Abstract
The cell has many ways to regulate the production of proteins. One mechanism is through the changes to the machinery of translation initiation. These alterations favor the translation of one subset of mRNAs over another. It was first shown that internal ribosome entry sites (IRESes) within viral RNA genomes allowed the production of viral proteins more efficiently than most of the host proteins. The RNA secondary structure of viral IRESes has sometimes been conserved between viral species even though the primary sequences differ. These structures are important for IRES function, but no similar structure conservation has yet to be shown in cellular IRES. With the advances in mathematical modeling and computational approaches to complex biological problems, is there a way to predict an IRES in a data set of unknown sequences? This review examines what is known about cellular IRES structures, as well as the data sets and tools available to examine this question. We find that the lengths, number of upstream AUGs, and %GC content of 5′-UTRs of the human transcriptome have a similar distribution to those of published IRES-containing UTRs. Although the UTRs containing IRESes are on the average longer, almost half of all 5′-UTRs are long enough to contain an IRES. Examination of the available RNA structure prediction software and RNA motif searching programs indicates that while these programs are useful tools to fine tune the empirically determined RNA secondary structure, the accuracy of de novo secondary structure prediction of large RNA molecules and subsequent identification of new IRES elements by computational approaches, is still not possible.
Keywords: IRES, RNA, secondary structure, prediction software
INTRODUCTION
The cell has many ways of regulating the production of a protein from a gene. In this review, we focus on one mechanism of initiating the translation of messenger RNA without following the standard pathway used by the majority of mRNAs. Translation initiation via the internal ribosome entry site (IRES) is a mechanism by which the cell allows translation of specific mRNAs because of unique RNA sequences in their untranslated regions (UTRs), which recruit ribosomes. Although some viral IRESes share primary sequence or secondary structure similarity, this similarity has not yet been found between known cellular IRESes, which has raised some questions regarding the existence of cellular IRESes (Kozak 2001, 2003). A sequence containing a cellular IRES can be cloned and tested for function outside of its native gene context, but it is not known what commonality exists to allow various IRESes to recruit ribosomes without using the standard protein translation initiation mechanism.
There have been many very good reviews on IRESes over the years that are helpful in understanding the different facets of this mechanism of translation initiation. Some favorites are: Hellen and Sarnow (2001), Jackson et al. (1995), on Picornavirus (Belsham and Sonenberg 1996), FMDV IRES structure/function (Martinez-Salas et al. 2002), structural aspects relevant to medical intervention (Gallego 2002), with respect to cancer (Holcik 2004; Stoneley and Willis 2004), the very critical and controversial Kozak (2001, 2003), and on stress-related IRES (Holcik et al. 2000; Holcik and Sonenberg 2005; Lewis and Holcik 2005). In this review, we examine the published data that could aid in the detection of unknown IRESes in an mRNA database and the RNA motif/structure-predicting and search programs presently available, which could be applicable to this search.
MECHANISM OF CAP-DEPENDENT TRANSLATION, INHIBITION, AND IRES
The textbook explanation of standard translation initiation has the cap-binding protein, the eukaryotic initiation factor eIF4E, recruited to the 5′-end of the mRNA, where it binds to the modified “cap” nucleotide, a methyl7GDP (guanadyldiphosphate) on the 5′-end of all cellular mRNAs. With this, the initiation factor eIF4G binds to both the cap-binding protein and the mRNA. eIF4G is called the “scaffold” protein as it also binds eIF4A, an ATP-dependent helicase, which is responsible for unwinding the secondary and tertiary structure of the RNA during translation, as well as the kinase MnkI, which regulates eIF4E-binding activity through phosphorylation. The initiation factors eIF4A, eIF4G, and eIF4E are also known collectively as the protein complex eIF4F. These factors are key to the recruitment of the ribosome to the 5′-cap structure of mRNA. The 40S small subunit of the ribosome, the initial part of the ribosome to bind the mRNA, binds the activated start codon tRNA, fMet-tRNAi fMet with eIF2 and GTP, to its P-site. This binding is promoted by eIF1A, the eIF4 factors, and eIF3. Together these proteins and tRNAs, now called the 43S complex, are believed to scan along the mRNA with the use of ATP to drive them, looking for the proper place to start translation. At the point where the proper start codon is found by the 43S scanning complex, the GTP with eIF2 is hydrolyzed to GDP in the presence of eIF5. Several factors dissociate, leaving the 40S subunit with the Met-tRNA anticodon base-paired to the start codon. The large 60S ribosomal subunit then joins the small subunit, and protein synthesis begins. For a review on translation initiation factors, see Dever (2002) and on structural aspects of initiation factors, see Sonenberg and Dever (2003).
