Abstract
Recent data suggest that small differences in editing efficiency can have significant functional consequences. Here we present a fluorescent poisoned primer extension assay that is capable of distinguishing editing efficiency differences as low as 5%. For a poison-primer extension assay to be accurate, the extension product must stop at the intended base. Sometimes, however, it runs beyond. We tested the effect of specific enzyme–terminator combinations on the amount of run through. In the worst cases it accounted for 70% of the total signal, and in the best cases <5%. In addition, the specific base can affect run through, with G producing the least. The accuracy of the assay was demonstrated on templates derived from mixed plasmids and then verified on two biological substrates. Using either a K+ channel mRNA that contains a site for adenosine deamination or an ndhB mRNA that contains a site for cytidine deamination, the editing efficiency predicted by the assay closely matched that predicted by bulk sequencing of individual cDNA clones. This assay should prove useful for analyzing small changes in editing efficiency or for quantifying single nucleotide polymorphisms.
Keywords: RNA editing, poisoned primer extension, cytosine deamination, adenosine deamination, thermostable DNA polymerases, chain terminators
INTRODUCTION
RNA editing by base substitution is widespread in both plants and animals. The most common forms involve the hydrolytic deamination of either cytosine (C) to uracil (U) or of adenosine (A) to inosine (I). C → U editing is thought to occur in the organelles of all higher land plants (Freyer et al. 1997), and is critical for proper chloroplast and mitochondria function (Zito et al. 1997; Kofer et al. 1998; Horvath et al. 2000; Schmitz-Linneweber et al. 2005). In mammals, C → U editing regulates plasma cholesterol levels (Chen et al. 1987; Powell et al. 1987). Thus far, A → I editing has been found only in the nervous system of a wide range of animals, including nematodes (Morse and Bass 1999; Morse et al. 2002), insects (Hoopengardner et al. 2003), squid (Patton et al. 1997; Rosenthal and Bezanilla 2002), and mammals (Seeburg 1996; Burns et al. 1997). It is now becoming clear that the precise regulation of editing efficiency is critical for proper nervous system function. For example, some forms of epilepsy are associated with a reduced percentage of edited kainate receptor mRNAs (Sprengel et al. 1999; Vissel et al. 2001). Other mental disorders, such as schizophrenia and depression, are linked to reduced editing of the 5HT2C serotonin receptor mRNAs (Akbarian et al. 1995; Sodhi et al. 2001; Iwamoto and Kato 2003; Schmauss 2003). In all of these cases, the changes are subtle, and large numbers of individual cDNAs were sequenced in order to quantify the differences. It would be useful, therefore, to have an accurate and simple assay for measuring editing efficiency.
Assays that measure RNA editing efficiencies have relied on two basic approaches: DNA sequencing, or primer extension. Using DNA sequencing, the most accurate method is to sequence individual clones from RT-PCR reactions. A major drawback to this approach is that it is labor intensive and expensive, especially when comparing multiple samples. Direct sequencing of PCR products using standard dye-labeled terminators is not accurate, and can only be used to measure qualitative differences. In general, poisoned primer extension (PPE) assays have been the method of choice for comparing editing efficiencies between multiple samples. These assays are based on using a polymerase to extend a 32P end-labeled primer through the editing site in the presence of three deoxynucleotide triphosphates and one dideoxynucleotide triphosphate. The dideoxynucleotide, or chain terminator, normally contains the same base as the edited ribonucleotide. Thus, if a transcript is edited, the extension stops at the editing site. If not, it extends to the next instance of the edited nucleotide in the chain. To estimate editing efficiency, reactions are separated on an acrylamide gel and the relative proportions of the two extension products are quantified.
The first PPE assays used RNA templates and reverse transcriptase as a polymerase (Dabiri et al. 1996; Maas et al. 1996; O'Connell et al. 1997). This approach required large amounts of template, high specific activity primers, and often generated extraneous bands that made accurate quantification difficult. The sensitivity of the assay was greatly improved by using PCR products as a template and thermostable polymerases (Burns et al. 1997; Schiffer and Heinemann 1999; Peeters and Hanson 2002). By amplifying the template with RT-PCR, tiny amounts of mRNA could be assayed. The factors that contribute to the accuracy of the assay, however, have not been examined in detail. In this study, we make further improvements to the assay by using fluorescently labeled primers and by directly quantifying how a variety of different polymerases, chain terminators, and templates affect accuracy.
