Abstract
Adenovirus (Ad) and adeno-associated virus (AAV) have attractive and complementary properties that can be exploited for gene transfer purposes. Ad vectors are probably the most efficient vehicles to deliver foreign genes both in vitro and in vivo. AAV exhibits the unique ability to establish latency by efficiently integrating at a specific locus of human chromosome 19 (AAVS1). Two viral elements are necessary for the integration at AAVS1: Rep68/78 and the inverted terminal repeats (AAV-ITRs). In this study, we report the development of two helper-dependent adenoviral (HD) vectors, one carrying the Rep78 gene, the other an AAV-ITR-flanked transgene. Although Rep proteins have been demonstrated to interfere with Ad replication, HD Rep78 vector was successfully amplified on serial passages in 293CRE4 cells with a yield of 50–100 transducing units per cell. DNA integration at the AAVS1 site also was demonstrated in hepatoma cells coinfected with the HD-expressing Rep78 and with the second HD vector carrying a transgene flanked by AAV-ITRs. The high transduction efficiency, large cloning capacity, and high titer of the HD, combined with the site-specific integration machinery provided by AAV-derived components, make the Ad/AAV hybrid viruses a promising vehicle for gene therapy.
Vectors based on different viruses have been developed to embrace a wide range of strategies for the gene therapy of a variety of diseases (1). Preclinical evaluation of many of these approaches has revealed a number of fundamental problems. These limitations are driving vector technology efforts toward a further evolution of the present systems and the generation of new classes of vectors that combine the best features of different viruses (2–5). The modification of recombinant adenovirus (Ad) vectors is an example of vector engineering directed to solving problems identified by preclinical studies. Ad is considered an attractive vehicle for several reasons. The viral life cycle is well characterized, its genome is easy to manipulate, and the resulting vector can be grown to high titers (6). In addition, Ad has a broad tropism and can infect both dividing and nondividing cells. However, studies with E1-deleted Ad vectors have shown that (generally) only short-term expression of the transgene can be achieved (7). Administration of high titer Ad vectors to nonhuman primates has been associated with severe host inflammatory responses (8) and immune clearance of transduced cells thought to be, at least in part, a consequence of leaky expression of Ad early and late genes (9, 10). Deletion or mutation of additional viral genes and parallel development of complementing cell lines is one approach proposed to overcome the problem of vector immunogenicity (11, 12). The most recent advance is the development of a helper-dependent (HD) Ad gutless vector (13–18). In this system a helper virus provides in trans all of the viral proteins required for propagation of the vector, which contains only the inverted terminal repeats (ITRs) and packaging signal required for DNA replication and virus assembly. These two elements are contained within about 500 base pairs located at the ends of viral genome allowing the cloning of up to 37 kb of foreign DNA. Recently, Graham and coworkers (17) developed an efficient system for HD vector propagation based on a helper virus that includes a packaging signal flanked by loxP sites and a 293 cell line expressing Cre recombinase.
Although deletion of all coding regions should solve many of the problems related to immunogenicity of Ad vectors (19, 20), vector DNA is expected to persist extrachromosomally in nuclei of transduced cells. Therefore, Ad-mediated gene delivery to tissues with rapid regeneration is likely to lead to loss of the transgene. The adeno-associated virus (AAV) life cycle suggests a potential solution to this problem. AAV is a human parvovirus dependent on Ad for replication and propagation (21) and, in the absence of helper virus, AAV establishes latency by frequently integrating into a specific locus of chromosome 19, called AAVS1 (22, 23). Two AAV elements are required for viral DNA integration: the ITRs and either of the two larger Rep polypeptides, Rep68 and Rep78 (24–26). These two polypeptides are derived by transcription from the P5 promoter, whereas the smaller Rep proteins, Rep52 and Rep40, are derived from transcription from the P19 promoter (21). This unique property of AAV has stimulated considerable efforts toward the development of AAV-based vectors in which the viral genes encoding the replication functions as well as the structural polypeptides have been substituted by the gene of interest. Recombinant AAV vectors (rAAV) in which rep has been deleted cannot, however, integrate in the AAVS1 locus (27), but several groups have recently demonstrated that site-specific integration can occur if Rep protein(s) is provided in trans (24–26, 28, 29).
Incorporating the AAV integration machinery into Ad vectors would combine advantages of both viral systems. Although transgenes flanked by AAV-ITRs can be easily inserted into Ad vectors, the rescue of Ad viruses carrying rep gene is more problematic. Efforts to construct an Ad vector expressing Rep protein(s) have been unsuccessful so far, presumably because Rep inhibits Ad replication (30). We report here the successful construction of an Ad/AAV hybrid virus system consisting of the combination of two HD vectors: the first expressing AAV Rep78 gene under control of either the T7 or α1antitrypsin (α1at) liver-specific promoter, the second carrying an AAV-ITR-flanked DNA.
MATERIALS AND METHODS
Tissue Culture and Virus Growth.
The 293CRE4 cell line (31) was propagated in minimal essential medium (MEM) supplemented with 10% fetal calf serum and 0.4 mg/ml G418. HepG2, Hep3B, Huh7, and HeLa cells were grown in DMEM supplemented with 10% fetal calf serum. Ad2 virus and AdLC8cluc helper virus (17) was amplified and titrated on 293 cells as described (32).