At some points of a cell's life, this standard mechanism of translation initiation is compromised, but at these times, some mRNAs use an alternative form of initiation that does not require the cap nucleotide as a congregation site for initiation factors. This was first observed in Picornavirus infections, where the uncapped RNA viral genomes of the polio and encephalomyocarditis virus were efficiently translated in eukaryotic cells through the binding of the ribosome to an internal portion of the 5′-UTR of the viral RNA (Jang et al. 1988; Pyronnet et al. 2001). A viral protease cleaves the two forms of eIF4G, shutting down host protein (Gradi et al. 1998a,b; Svitkin et al. 1999). Even though the cleaved form of eIF4GI does not bind to eIF4E, it has been shown to translate capped mRNA but much less efficiently than viral RNA (Ali et al. 2001).
There are several other mechanisms that lower the efficiency of cap-dependant translation initiation inside a cell besides viral infection. During mitosis, the eIF4E-binding proteins (4E-BPs) are hypophosphorylated and competitively bind onto the cap binding protein, eIF4E, preventing eIF4E from forming the eIF4F initiation complex (Pyronnet et al. 2001). The phosphorylation states of eIF4E and the 4E-BPs in different cellular conditions have been well reviewed by Gingras et al. (1999).
During times of cellular perturbation, changes in protein levels and mRNA levels do not always correlate (Ideker et al. 2001; Nishizuka et al. 2003). Cellular stresses and the induction of apoptosis cause inhibition of standard translation initiation through the phosphorylation of the eIF2α subunit by one of the four known eIF2α kinases in mammalian cells: HRI, PKR, PERK, and GCN2 (Proud 2005). The initiation factor eIF2 is the adapter protein that binds Met-tRNA and GTP as part of the 43S preinitiation complex. Phosphorylation of the α-subunit of eIF2 creates tighter binding to eIF2B, which prevents the GDP-GTP exchange activity of eIF2B needed for the recycling of eIF2 for the successive rounds of initiation of protein synthesis. During apoptosis, the cell still requires the de novo synthesis of proteins required for the orderly breakdown of the cell, but the standard protein translation initiation machinery is also slowed down with changes to the phosphorylation of eIF4G (Ling et al. 2005), eIF4E, and 4E-BPs (Clemens 2001), as well as caspase cleavage of several canonical initiation factors, eIF4B, eIF3, eIF2α, and proteins of the eIF4G family (Clemens et al. 2000). Other molecular events like the hyperphosphorylation of eIF4GII (Pyronnet et al. 2001) or Hsp27 binding to eIF4G during heat shock may hinder the formation of eIF4F (Cuesta et al. 2000).
Despite these cellular conditions that change the normal translation initiation machinery, some cellular mRNAs and viral RNA still retain the ability to recruit ribosomes to a region of their 5′-UTR to initiate translation. There have been at least 85 cellular IRESes and 39 viral IRESes described in the literature so far, as given in Tables 1 and 2. These sequences have been shown to exhibit cap-independent translation initiation. The standard method of defining this activity has been the ability of the sequence to initiate translation of the second open reading frame (ORF) or cistron in a bicistronic construct. There have been some criticism and caveats attached to the use of bicistronic constructs for measuring IRES activity (Hellen and Sarnow 2001; Kozak 2001; Sherrill et al. 2004), and checks need to be made for promoter activity in the UTR, reinitiation of ribosome on the second ORF, aberrant splicing (Holcik et al. 2005), and inconsistent values (Hennecke et al. 2001) of the dual luciferase reporter gene construct. Re-evaluation of the 5′-UTRs of PDGF (Han et al. 2003), PIM-1 (Wang et al. 2005), and the human cyclin-dependent kinase inhibitor, p27kip1 (Liu et al. 2005), has shown that they do not have IRESes as was initially thought (Bernstein et al. 1997; Johannes et al. 1999; Miskimins et al. 2001), but the sequences are able to function as promoters. A statistically rigorous methodology has been published to evaluate the output values of the bicistronic constructs (Jacobs and Dinman 2004). The only drawback to this method becoming universally used is that the minimal sample size might require 25–50 measurements, which is more than the three to nine sample measurements that are usually done.
TABLE 1.
Reported cellular IRES
TABLE 1.
Continued
TABLE 1.