RESULTS
Our overall aim was to make an accurate, sensitive assay without using radioisotopes. Here we show that fluorescently labeled primers in conjunction with a thermostable polymerase produce a sensitive assay. Accuracy, however, is affected by the polymerase and terminator used. For the assay to function properly, it is essential that the extension stops at the intended base. Yet, under many conditions, this is not the case. Frequently, the extension runs through to the next instance of the terminator, or even beyond. The extent of this run through depends on the specific polymerase and terminator combination.
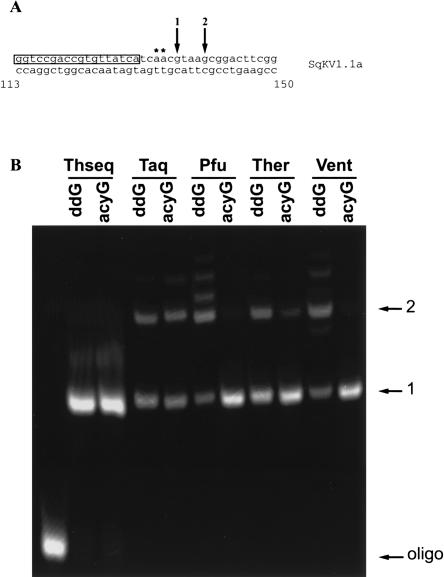
Figure 1 shows results of the PPE assay using five different polymerases and two different terminators. As a template, a PCR amplification of a squid giant axon K+ channel clone (SqKV1.1A) was used. Because this clone was fully genomic, no stops are expected at the editing sites (Fig. 1A, asterisks). Instead, all products should terminate at nt 136, the first instance of the poisoned nucleotide (nt) after the primer (arrow 1). Figure 1B indicates that for all polymerase–terminator combinations, a proportion of the product runs through to the next G (nt 140, arrow 2), and in some cases, beyond. However, the extent of run through varied greatly depending on the polymerase–terminator combination (Table 1). For example, in the worst cases [Pfu and Vent exo(−) coupled with ddGTP], the run through is ∼70%. For Taq, it was about 40% for both terminators. The combinations of Vent exo(−) or Pfu with acycloGTP, produced only 6% run through. Thermosequenase run through was even lower (3%), but an additional, unexplained band also was present. This band was smaller than that predicted for the run through, and constituted 7% of the total intensity. Based on these results and cost, we selected Vent exo(−) and acyclonucleotides for subsequent analyses. It should be noted that for all polymerases, even small amounts of dNTP contamination generate significant run through that renders the assay useless (data not shown). For this reason, the method of template purification is critical. In this study, PCR products were gel purified. Ethanol precipitation or column purification on glass fibers (Qiagen PCR purification kit) were not adequate.
FIGURE 1.
Effect of enzyme and terminator on chain termination. (A) Map of relevant SqKV1.1A sequence used as a template for the assay. Primer sequence is boxed. Asterisks denote editing sites (A → I) relevant in Figure 4. Arrow 1 indicates the first instance of the terminator (intended stop) and arrow 2 indicates the second instance (run through). (B) Fluorescence scan of a polyacrylamide gel containing SqKV1.1A assay products resulting from different polymerase–terminator combinations. Theseq = Thermosequenase; Ther = Therminator; Vent = Vent exo(−); ddG = dideoxy guanosine. Arrows correspond to fragments generated by stops indicated in A.
TABLE 1.
Quantification of the effect of enzyme and terminator on chain termination
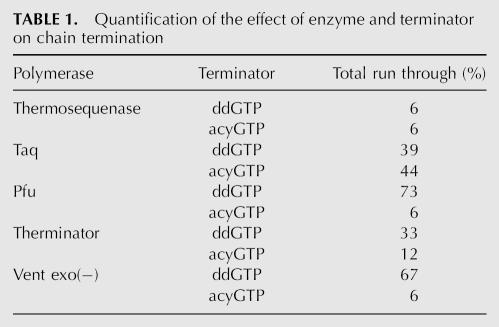
In addition to the polymerase and terminator, the specific base can affect the amount of run through. Figure 2 shows the unedited seagrass ndhB cDNA plasmid clone assayed with acycloG, C, A, and T. The template was changed to demonstrate the robustness of the assay on different substrates. AcycloG produced no run through, while acycloA produced the highest amount of run through (7%). The other bases were intermediate. This same pattern also was observed in the SqKV1.1A template (data not shown).