Construction of Ad/AAV Hybrid Viruses.
Plasmids were constructed according to standard protocols (33). The rep 78 gene was derived as described (34) by mutating the ATG start codon for Rep 52/40 to GGA (methionine → glycine, amino acid 225) and creating a G → A mutation at nucleotide 1,907 to eliminate the splice donor site required for expression of spliced versions Rep 68/40. RepΔATG mutants were obtained by PCR designed to delete the first ATG of the rep ORF.
A T7 promoter fused to an upstream synthetic transcription pause site was isolated by PCR amplification from pCAT-3 (Promega) then ligated to Rep or Rep78 (nucleotides 321–2,252 of AAV-2) and to the corresponding ΔATG mutant cassettes and inserted in the shuttle plasmid pABS-4 (35), generating pABT7-Rep78, pABT7-Rep, pABT7-Rep78ΔATG, and pABT7-RepΔATG. To construct the first generation vectors carrying rep genes, a PacI fragment from these four plasmids containing the T7-Rep cassettes was ligated into the unique PacI site located in the E3 region of pLBG40. Plasmid pLBG40 (N. Louis and F. L. Graham, unpublished results) contains an Ad5 genome deleted in E1 and E3 regions allowing insertion of up to 6 kb of foreign DNA by direct cloning into either the E1 or E3 region and pLBG40 is infectious when transfected in 293 cells as the parental plasmid pFG140 (36).
HD vectors carrying the Rep78 gene were constructed in the context of pRP1030 Ad helper-dependent plasmid. pRP1030 was derived from pRP1001 (17) by deleting all Ad5 coding sequences and substituting them with λ phage DNA (22,425-bp BglII fragment) to produce a genome of packageable size (Fig. 1) (37). T7-Rep78 or T7-Rep78ΔATG cassettes were ligated into the unique StuI site of the λ DNA stuffer, generating the plasmids pRS1033 and pRS1032, respectively. The cassette comprising Δ137α1at promoter (38) fused to Rep78 gene was constructed in the shuttle plasmid pABS-4 following the reported cloning strategy and then inserted in the StuI site of pRP1030 obtaining pRP1034. Finally, HDFB1 was derived from pSTK120 (a gift of S. Kochanek, University of Cologne) following the strategy reported below. A transgene cassette containing the green fluorescent protein (GFP) and hygromicin B resistance genes flanked by AAV-ITR sequences was derived from pBac-ITR plasmid (4) by replacing lacZ with GFP/UF5 gene excised from pITR-UF5 plasmid (kindly obtained by N. Muzyczka, University of Florida). The AAV-ITR-flanked cassette was then inserted into the SmiI site of pSTK120, generating pSTK-FB1. HDFB1 was rescued by PmeI restriction from pSTK-FB1 before transfection. STK120 plasmid is a pBluescript II KS that contains (in the following order) the Ad5 ITR sequences and the packaging signal Ψ, 440 bp (nucleotides 1–440 of Ad5); a 16,054-bp fragment of hypoxanthine guanine phosphoribosyltransferase (nucleotides 1,799–17,853 in gb:humhprtb); a HindIII 9,063-bp fragment of C346 cosmid (nucleotides 12,421–21,484 in gb:L31948); the right-end terminus of Ad5, 117 bp (nucleotides 35,818–35,935 of Ad5); and the pBluescript II KS plasmid sequence. Further details of vector construction are available on request.
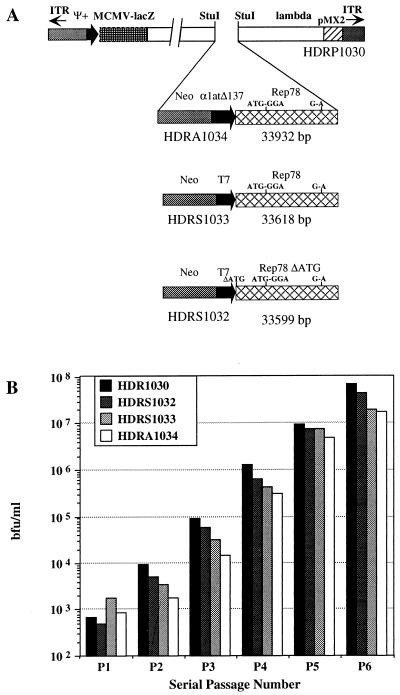
Figure 1.
(A) Schematic representation of vector structures. All rep cassettes were constructed and inserted in the helper-dependent plasmid pRP1030 as outlined in Materials and Methods. Mutation of Rep52/40 ATG to GGA, G → A mutation of splice donor site, and deletion of first Rep78 ATG (respectively nucleotides 993–996, 1,907, and 321–323 of AAV-2) were reported on the vector diagrams. pRP1030 (shown in linear form) contains sequences corresponding to left ends (base pairs 3–466) and to the right end (base pairs 35,464–35,924) of Ad5 genome including ITRs and packaging signal. All other Ad5 genomic sequences were deleted and substituted with a mouse cytomegalovirus immediate early promoter (MCMV)-lacz cassette that corresponds to nucleotides 467–4,920 of pRP1030, a 22,425-bp BglII fragment of λ DNA (nucleotides 5,128–27,138). A bacterial plasmid (pMX2) is present between nucleotides 27,138 and 29,373. (B) Serial amplification of Hd-Rep vectors. pRP1030 was amplified as positive control. All plasmids were converted to linear molecules and packaged into infectious virions after transfection of 293CRE4 cells infected with AdLC8 helper virus. Amplification was measured by 293 infection with aliquots of 293CRE lysate and β-galactosidase staining.