Continued
TABLE 2.
Reported viral IRES
TABLE 2.
Reported viral IRES
The problems in assessing IRES activity must be kept in mind when using the published IRES sequences as a data set for bioinformatics tools. Like all biological databases, the data are not perfect, as some sequence activity may have been inadvertently misinterpreted.
mRNA binding proteins
When the function of some canonical translation initiation factors have been disabled or limited in the cell to make cap-dependent translation less efficient, other RNA binding proteins have been found to be required or enhance IRES-mediated translation initiation. RNA binding proteins have many functions that affect translation from localization of mRNA in the cytoplasm (zipcode), stabilization of message (AREs), metabolite riboswitch (Sudarsan et al. 2003), and translation repression. Our interest in this case is in IRES-specific cellular trans-acting factors (ITAFs). Some of the mRNA binding proteins implicated in IRES-mediated translation are polypyrimidine tract binding protein PTB/hnRNP I (Giraud et al. 2001; Mitchell et al. 2001, 2003, 2005; Pickering et al. 2003; Cho et al. 2005); La autoantigen (Holcik and Korneluk 2000; Bhattacharyya and Das 2005; Marash and Kimchi 2005); hnRNP A1 (Bonnal et al. 2005); hnRNP C1/C2 (Sella et al. 1999; Millard et al. 2000; Holcik et al. 2003); hnRNP E, hnRNP K, DAP5/p86 (Henis-Korenblit et al. 2000; Nevins et al. 2003; Warnakulasuriyarachchi et al. 2004; Marash and Kimchi 2005); Unr (Mitchell et al. 2001, 2003; Tinton et al. 2005); p60 (Vagner et al. 1996), HuR (Millard et al. 2000), and PCBP1 (Pickering et al. 2003). For a more comprehensive list, see the online IRES database (http://ifr31w3.toulouse.inserm.fr/IRESdatabase/) or the list in Stoneley and Willis (2004).
One group of ITAFs is the many heterogeneous nuclear ribonucleoproteins, hnRNPs that bind onto transcripts and form ribonucleoprotein complexes. They play a key role in pre-mRNA processing as well as mRNA export, localization, stability, and translation (Dreyfuss et al. 2002). As an example, PTB has been connected with several functions such as splicing repression, pre-mRNA 3′-end processing, mRNA localization, and mRNA stability. It has also been shown to be involved in IRES activation with both viral (Sanderbrand et al. 2000; Wollerton et al. 2001; Bieleski et al. 2004) and cellular (Giraud et al. 2001; Mitchell et al. 2003, 2005; Pickering et al. 2003) IRES but has been also shown to be inhibitory to IRES activity in Unr (Cornelis et al. 2005) or Bip (Kim et al. 2000). Several IRESes have a polypyrimidine tract at the 3′-end that has been shown to be important for activity (Kaminski et al. 1994) and a recognition site for PTB (Kolupaeva et al. 1996). The PTB consensus recognition sequence shown to be important in the HCV IRES sequence is CYYYYCYYYY(G|Y)G, where Y is a pyrimidine (Anwar et al. 2000). It is not known if the location of this sequence within the HCV IRES structure is important as only the first four bases are consistently in single-stranded regions, but some believe the binding site needs to be within double-stranded regions (Mitchell et al. 2005). PTB has at least three splicing isoforms (Wollerton et al. 2001) and caspase (Back et al. 2002b) or viral protease cleavage products (Back et al. 2002a), which affect IRES activity to differing degrees, as well as tissue-specific paralogs (Pilipenko et al. 2001; Gooding et al. 2003). The protein also contains four RNA binding motifs, thus it is not surprising that other recognition sites for PTB/RNA interactions have also been found that do not match this consensus sequence (Wollerton et al. 2001). It is understandable from the variety of PTB forms available that there does not seem to be an unequivocal consensus sequence for PTB.
The ITAF La is very promiscuous as well with its binding site recognition, and the requirement for IRES function is not always clear. For example, an early in vitro study had shown that the HCV IRES could function without any noncanonical factors, but recent in vivo studies show that La is required for HCV IRES translation (Shimazaki et al. 2002; Costa-Mattioli et al. 2004). The RNA binding protein Unr is required for HRV IRES activity, and although all five cold shock domains of Unr are necessary for RNA binding, the binding seems to be a nonspecific sequence interaction (Brown and Jackson 2004).