FIGURE 2.
Effect of base choice on run through. (A) Map of relevant Syr ndhB sequence used as a template for the assay. Primer sequence is boxed. Arrows indicate the first instance of each base. (B) Fluorescence scan of a polyacrylamide gel containing assay products of Syr ndhB unedited plasmid template using different acyclo bases. Numbers in parentheses above lanes are the number of bases to the second instance of the respective bases (run through; marked by asterisks on gel). (C) Quantified percent run through (RT) for each base.
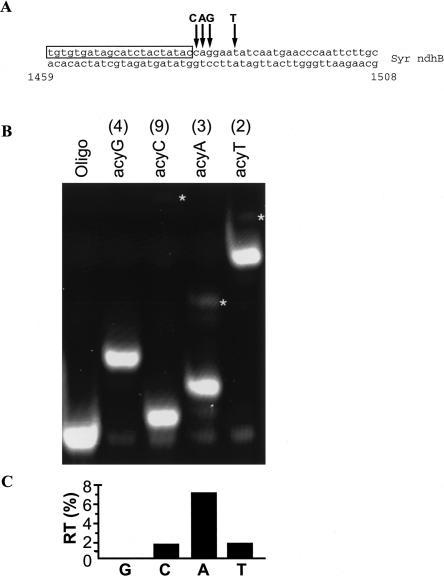
To determine the assay's accuracy and resolution, we mixed edited and unedited ndhB cDNA plasmid clones to cover a wide range of editing efficiencies (Fig. 3). The two plasmids contained either a C or T at nt 1481, but were otherwise identical. Position 1481 is immediately adjacent to the primer and the next C is 9 nt downstream (Fig. 3A). A visual inspection of the gel indicated that there was little run through with acycloC (<2%; Fig. 2B). Band intensities from this gel were quantified, and are presented in Figure 3C. Clearly, they accurately predict the ratios of the plasmid mixes with an R 2 value for a linear fit to the data of 0.995. The discrepancy between predicted and actual results was between 1.5% and 5.5% error (95% confidence interval), and represents an unknown combination of pipeting and assay error. Reproducibility was tested by repetitive runs on a template derived from a 1:1 plasmid mix (50% editing, see Materials and Methods). In this case, the SD was ±1.2% (N = 8). Although we could detect the presence of a base that made up only 2% of the total, because of the error described above, only differences of 5% or greater could be resolved with accuracy.
FIGURE 3.
Accuracy of the assay on known template mixes. (A) Map of relevant Syr ndhB sequence used as a template for the assay. Primer sequence is boxed. Asterisk denotes editing site (C → U). Arrow 1 indicates the first instance of the terminator after the editing site (intended stop if editing site is edited). (B) Fluorescence scan of a polyacrylamide gel containing assay products of Syr ndhB plasmid templates. Plasmid clones of edited or unedited templates were mixed prior to PCR amplification to produce heterogenous templates for the assay. Numbers above the lanes indicate the percent of edited template in the mixture. (C) Quantification of the known template mix assay. The known ratio of edited to unedited plasmid is plotted against the values measured in the assay from B. The line represents the expected results if known and measured values are identical (y = x). The least-squares fit of the actual data is defined by the equation y = x + 3, F(1,14) = 2871, p << 0.001).