Propagation of Ad/AAV Hybrid Viruses.
To rescue the first generation Ad/AAV hybrid vectors, 60-mm dishes of a semiconfluent monolayer of 293 cells were transfected with 5 μg each of pLBG40 derivatives, and plaques were isolated and processed by standard methods (32).
HD vectors were propagated and purified as described by Parks et al. (17). Rescue and amplification of HD vector were performed on 293CRE4 cells preinfected with AdLC8cLucl helper virus. Amplification of β-gal-expressing viruses was monitored as described (17). Large-scale virus preparations were obtained by infecting 150-mm dishes of 293CRE4 cells at a multiplicity of infection (moi) of 1 transducing unit per cell of HDs on preinfection with the same moi of helper virus. The virus was purified as described (32).
To evaluate the efficiency of Ad/AAV hybrid viruses, rescue and replication assay was performed by infecting 293 or Hep3B cells with Rep-expressing viruses in combination with HDFB1. Wild-type Ad2 was included as helper virus when the experiments were performed with HD vectors in Hep3B cells. Total cellular DNA was extracted 48 hours postinfection with standard techniques and analyzed by Southern blot using a hygromicin B DNA-specific probe (see Fig. 3B).
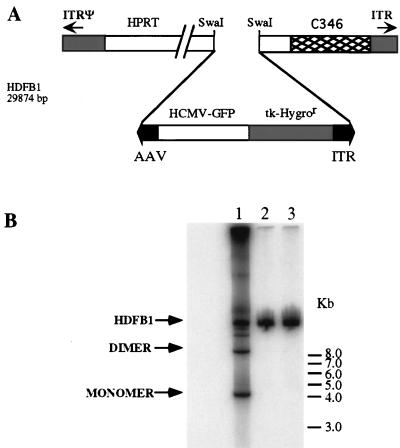
Figure 3.
Structure of HDFB1 and evaluation of Rep78 expression by AAV-ITR-flanked transgene rescue and replication. (A) HDFB1 carries an integration cassette constituted by hygromicin resistance and GFP genes flanked by AAV-ITRs and was constructed as outlined in Materials and Methods. (B) Hep3B cells were infected with an moi of 10 transducing units per cell of HDFB1 (lanes 1–3) in combination with HDRA1034 (lane 1) or HDRS1032 (lane 2) at an moi of 10. Infection with HDFB1 alone served as negative control (lane 3). Rescue and replication experiment was carried out infecting the cells with Ad2 wild type (moi = 10) as helper virus. Total cellular DNA was separated on agarose gel, transferred on nylon membrane, and hybridized with an HygroR DNA-specific probe. Signals corresponding to ITR(GFP/Hygro) cassette monomer, dimer, and HDFB1 whole genomic DNAs are indicated.
PCR Assay for AAVS1 Integration.
HepG2, Hep3B, and Huh7 cells were coinfected at an moi of 10 blue-forming units (bfu)/cell with HDRS1032 or HDRA1034 in combination with HDFB1 at an moi of 10 transducing units per cell. Cells were harvested by scraping 48 hours after infection, and total DNA was extracted by using standard techniques (33). Nested PCR was performed as described (25). One-tenth of the amplified DNA was loaded in duplicate on a 1.3% agarose gel, electrophoresed, transferred to Hybond N+ membrane (Amersham Pharmacia), and hybridized with AAVS1 or AAV-ITR-specific probes.
Fluorescence in Situ Hybridization.
A 3.7-kb DNA fragment corresponding to the ITR(GFP/Hygro) cassette and a 80-kb AAVS1 DNA fragment isolated from a genomic DNA library were labeled using nick-translation kit (Boehringer Mannheim) according to the manufacturer’s instruction and used as probes in chromosome analysis. The chromosome spreads from a pool of hygromicin-resistant HepG2 cells were prepared by standard cytogenetic techniques (39). Cytogenetic preparations were treated as described (4). Images were processed by using Adobe Photoshop on a Power Macintosh computer (Apple).
RESULTS AND DISCUSSION
Rescue and Amplification of Ad/AAV Vectors Expressing rep Gene.