Although there is no overall consistency as to which RNA binding proteins are required for IRES activity, they may be consistently involved in ribonucleoprotein complexes that exist during specific cellular contexts of stress, cell cycle, or particular mechanism of viral control over cellular functions. Using microarrays, some RNA binding proteins have been shown to interact with a specific group of transcripts during times of cellular perturbation (Tenenbaum et al. 2002, 2003). It has been postulated that RNA binding proteins play a role in coregulating the translation of groups of proteins analogous to bacterial operons (Keene and Tenenbaum 2002). Therefore, knowing an RNA binding protein that binds an IRES in a specific cellular context would suggest that other IRESes in that context may also be bound to the same protein.
The binding of the canonical initiation factors also affect IRES activity. When eIF4E, the cap binding protein, has been removed (Hernandez et al. 2004), the Drosophila reaper IRES initiates translation more efficiently than capped messages. The eIF4G family member DAP5/p97 is cleaved to DAP5/p86 and enhances IRES-mediated translation (Henis-Korenblit et al. 2002; Nevins et al. 2003; Warnakulasuriyarachchi et al. 2004; Marash and Kimchi 2005). The importance of ribosomal proteins in IRES translation initiation was shown using a genome-wide RNAi screen of Drosophila genes. One hundred twelve cellular genes were found that were required for infection by the IRES-dependent Drosophila C virus (Cherry et al. 2005). More than 50% of these were genes of ribosomal proteins, two of which when deleted affected IRES but not cap-dependent translation. This suggests that some ITAFs will be not be unique to only IRES translation machinery and will include components of the ribosome as well.
The ITAFs are abundant in the cell and seem quite ubiquitous but are not required in all examples of known IRES activity. Specific initiation factors and ribosomal proteins will also play a role. They can also exist in several forms of post-translational modification and bind a wide range of sequence motifs. Their own regulation and regulated cellular localization would control the IRES function. Do the mRNA binding proteins that are used for induction of IRES activity in the subsets of the published IRES possibly have shared binding motifs? Although the RNA binding protein data are not as well defined to use for database searching alone, they could still be used in a search algorithm as an added weight in a search.
Functional classes
There appear to be differences among IRESes as to which proteins are necessary to bind to the UTR in order to recruit the ribosome for translation. When this is coupled with the results of different IRESes initiating translation with varying efficiency dependent on which cell type or cellular context they are measured in (Nevins et al. 2003), the results suggest that there exist several IRES classes. Many groups have pointed out this observation already. Several groups have examined their characterized IRESes in several cell lines, comparing it to other IRESes, and found specific IRESes will have more activity in one specific cell line relative to another IRES (Stoneley et al. 2000; Jopling and Willis 2001; Nevins et al. 2003; Jopling et al. 2004). This may be due to different available protein factors in each cell line and, therefore, the different subsets of mRNAs that different cells or tissues are able to translate at any one time. This may also explain the lack of primary sequence similarity between the cellular IRES. For example, the 5′-UTRs containing IRES from c-Myc and cyclin D are dependent on the activity of AKT through p38 MAPK and ERK signaling to initiate translation (Shi et al. 2005) but not the 5′-UTR of P27kip1. The link may be the ITAFs PCBP1, PCBP2, and hnRPK, which are known to be required by the c-Myc IRES (Evans et al. 2003) and are regulated by phosphorylation (Shi et al. 2005).
In investigation of the FGF1 IRES activity in muscle and cell culture (Martineau et al. 2004), FGF1 has four separate 5′-UTRs, each exhibiting some IRES activity. FGF1A and C have similar activity to each other and that of FGF2 IRES but much less than the EMCV IRES in cell culture. The same IRES constructs electrotransferred into mouse muscle cells showed FGF1A to be much more active than FGF1C and similar to EMCV, while the FGF2 IRES seemed to exhibit no activity at all. Clearly the context of available ITAFs must favor the translation of one mRNA over another. A very similar contextual difference is seen where IRES from the muscle-relevant genes SMAD and utrophin are active in myoblast C2C12 muscle cells but not at all in 293T renal epithelial cells for SMAD (Shiroki et al. 2002) or differentiated muscle cells for utrophin (Miura et al. 2005). Other examples are the IRES from transcripts of the calcium channel proteins like Scamper exhibiting tissue-specific activity in kidney cells (De Pietri Tonelli et al. 2003) and the Nkx6.1 IRES being most active in β-cells (Watada et al. 2000). The Apaf1 IRES has been found to be more active in neuronal cell types possibly due to the presence of a neuronal isoform of PTB, which seems to confer greater activity than PTB-1 (Mitchell et al. 2003), and this may be where the APAF IRES is most physiologically relevant. This correlates well with the developmental problems found in the brains of Apaf-1 knockout mice (Cecconi et al. 1998). In contrast, the IRES of HRV is repressed in neuronal cells because of the presence of the mRNA binding protein DRBP76/NF90 (Merrill et al. 2006). A list of IRESes that are regulated can be found in the review by Komar and Hatzoglou (2005). Whereas viral IRES might share a more universal context of translation regulation and therefore some similarity has been found, the larger number of cellular contexts that would require different regulation infers a large number of classes of IRES with many different sequence and structural components. Even saying there are regulatory classes of IRESes may be too rigid, as there may be a loose overlap of some mRNA translation.