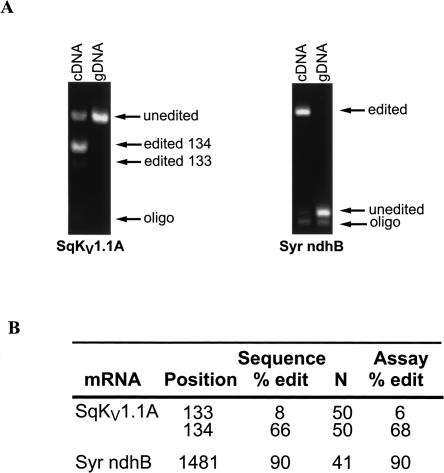
Finally, the assay's ability to accurately measure editing percentages in two biological samples was tested (Fig. 4). The general strategy was to compare assay results with those obtained by sequencing multiple cDNA clones from the same preparation. For SqKV1.1A, the entire coding sequence was amplified from giant axon-specific cDNA and cloned. A total of 50 individual clones were sequenced to completion. Two editing sites, nt 133 and 134, were used for the present purposes (see Fig. 1A for map). Nucleotide 134 has been previously identified as an editing site (Rosenthal and Bezanilla 2002) using a direct sequencing method. In the present study, 66% of the clones were edited at that position (Fig. 4B). At nt 133, only 8% of the clones were edited. This position was not identified in the study cited above, probably because the low editing percentage was below the threshold of the direct sequencing assay used in that study. As a template for the assay, SqKV1.1A PCR product was used. Both editing sites were easily detected and for both sites the editing efficiency estimated by the assay closely matched that measured by sequencing (68% and 6%). As expected, the same PCR product using genomic DNA as a template showed no sign of editing. It should be noted that since we are measuring two editing sites simultaneously, the editing percentage at nt 134 is a measurement of only those templates not edited at nt 133. If editing at nt 134 influences editing at nt 134 (either negatively or positively), then an error would result. For example, if 100% of the templates that are edited at nt 133 were also edited at nt 134, then the assay measurements would be 2% lower than the actual value. At the opposite extreme, if 0% of the templates that are edited at nt 133 were also edited at nt 134, then the assay measurements would be 4% higher than the actual value. In the case of SqKV1.1A, the error is very small because of the low editing percentage at nt 133. For more general applications, two sites can be measured at the same time only when the first site is edited at a low percentage (i.e., <10%).
FIGURE 4.
Utility of assay on natural substrates. (A) Fluorescence scan of a polyacrylamide gel containing assay products of cDNA and gDNA templates. SqKV1.1A was amplified from squid giant fiber lobe cDNA and from gill tissue gDNA. Syr ndhB was amplified from green leaf tissue cDNA and gDNA. (B) Quantification of the natural substrate assay in A compared with sequencing individual cDNAs.
The seagrass templates produced similar results (Fig. 4B). Sequences from 41 individual clones revealed that 90% contained a T at position 1481, whereas 10% contained a C. In contrast, 100% of clones from the same PCR reaction amplified from genomic DNA showed a C at the same position. The assay results closely mirrored the sequencing data. Analysis of the assay showed the genomic template to have 0% editing (100% C) while the cDNA template showed a T 90% of the time. From these results we conclude that position 1481 is edited and that the assay accurately predicts the efficiency.
DISCUSSION
In this study we present a sensitive and accurate PPE assay. Sensitivity is limited only by the RT-PCR reaction used to generate the template, and therefore it should be possible to quantify editing in single cells. Because the assay relies on fluorescently labeled primers, multiple editing sites can be measured simultaneously by using primers tagged with different fluors. The accuracy of the assay is dependent mainly on the specific polymerase, terminator, and base combination used. These factors have a significant impact on the degree of run through (up to 70% in some cases). We found that Vent exo(−) and acycloG yielded run through of 0%–6%. Pfu polymerase and acycloG gave comparable results. Thermosequenase in combination with ddNTPs also has low run through, probably due to its enhanced ability to incorporate modified nucleotides (Tabor and Richardson 1987). However, it also has a pyrophosphorolysis activity, necessitating the addition of pyrophosphatase to the reaction mixture (Tabor and Richardson 1990; Vander Horn et al. 1997). In our hands, Thermosequenase generated a diffuse band that was slightly larger than the correct product (see Fig. 1B) but too small to be due to run through. The genesis of this band is unknown. Because Vent exo(–) did not produce this band, and because it is significantly less expensive than Thermosequenase, it was selected.