Different studies on the relationship between Ad and AAV demonstrated a strong interference of AAV on Ad life cycle (40–42). Although the molecular mechanism of AAV-mediated inhibition is not clear, Wilson and coworkers (30) have demonstrated that Rep expression is sufficient to suppress the maturation of Ad replication centers. In view of the observation that the P5 promoter, which drives the expression of Rep68 and Rep78 (21), is transactivated by Ad E1A proteins (43, 44), we reasoned that replacement of P5 with the phage T7 promoter could reduce Rep expression during virus growth, thus minimizing Rep-mediated interference on Ad vector replication. The T7 promoter has been used to rescue a recombinant Ad expressing the highly toxic virion host shutoff protein of Herpes simplex (R. Tomanin, M. Rosa, and F.L.G., unpublished results). Rep expression cassettes were constructed by fusing the T7 promoter to the wild-type rep and rep78 genes and inserting them into the infectious Ad plasmid pLBG40 (see Materials and Methods). As control, the corresponding ΔATG mutant plasmids were also constructed. Surprisingly, as shown in Table 1 no plaques were obtained by transfecting either pLBT7-Rep or pLBT7-RepΔATG. It is worth noting that the deletion of the first ATG introduced into the rep coding sequence is expected to abolish expression of full-length Rep78 and Rep68, but not of p19-promoted Rep52 and Rep40 (45). In contrast, pLB-Rep78 was successfully rescued, albeit with a lower efficiency than pLBT7-Rep78ΔATG and pLBG40 control plasmid. No rearragement of the genomic DNA was detected in LBT7-Rep78 virions. Furthermore, the functionality of the Rep78 polypeptide was demonstrated by coinfection of 293 cells with AdLBT7-Rep78 and the HD vector HDFB1 (Table 1). This virus carries an AAV-ITR-flanked DNA (Fig. 3A) that was rescued and amplified on expression of Rep78. Taken together, these data indicate that Rep78 is compatible with Ad replication and that Rep mediated interference on Ad replication could be reduced by replacing the P5 promoter and abolishing the expression of other Rep polypeptides.
Table 1.
Infectivity of Ad5 genomic plasmid carrying the rep gene
| Vector | Experiment
|
Rep activity | |
|---|---|---|---|
| 1 | 2 | ||
| pLBT7-Rep | 0 | 0 | ND |
| pLBT7-Rep78 | 4 | 6 | + |
| pLBT7-RepΔATG | 0 | 0 | ND |
| pLBT7-Rep78ΔATG | 15 | 18 | − |
| Control | 22 | 26 | − |
Data given as number of plaques obtained. Plaques were observed 10 days after 293 transfection. Plaque isolates were amplified through four serial passages on 293 cells and then evaluated for Rep activity in rescue and replication experiments as described in Materials and Methods. All AdLBT7-Rep78 isolates were positive for Rep enzymatic activity. ND, not determined.
To exploit the potential of the HD vectors for gene transfer, the T7Rep78 expression cassette was inserted into the Ad helper dependent plasmid pRP1030 (Fig. 1A), generating pRS1033. Similarly, plasmid pRA1034 was constructed by fusing the Rep78 coding sequences to the Δ137α1at promoter (38). This promoter is poorly active in 293CRE cells (C.T., unpublished data) and thus is expected to minimize Rep78 expression during vector amplification. As control, plasmid pRS1032 carrying T7Rep78ΔATG was also constructed. Fig. 1B shows the efficiency of amplification of the HD-Rep vectors evaluated as bfu/ml. Little or no difference in rescue and amplification efficiency was observed between the control vector HDRP1030 and its rep derivatives. Additionally, the time course of CPE during vector propagation was not affected by infection with HD-Rep viruses and was usually complete by 48 to 72 hrs postinfection, suggesting that the overall Ad life cycle had not been perturbed. A large-scale preparation of HDRA1034 resulted in the production of 3 × 109 bfu from 5 × 107 cells, indicating that 50–100 Rep-expressing viruses per cell could be produced without interference. Furthermore, no apparent rearrangement of virus genome had occurred on vector amplification, as indicated by Southern blot analysis of HDRS1034 and HDRS1032 genomic DNA (data not shown).
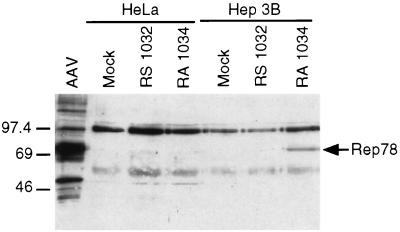
The expression of Rep78 was assessed by infecting Hep3B and HeLa cells with an moi of 50 bfu/cell of HDRA1034 or HDRS1032. Cells were harvested 36 hours postinfection, and Western blot analysis by using a rabbit anti-Rep antiserum was performed on infected cell lysates (Fig. 2). A cell extract of 293 cells infected with AAV was used as positive control. As expected, Rep78 expression was detected only in Hep3B infected with HDRA1034. The functionality of Rep78 expressed by HDRA1034 was evaluated by exploiting the ability of this protein to rescue an ITR-flanked DNA and to support its replication in Ad-infected cells (46). Hep3B cells were coinfected with HDFB1 (Fig. 3A) in combination with HDRA1034 by using as helper Ad2 wild-type virus. Infection was carried out using an moi of 10 for each virus. Total DNA was extracted and analyzed 48 hr postinfection by Southern blot by using a DNA probe specific for the transgene. As shown in Fig. 3B, the presence of low molecular weight DNA bands corresponding in size to the monomer and dimer forms of the replicated AAV-ITR/transgene cassette were detected in HDFB1-infected cells coinfected with HDRA1034 vector (Fig. 3B, lane 1). No rescue of the ITR-flanked cassette was detected in cells that were infected with HDFB1 alone or coinfected with HDRS1032 (Fig. 3B, lanes 2 and 3). Thus, these data indicated that HDRA1034 encodes a fully functional Rep78 protein.
Figure 2.
