B., Wu Y., Green R.D., Ren B., Green R.D., Ren B., Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landegren U., Kaiser R., Sanders J., Hood L., Kaiser R., Sanders J., Hood L., Sanders J., Hood L., Hood L. A ligase-mediated gene detection technique. Science. 1988;241:1077–1080. doi: 10.1126/science.3413476. [DOI] [PubMed] [Google Scholar]
- Li Q., Peterson K.R., Fang X., Stamatoyannopoulos G., Peterson K.R., Fang X., Stamatoyannopoulos G., Fang X., Stamatoyannopoulos G., Stamatoyannopoulos G. Locus control regions. Blood. 2002;100:3077–3086. doi: 10.1182/blood-2002-04-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chu X., Liu Y., Jiang J.H., He Z., Zhang Z., Shen G., Yu R.Q., Chu X., Liu Y., Jiang J.H., He Z., Zhang Z., Shen G., Yu R.Q., Liu Y., Jiang J.H., He Z., Zhang Z., Shen G., Yu R.Q., Jiang J.H., He Z., Zhang Z., Shen G., Yu R.Q., He Z., Zhang Z., Shen G., Yu R.Q., Zhang Z., Shen G., Yu R.Q., Shen G., Yu R.Q., Yu R.Q. A colorimetric method for point mutation detection using high-fidelity DNA ligase. Nucleic Acids Res. 2005;33:e168. doi: 10.1093/nar/gni163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J.Q., Li T., Hu J.F., Vu T.H., Chen H.L., Qiu X.W., Cherry A.M., Hoffman A.R., Li T., Hu J.F., Vu T.H., Chen H.L., Qiu X.W., Cherry A.M., Hoffman A.R., Hu J.F., Vu T.H., Chen H.L., Qiu X.W., Cherry A.M., Hoffman A.R., Vu T.H., Chen H.L., Qiu X.W., Cherry A.M., Hoffman A.R., Chen H.L., Qiu X.W., Cherry A.M., Hoffman A.R., Qiu X.W., Cherry A.M., Hoffman A.R., Cherry A.M., Hoffman A.R., Hoffman A.R. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- Liu Z., Garrard W.T., Garrard W.T. Long-range interactions between three transcriptional enhancers, active Vκ gene promoters, and a 3′ boundary sequence spanning 46 kilobases. Mol. Cell. Biol. 2005;25:3220–3231. doi: 10.1128/MCB.25.8.3220-3231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Berka J., Braverman M.S., Chen Y.J., Chen Z., Braverman M.S., Chen Y.J., Chen Z., Chen Y.J., Chen Z., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele A., Gheldof N., Tabuchi T.M., Dostie J., Dekker J., Gheldof N., Tabuchi T.M., Dostie J., Dekker J., Tabuchi T.M., Dostie J., Dekker J., Dostie J., Dekker J., Dekker J. Mapping chromatin interactions by chromosome conformation capture (3C) In: Ausubel F.M., et al., editors. Current protocols in molecular biology. John Wiley & Sons; New York: 2006. pp. 21.11.21–21.11-20. [DOI] [PubMed] [Google Scholar]
- Murrell A., Heeson S., Reik W., Heeson S., Reik W., Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- Ning Z., Cox A.J., Mullikin J.C., Cox A.J., Mullikin J.C., Mullikin J.C. SSAHA: A fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuwaysir E.F., Huang W., Albert T.J., Singh J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Huang W., Albert T.J., Singh J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Albert T.J., Singh J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Singh J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., Richmond T., Gorski T., Berg J.P., Ballin J., Gorski T., Berg J.P., Ballin J., Berg J.P., Ballin J., Ballin J., et al. Gene expression analysis using oligonucleotide arrays produced by maskless photolithography. Genome Res. 2002;12:1749–1755. doi: 10.1101/gr.362402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill D., Yang J., Erdjument-Bromage H., Bornschlegel K., Tempst P., Bank A., Yang J., Erdjument-Bromage H., Bornschlegel K., Tempst P., Bank A., Erdjument-Bromage H., Bornschlegel K., Tempst P., Bank A., Bornschlegel K., Tempst P., Bank A., Tempst P., Bank A., Bank A. Tissue-specific and developmental stage-specific DNA binding by a mammalian SWI/SNF complex associated with human fetal-to-adult globin gene switching. Proc. Natl. Acad. Sci. 1999;96:349–354. doi: 10.1073/pnas.96.