To our knowledge, the most accurate assay described to date is based on Thermosequenase, ddNTPs, and radiolabeled primers (Schiffer and Heinemann 1999; Peeters and Hanson 2002). Although the total error was not quantified in these cases, Peeters and Hanson (2002) estimated that differences in editing efficiency as low as 20% could be resolved accurately. In our version of the assay, we were able to accurately distinguish differences in editing efficiency as low as 5% (total error). Although we could detect editing at lower percentages, the limit for comparisons is based on the sum of human error and the assay's accuracy and reproducibility. The ability to measure subtle differences in editing efficiency is becoming increasingly important, particularly for clinical studies. For example, subtle reductions in editing efficiency have been associated with clinical depression, suicide, certain forms of epilepsy, and schizophrenia (Sprengel et al. 1999; Sodhi et al. 2001; Vissel et al. 2001; Gurevich et al. 2002; Schmauss 2003). At certain editing sites, the difference between healthy and diseased samples is on the order of 5%. The assay presented in this paper will help resolve these differences.
MATERIALS AND METHODS
We optimized our PPE assay using two different mRNA templates, SqKV1.1A and Syr ndhB. SqKV1.1A (U50543) encodes the delayed rectifier K+ channel for the giant axon of the squid Loligo opalescens and is edited at multiple positions (Rosenthal and Bezanilla 2002). Here we focus on nt 133 and 134 (codon N45), both A → I substitutions. Position 133, uncovered in this work, is edited at a low percentage and was not identified as an editing site in the original publication. The second mRNA template used in this work, Syr ndhB, encodes the ndhB subunit of NAD(P)H dehydrogenase from the chloroplasts of the seagrass Syringodium filiforme. Position 1481 (codon P494 or edit site ndhB-10, as described by Tsudzuki et al. 2001) is edited in Arabadopsis (Tillich et al. 2005), corn (Maier et al. 1995), and tobacco (Tsudzuki et al. 2001), but has not been studied previously in seagrasses.
For SqKV1.1A, adult Loligo opalescens were collected in Monterey, California. Both giant fiber lobes and one gill were dissected from individual specimens. The giant fiber lobes were immersed in RNA Later (Ambion) and the gill was immediately frozen in liquid nitrogen. Total RNA was extracted from the giant fiber lobes using the RNAqueous Micro kit from Ambion. The entire RNA sample was then transcribed into cDNA using the Superscript III RT kit from Invitrogen using the oligo dT primer. The final product was diluted 5× with H2O prior to amplification. Genomic DNA was isolated from gill tissue using Genomic-Tip 500/G columns from Qiagen according to the supplied protocol. Either 1 μL of cDNA, 200 ng of genomic DNA, or 100 pg of cloned SqKV1.1A plasmid was used as a template to amplify nt 20–477 of SqKV1.1A using oligonucleotides SqKVPR11 (CATTGCTGCTTCCAAGGTCTCTTG) and SqKVPR12 (CAAGAGCCATACACGTCGCTGG) and Hotstar Taq (Qiagen). After being gel purified using the MinElute Gel Extraction Kit (Qiagen), this PCR product served as a template for the assay. Full-length cDNAs for DNA sequencing were amplified using primers SqKVPR3 (AGTCGTGGGGTGACAACCG) and SqKVPR4 (GCATGTATCAAGGCCATAGTCTCTCA). DNA sequencing was performed by the Bioresource Center of Cornell University using ABI sequencers.
The seagrass, Syringodium filiforme, was collected from Escambron, Puerto Rico. Total RNA was immediately extracted from cleaned leaf tissue using the RNAqueous kit with Plant RNA Isolation Aid from Ambion and transcribed into cDNA as described above; however, oligonucleotide ndhBcomp 1R (CTGATGATCGAGTCGATTCCATG) was used to prime synthesis. Genomic DNA was immediately isolated from leaf tissue of the same individual used for the RNA extraction using the DNeasy Plant Mini Kit from Qiagen. For amplification of the ndhB template, 1 μL of cDNA or DNA was used with oligonucleotides ndhB 9F (ATGGTTTCTCTTGGCTATATGG) (Heinze 2005) and ndhBcomp 2R (CCAGTTAGTAAGAGGGATCTTG) with Hot StarTaq (Qiagen). These oligos span the 699 bp intron of ndhB, and therefore allowed easy separation of cDNA from gDNA or unspliced cDNA. PCR products were gel purified as described above before use in the assay.