Western blot analysis of protein extracted from HeLa and Hep3B cells infected with Rep78-expressing Ad vectors. Cells (1.2 × 106) were infected with HDRA1034 or HDRS1032 at an moi of 50 bfu/cell. An AAV-infected 293 cell extract was included in the experiment as positive control. Cell extracts equivalent to 3 × 105 cells were separated on 7.5% SDS/PAGE and transferred to nitrocellulose membrane. Rep polypeptides were detected with a polyclonal rabbit antiserum and chemiluminescence kit (ECL; Amersham Pharmacia). Rep78 polypeptide is indicated on the right margin. A cross-reacting protein of slightly lower molecular weight of Rep78 was detected in some experiments.
Site-Specific Integration of an AAV-ITR Transgene.
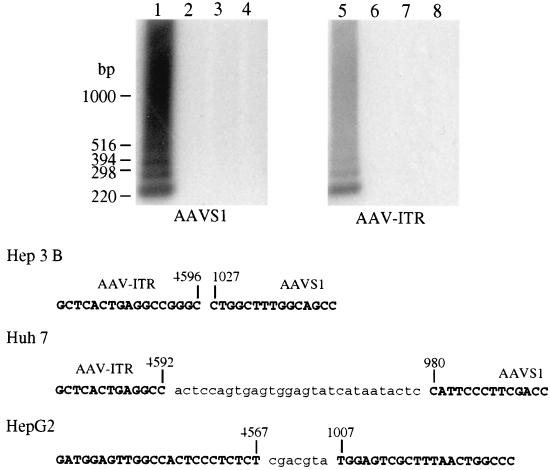
To assess whether Rep-mediated targeted integration could be observed on infection with HD vector expressing Rep78, Hep3B cells were coinfected with HDRA1034 and HDFB1. Forty-eight hours postinfection, cells were harvested, genomic DNA was extracted, and a nested PCR-based assay on genomic DNA was performed to detect junctions between AAVS1 and AAV-ITRs (24, 25). Two pairs of primers specific for AAVS1 and AAV-ITRs were used, the AAVS1 primers annealed to a region located downstream the 100-bp AAVS1 region identified as “hot spot” for site-specific integration (23). The amplification products were identified as the junction fragments by virtue of their hybridization to AAVS1- and ITR-specific probes. Fig. 4A shows the results obtained on coinfection of Hep3B cells. Similar results were obtained by using other cell line of hepatic origin (data not shown). Three bands between 400 bp and 220 bp superimposed on a smear (with the major species being approximately 220 bp) were detected only in cells infected with HDFB1 and HDRA1034 (Fig. 4A, lanes 1 and 5). No junctions were amplified from mock-infected cells (Fig. 4A, lanes 4 and 8) or from cells infected with HDFB1 alone or with HDRA1032 (Fig. 4A, lanes 2 and 6 and 3 and 7, respectively). To better characterize the AAV-ITR/AAVS1 junctions, amplified DNA was cloned and sequenced. The sequences of the most frequently isolated clones are shown in Fig. 4B. The junctions analyzed indicated that insertion of the ITR-flanked DNA had occurred at nucleotids 980, 1,027, and 1,034 of AAVS1 in Huh7, Hep3B, and HepG2, respectively. Additionally, deletions within the ITRs and insertions of nucleotides were detected.
Figure 4.
PCR amplification of AAVS1-AAV-ITR junction. (A) Southern blot analysis of amplification products obtained by using DNA extracted from infected cells as template. Total DNA was isolated from Hep3B cells infected with HDFB1 (moi = 10 transducing units per cell) alone (lanes 3 and 7) and in combination with HDRA1034 (lanes 1 and 5) or HDRS1032 (lanes 2 and 6) (moi = 10 bfu/cell). DNA from mock-infected cells is shown in lanes 4 and 8. Nested PCR amplification was performed as described in ref. 25. One-tenth of the PCR reaction was loaded on 1.2% agarose gel in duplicate, transferred to nylon membrane, and hybridized with two different probes derived from AAVS1 or AAV-ITR DNAs. (B) Amplified DNA was cloned and sequenced. The junction DNA sequences reported represent the most frequent clones obtained in different experiments performed in Hep3B, Huh7, and HepG2 cell lines. Sequences not belonging to ITR or AAVS1 are in lower case letters.
Different studies demonstrate that transduction of 293 cells with vectors carrying the rep gene and a reporter gene inserted between AAV-ITRs lead to a highly efficient integration of the ITR-flanked DNA (4, 24, 28). To verify whether the delivery of AAV components mediated by the HD vectors can establish more stable cell clones also in hepatic cells, 1 × 105 Hep3B cells were infected with HDFB1 alone or in combination with HDRA1034 at a moi of 10. Cells were diluted 48 hr postinfection and cultivated in the presence of 100 μg/ml hygromicin B. Stable integration frequency was evaluated by comparing the number of clones obtained in the presence and absence of Rep78 expression. An average of 400 stable transformants were obtained on coinfection of the two vectors, whereas 600 hygromicin B-resistant clones were detected after infection with HDFB1. Similar results were obtained on infection of HepG2 and Huh7 cells (data not shown). The lack of a Rep78-mediated increase in stable transformants in hepatic cells was apparently not caused by any toxic effects on host cells mediated by constitutive expression of Rep78. In fact, no differences in Hep3B viability were noted in a cell proliferation assay between uninfected Hep3B and cells infected with HDRA1034 at a moi of 1, 10, and 100. These results suggested that the increased integration efficiency of an ITR-flanked DNA mediated by Rep is restricted to 293 cells. It is very likely that specific host factors preferentially expressed in these cells may influence the frequency of Rep-mediated integration.