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palstra R.J., Tolhuis B., Splinter E., Nijmeijer R., Grosveld F., de Laat W., Tolhuis B., Splinter E., Nijmeijer R., Grosveld F., de Laat W., Splinter E., Nijmeijer R., Grosveld F., de Laat W., Nijmeijer R., Grosveld F., de Laat W., Grosveld F., de Laat W., de Laat W. The β-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- Patrinos G.P., de Krom M., de Boer E., Langeveld A., Imam A.M., Strouboulis J., de Laat W., Grosveld F.G., de Krom M., de Boer E., Langeveld A., Imam A.M., Strouboulis J., de Laat W., Grosveld F.G., de Boer E., Langeveld A., Imam A.M., Strouboulis J., de Laat W., Grosveld F.G., Langeveld A., Imam A.M., Strouboulis J., de Laat W., Grosveld F.G., Imam A.M., Strouboulis J., de Laat W., Grosveld F.G., Strouboulis J., de Laat W., Grosveld F.G., de Laat W., Grosveld F.G., Grosveld F.G. Multiple interactions between regulatory regions are required to stabilize an active chromatin hub. Genes & Dev. 2004;18:1495–1509. doi: 10.1101/gad.289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck D., Crawford E.D., Ross K.N., Stegmaier K., Golub T.R., Lamb J., Crawford E.D., Ross K.N., Stegmaier K., Golub T.R., Lamb J., Ross K.N., Stegmaier K., Golub T.R., Lamb J., Stegmaier K., Golub T.R., Lamb J., Golub T.R., Lamb J., Lamb J. A method for high-throughput gene expression signature analysis. Genome Biol. 2006;7:R61. doi: 10.1186/gb-2006-7-7-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson K.R., Clegg C.H., Navas P.A., Norton E.J., Kimbrough T.G., Stamatoyannopoulos G., Clegg C.H., Navas P.A., Norton E.J., Kimbrough T.G., Stamatoyannopoulos G., Navas P.A., Norton E.J., Kimbrough T.G., Stamatoyannopoulos G., Norton E.J., Kimbrough T.G., Stamatoyannopoulos G., Kimbrough T.G., Stamatoyannopoulos G., Stamatoyannopoulos G. Effect of deletion of 5′HS3 or 5′HS2 of the human β-globin locus control region on the developmental regulation of globin gene expression in β-globin locus yeast artificial chromosome transgenic mice. Proc. Natl. Acad. Sci. 1996;93:6605–6609. doi: 10.1073/pnas.93.13.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe K. Making contacts on a nucleic acid polymer. Trends Biochem. Sci. 2001;26:733–740. doi: 10.1016/s0968-0004(01)01978-8. [DOI] [PubMed] [Google Scholar]
- Selzer R.R., Richmond T.A., Pofahl N.J., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Richmond T.A., Pofahl N.J., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Pofahl N.J., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Green R.D., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Eis P.S., Nair P., Brothman A.R., Stallings R.L., Nair P., Brothman A.R., Stallings R.L., Brothman A.R., Stallings R.L., Stallings R.L. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005;44:305–319. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- Singh-Gasson S., Green R.D., Yue Y., Nelson C., Blattner F., Sussman M.R., Cerrina F., Green R.D., Yue Y., Nelson C., Blattner F., Sussman M.R., Cerrina F., Yue Y., Nelson C., Blattner F., Sussman M.R., Cerrina F., Nelson C., Blattner F., Sussman M.R., Cerrina F., Blattner F., Sussman M.R., Cerrina F., Sussman M.R., Cerrina F., Cerrina F. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotechnol. 1999;17:974–978. doi: 10.1038/13664. [DOI] [PubMed] [Google Scholar]