Complete ndhB transcripts for sequencing were amplified using oligonucleotides ndhcomp 2F (GGGACTTTTTCGGAGATTGG) and ndhBcomp 2R and sequenced as described above. Edited and unedited plasmids were mixed prior to amplification to calibrate the assay. To determine the standard deviation of repetitive runs of the same sample, a 1:1 plasmid mix was run in eight separate reactions on eight different gels.
The basic assay consisted of 0.5 pmol of flourescently labeled oligonucleotide, 0.1–2 pmol template, 0.05 mM dNTP (each of three dNTPs), 0.05 mM appropriate terminator (Acyclonucleotide, New England Biolabs) or ddNTP (Invitrogen), 20 mM Tris-HCl, 10 mM (NH4)2SO4, 10 mM KCl, 2 mM MgSO4, 0.1% Triton X-100, pH 8.8, and 1 unit Vent exo(−) DNA polymerase (New England Biolabs) in a total volume of 20 μL. In addition to Vent exo(−), Thermosequenase (USB), Taq (Qiagen), Pfu (Stratagene), and Therminator (New England Biolabs) were tested. When other polymerases were used, the recommended buffer was substituted. Reactions were cycled between 10 and 40 times, depending on the amount of template, through the following steps: 94°C for 15 sec, 55°C for 20 sec, and 72°C for 30 sec. The total number of cycles required to incorporate most of the primers into extension products was determined empirically. IE-HPLC purified 5′ Hexachlorofluorecein (HEX) labeled primers were from Integrated DNA Technologies. Assay products were electrophoresed on a 38 cm long, 15% acrylamide gel at 25 V/cm in 1× TBE. Following electrophoresis, gels were scanned directly at 532 nm using a Typhoon 9200 imager (GE Healthcare). Green fluorescence was collected using a 555 nm bandpass-20 filter and the photomultiplier tube voltage was set at 550.
Scans were analyzed using ImageQuant software. Band intensities were compared using the maximal value of the integrated pixel intensity for a two-pixel line drawn through the middle of each lane. The assay products that extended past the intended stop were considered “run through.” Run through percentage was measured for every assay using 100% unedited or edited templates, and was calculated as the ratio of run through band (or bands) intensity to the sum of all bands (excluding the primer). In the case of multiple run through bands, only those bands with intensities ≥10% of the total intensity were considered. Assays were corrected for run through by using the following equations:
where CI1 = corrected intensity for stop 1 and CI2 = corrected intensity for stop 2, BI1 = measured band intensity at stop 1, BI2 = measured band intensity at stop 2, and RT = % run through. The corrected values were then used to calculate the editing efficiency (% edit) as the ratio between the corrected edit site intensity (either CI1 or CI2, depending on the edit site or terminator used) to the total intensity (sum of CI1 and CI2).
ACKNOWLEDGMENTS
This work was directly supported by IBN-0344070 (NSF) and NS039405-06 (NIH-SNRP). The research infrastructure was partially supported by the RCMI Program, UPR Medical Sciences Campus, grant number G12RR03051.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.166906.