To determine the efficiency of site-specific integration, in situ hybridization studies were performed on chromosomes of infected cells. Stable clones were obtained by infecting HepG2 cells with 5 bfu/cell of HDRA1034 or HDRS1032, the lowest moi compatible with infection of 80–90% of the cell population, in combination with the same moi of HDFB1. Infected cells were passaged several times in presence of 150 μg/ml hygromicin B, and fluorescence in situ hybridization analysis was then performed on metaphase spreads by using AAVS1- and hygromicin B-specific probes. Double labeling on both sister chromatids was scored as a positive signal for site-specific integration. In HepG2 cells infected with HDRA1034 and HDFB1, targeted integration to the AAVS1 site was observed in 14 of 39 (35%) metaphases analyzed, whereas only one integration in chromosome 19 was observed in 34 metaphases (3%) of cells infected with HDRS1032 and HDFB1. Fig. 5 shows a metaphase spread in which the transgene probe colocalized with the AAVS1 probe in one of the three chromosomes 19 present in this cell line. The transgene was always associated with only one chromosome 19 in all positive metaphase spreads examined, whereas in the remaining metaphases the transgene probe was located on different chromosomes that were not detected by the AAVS1 probe (data not shown). Taken together, the analysis of the stable transduction capacity of the Ad/AAV hybrid vectors and the in situ hybridization study indicate that Rep expression increases targeted insertion of AAV-ITR-flanked DNA without affecting the overall integration frequency in cells of hepatic origin.
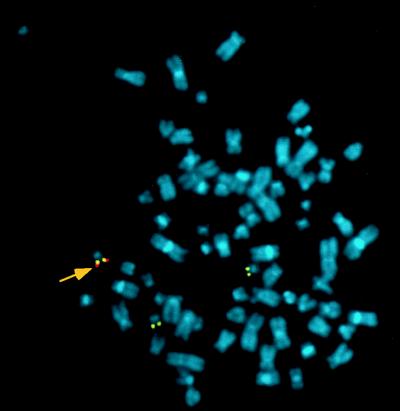
Figure 5.
Fluorescence in situ hybridization of metaphase spreads from a pool of HepG2 selected in the presence of hygromicin B. Cells were selected on coinfection with HDRA1034 and HDFB1, a helper-dependent vector carrying ITR(GFP/Hygro) cassette. Metaphase spreads were hybridized following the conditions described in ref. 4 with a GFP/hygromicin (red) and an AAVS1 probes (yellow). Colocalization of both probes on chromosome 19 is indicated by an arrow.
We report in this study the construction of Ad/AAV hybrid vectors that express functional Rep78 protein, the major trans-acting component of the AAV integration machinery. Several features might have contributed to reach this goal: (i) the Rep cassette used was designed to express only Rep78; (ii) the substitution of P5 with promoters poorly expressed in 293CRE cells have further reduced Rep78 production in the packaging cell line; and (iii) the helper-dependent packaging system has introduced amplification conditions that are likely to minimize Rep interference of vector production. Indeed, AAV-mediated inhibition of Ad replication depends on the ratio of the moi of the two viruses and on the temporal order in which each virus is added (35, 36). Furthermore, it has been reported that if AAV infection is delayed until Ad DNA replication has begun, AAV does not interfere with Ad replication (36). These observations may account, at least in part, for the fact that a HD vector carrying Rep78 could be amplified during consecutive rounds of coinfection with helper virus without significant differences from the control viruses. The mechanism by which rep gene expression interferes with Ad replication is not clear. However, our results with first-generation Ad vectors carrying rep gene suggest that P19-promoted Rep52 and Rep40 gave an important contribution to Ad replication inhibition. The biochemical properties of the Rep78–68 polypeptides and their role in AAV replication have been studied in detail. They are multifunctional proteins with DNA binding, site-specific endonuclease, helicase, and ATPase activity (47, 48). In contrast, little is known about the functions of Rep52/40 proteins. Recently, Rep52 was identified as a DNA helicase with ATPase activity displaying distinct biochemical characteristics with respect to those of Rep78 and Rep68 (49). The fact that rep gene encodes two different families of helicase proteins may suggest that Rep proteins are likely to influence Ad replication by interacting directly with the Ad genome. However, further investigation will be required to identify the step of Ad life cycle affected by Rep protein.
Recently, several hybrid viruses carrying AAV elements relevant for site-specific integration have been constructed, including baculovirus/AAV (4), herpes simplex/AAV (3), and E1-deleted Ad vector coupled to a plasmid expressing Rep (2). The HD/AAV hybrid vectors may offer several potential advantages over other vector systems, which include high titers and efficient in vivo gene transduction. The strategy we used to obtain site-specific integration was based on coinfection of target cells with two different viruses. We are now exploring the possibility of including both rep and AAV-ITR-flanked transgene in a single vector. According to the proposed model for rescue of AAV genome from integrated state (50), vector instability during amplification can be predicted if a constitutively active rep gene is used (4). We are currently modifying our vector system to provide a more stringent control of Rep expression, which may be crucial to improve production, transduction efficiency, and integration specificity. These new hybrid Ad/AAV vectors will broaden our spectrum of tools for the understanding of the molecular mechanisms that govern Rep-mediated targeted integration.