- Spilianakis C.G., Flavell R.A., Flavell R.A. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat. Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- Spilianakis C.G., Lalioti M.D., Town T., Lee G.R., Flavell R.A., Lalioti M.D., Town T., Lee G.R., Flavell R.A., Town T., Lee G.R., Flavell R.A., Lee G.R., Flavell R.A., Flavell R.A. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Splinter E., Grosveld F., de Laat W., Grosveld F., de Laat W., de Laat W. 3C technology: Analyzing the spatial organization of genomic loci in vivo. Methods Enzymol. 2004;375:493–507. doi: 10.1016/s0076-6879(03)75030-7. [DOI] [PubMed] [Google Scholar]
- Stamatoyannopoulos G. Control of globin gene expression during development and erythroid differentiation. Exp. Hematol. 2005;33:256–271. doi: 10.1016/j.exphem.2004.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B., Palstra R.J., Splinter E., Grosveld F., de Laat W., Palstra R.J., Splinter E., Grosveld F., de Laat W., Splinter E., Grosveld F., de Laat W., Grosveld F., de Laat W., de Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Vakoc C., Letting D.L., Gheldof N., Sawado T., Bender M.A., Groudine M., Weiss M.J., Dekker J., Blobel G.A., Letting D.L., Gheldof N., Sawado T., Bender M.A., Groudine M., Weiss M.J., Dekker J., Blobel G.A., Gheldof N., Sawado T., Bender M.A., Groudine M., Weiss M.J., Dekker J., Blobel G.A., Sawado T., Bender M.A., Groudine M., Weiss M.J., Dekker J., Blobel G.A., Bender M.A., Groudine M., Weiss M.J., Dekker J., Blobel G.A., Groudine M., Weiss M.J., Dekker J., Blobel G.A., Weiss M.J., Dekker J., Blobel G.A., Dekker J., Blobel G.A., Blobel G.A. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol. Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Wang Y., Moorhead M., Karlin-Neumann G., Falkowski M., Chen C., Siddiqui F., Davis R.W., Willis T.D., Faham M., Moorhead M., Karlin-Neumann G., Falkowski M., Chen C., Siddiqui F., Davis R.W., Willis T.D., Faham M., Karlin-Neumann G., Falkowski M., Chen C., Siddiqui F., Davis R.W., Willis T.D., Faham M., Falkowski M., Chen C., Siddiqui F., Davis R.W., Willis T.D., Faham M., Chen C., Siddiqui F., Davis R.W., Willis T.D., Faham M., Siddiqui F., Davis R.W., Willis T.D., Faham M., Davis R.W., Willis T.D., Faham M., Willis T.D., Faham M., Faham M. Allele quantification using molecular inversion probes (MIP) Nucleic Acids Res. 2005;33:e183. doi: 10.1093/nar/gni177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.G., Fraser P., Fraser P. Remote control of gene transcription. Hum. Mol. Genet. 2005;14:R101–R111. doi: 10.1093/hmg/ddi104. [DOI] [PubMed] [Google Scholar]
- Xu N., Tsai C.L., Lee J.T., Tsai C.L., Lee J.T., Lee J.T. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311:1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- Yuen T., Wurmbach E., Pfeffer R.L., Ebersole B.J., Sealfon S.C., Wurmbach E., Pfeffer R.L., Ebersole B.J., Sealfon S.C., Pfeffer R.L., Ebersole B.J., Sealfon S.C., Ebersole B.J., Sealfon S.C., Sealfon S.C. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 2002;30:e48. doi: 10.1093/nar/30.10.e48. [DOI] [PMC free article] [PubMed] [Google Scholar]