REFERENCES
- Akbarian, S., Smith, M.A., Jones, E.G. Editing for an AMPA receptor subunit RNA in prefrontal cortex and striatum in Alzheimer's disease, Huntington's disease and schizophrenia. Brain Res. 1995;699:297–304. doi: 10.1016/0006-8993(95)00922-d. [DOI] [PubMed] [Google Scholar]
- Burns, C.M., Chu, H., Rueter, S.M., Hutchinson, L.K., Canton, H., Sanders-Bush, E., Emeson, R.B. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- Chen, S.H., Habib, G., Yang, C.Y., Gu, Z.W., Lee, B.R., Weng, S.A., Silberman, S.R., Cai, S.J., Deslypere, J.P., Rosseneu, M., et al. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science. 1987;238:363–366. doi: 10.1126/science.3659919. [DOI] [PubMed] [Google Scholar]
- Dabiri, G.A., Lai, F., Drakas, R.A., Nishikura, K. Editing of the GLuR-B ion channel RNA in vitro by recombinant double-stranded RNA adenosine deaminase. EMBO J. 1996;15:34–45. [PMC free article] [PubMed] [Google Scholar]
- Freyer, R., Kiefer-Meyer, M.C., Kossel, H. Occurrence of plastid RNA editing in all major lineages of land plants. Proc. Natl. Acad. Sci. 1997;94:6285–6290. doi: 10.1073/pnas.94.12.6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich, I., Tamir, H., Arango, V., Dwork, A.J., Mann, J.J., Schmauss, C. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron. 2002;34:349–356. doi: 10.1016/s0896-6273(02)00660-8. [DOI] [PubMed] [Google Scholar]
- Heinze, B. 2005. A database for PCR primers for the study of the chloroplast genome in plants.http//bfw.ac.at/200/1859.html [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopengardner, B., Bhalla, T., Staber, C., Reenan, R. Nervous system targets of RNA editing identified by comparative genomics. Science. 2003;301:832–836. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- Horvath, E.M., Peter, S.O., Joet, T., Rumeau, D., Cournac, L., Horvath, G.V., Kavanagh, T.A., Schafer, C., Peltier, G., Medgyesy, P. Targeted inactivation of the plastid ndhB gene in tobacco results in an enhanced sensitivity of photosynthesis to moderate stomatal closure. Plant Physiol. 2000;123:1337–1349. doi: 10.1104/pp.123.4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto, K., Kato, T. RNA editing of serotonin 2C receptor in human postmortem brains of major mental disorders. Neurosci. Lett. 2003;346:169–172. doi: 10.1016/s0304-3940(03)00608-6. [DOI] [PubMed] [Google Scholar]
- Kofer, W., Koop, H.-U., Wanner, G., Steinmüller, K. Mutagenesis of the genes encoding subunits A, C, H, I, J and K of the plastid NAD(P)H-plastoquinone-oxidoreductase in tobacco by polyethylene glycol-mediated plastome transformation. Mol. Gen. Genet. 1998;258:166–173. doi: 10.1007/s004380050719. [DOI] [PubMed] [Google Scholar]
- Maas, S., Melcher, T., Herb, A., Seeburg, P.H., Keller, W., Krause, S., Higuchi, M., O'Connell, M.A. Structural requirements for RNA editing in glutamate receptor pre-mRNAs by recombinant double-stranded RNA adenosine deaminase. J. Biol. Chem. 1996;271:12221–12226. doi: 10.1074/jbc.271.21.12221. [DOI] [PubMed] [Google Scholar]
- Maier, R.M., Neckermann, K., Igloi, G.L., Kossel, H. Complete sequence of the maize chloroplast genome: Gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol. 1995;251:614–628. doi: 10.1006/jmbi.1995.0460. [DOI] [PubMed] [Google Scholar]
- Morse, D.P., Bass, B.L. Long RNA hairpins that contain inosine are present in Caenorhabditis elegans poly(A)+ RNA. Proc. Natl. Acad. Sci. 1999;96:6048–6053. doi: 10.1073/pnas.96.11.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse, D.P., Aruscavage, P.J., Bass, B.L. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc. Natl. Acad. Sci. 2002;99:7906–7911. doi: 10.1073/pnas.112704299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell, M.A., Gerber, A., Keller, W. Purification of human double-stranded RNA-specific editase 1 (hRED1) involved in editing of brain glutamate receptor B pre-mRNA. J. Biol. Chem. 1997;272:473–478. doi: 10.1074/jbc.272.1.473. [DOI] [PubMed] [Google Scholar]