Acknowledgments
We would like to thank Cindy Beauchamp and Uma Sankar for technical assistance, Antonella Sgura for helpful suggestions on in situ hybridization study, Stefan Kochanek for providing pSTK120, Nicholas Muzyczka for providing pITR-UF5, Janet Clench for editorial assistance, and Manuela Emili for graphics. We are also grateful to Linong Zhang for stimulating discussions during the course of this study. This work was partially supported by grants to FLG from the National Institutes of Health (US), the Medical Research Council of Canada, and the National Cancer Institute of Canada (NCIC). F.L.G. is a Terry Fox Research Scientist of the NCIC.
ABBREVIATIONS
- Ad
adenovirus
- AAV
adeno-associated virus
- HD
helper-dependent Ad vector
- bfu
blue-forming units
- moi
multiplicity of infection
- GFP
green fluorescent protein
References
- 1.Mulligan R C. Science. 1993;260:926–932. doi: 10.1126/science.8493530. [DOI] [PubMed] [Google Scholar]
- 2.Fisher K J, Kelley M W, Burda J F, Wilson J M. Hum Gene Ther. 1996;7:2079–2087. doi: 10.1089/hum.1996.7.17-2079. [DOI] [PubMed] [Google Scholar]
- 3.Fraefel C, Jacoby D R, Lage C, Hilderbrand H, Chou J Y, Alt F W, Breakefield X O, Majzoub J A. Mol Med. 1997;12:813–825. [PMC free article] [PubMed] [Google Scholar]
- 4.Palombo F, Monciotti A, Recchia A, Cortese R, Ciliberto G, La Monica N. J Virol. 1998;72:5025–5034. doi: 10.1128/jvi.72.6.5025-5034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conway J E, Zolotukhin S, Muzyczka N, Hayward G S, Byrne B J. J Virol. 1997;71:8780–8789. doi: 10.1128/jvi.71.11.8780-8789.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hitt M M, Addison C L, Graham F L. Adv Pharmacol. 1997;40:137–206. doi: 10.1016/s1054-3589(08)60140-4. [DOI] [PubMed] [Google Scholar]
- 7.Jaffe H A, Danel C, Longenecker G, Metzger M, Setoguchi Y, Rosenfeld M A, Gant T W, Thorgeirsson S S, Stratford-Perricaudet L D, Perricaudet M, et al. Nat Genet. 1992;1:372–378. doi: 10.1038/ng0892-372. [DOI] [PubMed] [Google Scholar]
- 8.Wilmott R H, Amin R S, Perez C R, Wert S E, Keller G, Boivin G P, Hirsch R, De Innocencio J, Lu P, Reising S F, et al. Hum Gene Ther. 1996;7:301–310. doi: 10.1089/hum.1996.7.3-301. [DOI] [PubMed] [Google Scholar]
- 9.Yang Y, Ertl H C, Wilson J M. Immunity. 1994;1:433–442. doi: 10.1016/1074-7613(94)90074-4. [DOI] [PubMed] [Google Scholar]
- 10.Yang Y, Nunes F A, Berencsi K, Furth E E, Gonczol E, Wilson J M. Proc Natl Acad Sci USA. 1994;91:4407–4411. doi: 10.1073/pnas.91.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q, Finer M H. Nat Med. 1996;2:714–716. doi: 10.1038/nm0696-714. [DOI] [PubMed] [Google Scholar]
- 12.Amalfitano A, Begy C R, Chamberlain J S. Proc Natl Acad Sci USA. 1996;93:3352–3356. doi: 10.1073/pnas.93.8.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitani K, Graham F L, Caskey C T, Kochanek S. Proc Natl Acad Sci USA. 1995;92:3854–3858. doi: 10.1073/pnas.92.9.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher K J, Choi H, Burda J, Chen S-J, Wilson J M. Virology. 1996;217:11–22. doi: 10.1006/viro.1996.0088. [DOI] [PubMed] [Google Scholar]
- 15.Kochanek S, Clemens P R, Mitani K, Chen H H, Chan S, Caskey C T. Proc Natl Acad Sci USA. 1996;93:5731–5736. doi: 10.1073/pnas.93.12.5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar-Singh R, Chamberlain J S. Hum Mol Genet. 1996;5:913–921. doi: 10.1093/hmg/5.7.913. [DOI] [PubMed] [Google Scholar]
- 17.Parks R J, Chen L, Anton M, Sankar U, Rudnicki M A, Graham F L. Proc Natl Acad Sci USA. 1996;93:13565–13570. doi: 10.1073/pnas.93.24.13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy S, Kitamura M, Harris-Stansil T, Dai Y, Phipps M L. J Virol. 1997;71:1842–1849. doi: 10.1128/jvi.71.3.1842-1849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schiedner G, Morral N, Parks R J, Wu Y, Koopmans S C, Langston C, Graham F L, Beaudet A L, Kochanek S. Nat Gen. 1998;18:180–183. doi: 10.1038/ng0298-180. [DOI] [PubMed] [Google Scholar]
- 20.Morsy M A, Gu M C, Motzel S, Zhao J, Lin J, Su Q, Allen H, Franlin L, Parks R J, Graham F L, et al. Proc Natl Acad Sci USA. 1996;95:7866–7871. doi: 10.1073/pnas.95.14.7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berns K I, Linden M R. BioEssays. 1995;17:237–245. doi: 10.1002/bies.950170310. [DOI] [PubMed] [Google Scholar]
- 22.Kotin R M, Siniscalco M, Samulski R J, Zhu X, Hunter L, Laughlin C A, McLaughlin S, Muzyczka N, Rocchi M, Berns K I. Proc Natl Acad Sci USA. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samulski R J, Zhu X, Xiao X, Brook J D, Housman D E, Epstein N, Hunter L A. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Surosky R T, Urabe M, Godwin S G, McQuiston S A, Kurtzman G J, Ozawa K, Natsoulis G. J Virol. 1997;71:7951–7959. doi: 10.1128/jvi.71.10.7951-7959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamartina S, Roscilli G, Rinaudo D, Delmastro P, Toniatti C. J Virol. 1998;72:7653–7658. doi: 10.1128/jvi.72.9.7653-7658.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pieroni L, Fipaldini C, Monciotti A, Cimini D, Sgura A, Fattori E, Epifano O, Cortese R, Palombo F, La Monica N. Virology. 1998;249:249–259. doi: 10.1006/viro.1998.9332. [DOI] [PubMed] [Google Scholar]
- 27.Flotte T R, Carter B J. Gene Ther. 1995;2:357–362. [PubMed] [Google Scholar]
- 28.Shelling A N, Smith M. Gene Ther. 1994;1:165–169. [PubMed] [Google Scholar]
- 29.Balagué C, Kalla M, Zhang W W. J Virol. 1997;71:3299–3306. doi: 10.1128/jvi.71.4.3299-3306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weitzman M D, Fisher K J, Wilson J M. J Virol. 1996;70:1845–1854. doi: 10.1128/jvi.70.3.1845-1854.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L, Anton M, Graham F L. Somatic Cell Mol Genet. 1996;22:477–488. doi: 10.1007/BF02369439. [DOI] [PubMed] [Google Scholar]
- 32.Hitt M, Bett A J, Addison C L, Prevec L, Graham F L. In: Methods in Molecular Genetics. Adolph K W, editor. Vol. 7. San Diego: Academic; 1995. pp. 13–30. [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 34.Horer M, Weger S, Butz K, Hoppe-Seyler F, Geisen C, Kleinschmidt J A. J Virol. 1995;69:5485–5496. doi: 10.1128/jvi.69.9.5485-5496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bett A J. Ph.D. thesis. Hamilton, ON, Canada: McMaster University; 1995. [Google Scholar]
- 36.Graham F L. EMBO J. 1984;3:2917–2922. doi: 10.1002/j.1460-2075.1984.tb02232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parks R J, Graham F L. J Virol. 1997;71:3293–3298. doi: 10.1128/jvi.71.4.3293-3298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Simone V, Ciliberto G, Hardon E, Paonessa G, Lundberg L, Cortese R. EMBO J. 1987;6:2759–2766. doi: 10.1002/j.1460-2075.1987.tb02570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawrence J B, Villnave C A, Singer R H. Cell. 1988;52:51–61. doi: 10.1016/0092-8674(88)90530-2. [DOI] [PubMed] [Google Scholar]
- 40.Casto B C, Atchison R W, Hammon W McD. Virology. 1967;32:52–59. doi: 10.1016/0042-6822(67)90251-6. [DOI] [PubMed] [Google Scholar]
- 41.Parks W P, Casazza A M, Alcott J, Melnick J L. J Exp Med. 1968;127:91–108. doi: 10.1084/jem.127.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carter B J, Laughlin C A, De la Maza L M, Myers M. Virology. 1979;92:449–462. doi: 10.1016/0042-6822(79)90149-1. [DOI] [PubMed] [Google Scholar]
- 43.Chang L S, Hi Y, Shenk T. J Virol. 1989;63:3479–3488. doi: 10.1128/jvi.63.8.3479-3488.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi Y, Seto E, Chang L S, Shenk T. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 45.Holscher C, Horer M, Kleinschmidt J A, Zentgraf H, Burkle A, Heilbronn R. J Virol. 1994;68:7169–7177. doi: 10.1128/jvi.68.11.7169-7177.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samulski R J, Chang L-S, Shenk T. J Virol. 1987;61:3096–3101. doi: 10.1128/jvi.61.10.3096-3101.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Im D-S, Muzyczka N. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 48.Ni T-H, Zhou X, McCarty D M, Zolotukhin I, Muzyczka N. J Virol. 1994;68:1128–1138. doi: 10.1128/jvi.68.2.1128-1138.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith R H, Kotin R M. J Virol. 1998;72:4874–4881. doi: 10.1128/jvi.72.6.4874-4881.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ward J, Berns K I. J Mol Biol. 1991;218:791–804. doi: 10.1016/0022-2836(91)90267-a. [DOI] [PubMed] [Google Scholar]