- Patton, D.E., Silva, T., Bezanilla, F. RNA editing generates a diverse array of transcripts encoding squid Kv2 K+ channels with altered functional properties. Neuron. 1997;19:711–722. doi: 10.1016/s0896-6273(00)80383-9. [DOI] [PubMed] [Google Scholar]
- Peeters, N.M., Hanson, M.R. Transcript abundance supercedes editing efficiency as a factor in developmental variation of chloroplast gene expression. RNA. 2002;8:497–511. doi: 10.1017/s1355838202029424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, L.M., Wallis, S.C., Pease, R.J., Edwards, Y.H., Knott, T.J., Scott, J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987;50:831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- Rosenthal, J.J., Bezanilla, F. Extensive editing of mRNAs for the squid delayed rectifier K+ channel regulates subunit tetramerization. Neuron. 2002;34:743–757. doi: 10.1016/s0896-6273(02)00701-8. [DOI] [PubMed] [Google Scholar]
- Schiffer, H.H., Heinemann, S.F. A quantitative method to detect RNA editing events. Anal. Biochem. 1999;276:257–260. doi: 10.1006/abio.1999.4369. [DOI] [PubMed] [Google Scholar]
- Schmauss, C. Serotonin 2C receptors: Suicide, serotonin, and runaway RNA editing. Neuroscientist. 2003;9:237–242. doi: 10.1177/1073858403253669. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber, C., Kushnir, S., Babiychuk, E., Poltnigg, P., Herrmann, R.G., Maier, R.M. Pigment deficiency in nightshade/tobacco cybrids is caused by the failure to edit the plastid ATPase α-subunit mRNA. Plant Cell. 2005;17:1815–1828. doi: 10.1105/tpc.105.032474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeburg, P.H. The role of RNA editing in controlling glutamate receptor channel properties. J. Neurochem. 1996;66:1–5. doi: 10.1046/j.1471-4159.1996.66010001.x. [DOI] [PubMed] [Google Scholar]
- Sodhi, M.S., Burnet, P.W., Makoff, A.J., Kerwin, R.W., Harrison, P.J. RNA editing of the 5-HT(2C) receptor is reduced in schizophrenia. Mol. Psychiatry. 2001;6:373–379. doi: 10.1038/sj.mp.4000920. [DOI] [PubMed] [Google Scholar]
- Sprengel, R., Higuchi, M., Monyer, H., Seeburg, P.H. Glutamate receptor channels: A possible link between RNA editing in the brain and epilepsy. Adv. Neurol. 1999;79:525–534. [PubMed] [Google Scholar]
- Tabor, S., Richardson, C.C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc. Natl. Acad. Sci. 1987;84:4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor, S., Richardson, C.C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Effect of pyrophosphorolysis and metal ions. J. Biol. Chem. 1990;265:8322–8328. [PubMed] [Google Scholar]
- Tillich, M., Funk, H.T., Schmitz-Linneweber, C., Poltnigg, P., Sabater, B., Martin, M., Maier, R.M. Editing of plastid RNA in Arabidopsis thaliana ecotypes. Plant J. 2005;43:708–715. doi: 10.1111/j.1365-313X.2005.02484.x. [DOI] [PubMed] [Google Scholar]
- Tsudzuki, T., Wakasugi, T., Sugiura, M. Comparative analysis of RNA editing sites in higher plant chloroplasts. J. Mol. Evol. 2001;53:327–332. doi: 10.1007/s002390010222. [DOI] [PubMed] [Google Scholar]
- Vander Horn, P.B., Davis, M.C., Cunniff, J.J., Ruan, C., McArdle, B.F., Samols, S.B., Szasz, J., Hu, G., Hujer, K.M., Domke, S.T., et al. Thermo Sequenase DNA polymerase and T. acidophilum pyrophosphatase: New thermostable enzymes for DNA sequencing. Biotechniques. 1997;22:758–762. 764–755. doi: 10.2144/97224pf02. [DOI] [PubMed] [Google Scholar]
- Vissel, B., Royle, G.A., Christie, B.R., Schiffer, H.H., Ghetti, A., Tritto, T., Perez-Otano, I., Radcliffe, R.A., Seamans, J., Sejnowski, T., et al. The role of RNA editing of kainate receptors in synaptic plasticity and seizures. Neuron. 2001;29:217–227. doi: 10.1016/s0896-6273(01)00192-1. [DOI] [PubMed] [Google Scholar]
- Zito, F., Zuras, R., Choquet, Y., Kossel, H., Wollman, F.A. Mutations of cytochrome b6 in Chlamydomonas reinhardtii disclose the functional significance for a proline to leucine conversion by petB editing in maize and tobacco. Plant Mol. Biol. 1997;33:79–86. doi: 10.1023/a:1005734809834. [DOI] [PubMed] [Google Scholar]