18S complementation and modular elements
It has been pointed out by Chappell et al. (2000) that partial IRES activity is still retained when the segments of a 5′-UTR ascribed to full IRES activity are partially deleted, and therefore some elements that help to recruit ribosomes must still exist in the remaining sequence. In some UTRs, nonoverlapping segments of sequence retain partial IRES activity, suggesting that different modules may act synergistically to provide full IRES activity in vivo. Nonoverlapping fragments of the Kv1.4 IRES that each retained partial activity showed different patterns of activity when tested in a variety of cell types (Jang et al. 2004). This suggests distinct modular elements with different modes of regulation. Some examples of postulated IRES elements are listed in Table 3. Chappell et al. (2000) had found an example of a distinct module with a 9-nt motif in the Gtx mRNA, complementary to 18S rRNA that can function as a site for internal initiation of translation. This is an attractive model for ribosome recruitment for internal initiation as it parallels the function of the bacterial Shine-Delgarno sequence and 16S rRNA. This complementation to the 18S rRNA is not new and has also been shown necessary for reattachment for scanning of the mRNA during “ribosome shunting,” where the ribosome is stalled because of a complex structure in the mRNA and must disengage and then re-engage the mRNA on the 3′-side of the structure (Yueh and Schneider 2000). Similar motifs with IRES activity exhibiting 18S rRNA complementation were found in a library of random nucleotides (Owens et al. 2001) and the plant potato virus Y (Akbergenov et al. 2004), and have been suggested to be in YAP1 and TIF4631 transcripts of yeast (Zhou et al. 2001). Additional copies of the elements from either Gtx or the potato virus Y arranged in tandem produced additive increases of IRES activity. A segment in the 3′-UTR of a hibiscus plant virus, although perhaps not an IRES, was shown to enhance translation through an 18S rRNA complementation (Koh et al. 2002). This is not a universal truth about sequences complementary to 18S, as several of the matches in the YAP1 and Tif4631 5′-UTRs are in segments that confer no IRES activity (Zhou et al. 2001). The rRNA/mRNA interaction cannot be too great, as increasing the degree of complementation increases the thermodynamic stability of this interaction, lowering the efficiency of translation, and if large enough can completely inhibit translation (Hu et al. 1999; Verrier and Jean-Jean 2000). This differential ability to translate an mRNA based on the rRNA and mRNA interactions as well as the interactions due to changes in the structure of ribosomal subunits from one cell type to another is postulated as an overall method of translation control in the “ribosome filter hypothesis” of Mauro and Edelman (2002). The last few years have seen the realization that rRNA is no longer just a scaffolding for the ribosomal proteins; the ribosome is a ribozyme, and translation is now more RNA centered (Woese 2001). It is, therefore, reasonable to believe with present-day evidence that interactions between mRNA and rRNA can enhance translation initiation.
TABLE 3.
Reported minimal IRES modules
Relatively small IRES elements have also been synthetically derived using a bicistronic vector with 50-nt-long randomly generated sequences inserted between two reporter genes (Venkatesan and Dasgupta 2001). Although two of the sequences that exhibited IRES activity showed no sequence homology with any transcripts in the public databases, the findings did show that a relatively small sequence is needed for IRES activity. In this case, the synthetic IRESes did not complement 18S rRNA but were able to compete for the trans-activating factors used by polio virus IRES. This procedure was repeated again in yeast, and 56 IRES functioning elements were found, with 10 having significant matches in the yeast 18S rRNA (Zhou et al. 2003).
Fine mapping of the c-Myc IRES sequence found a minimal 50-base sequence that was responsible for the bulk of the IRES activity (Cencig et al. 2004). The c-Myc sequence did not seem to map to 18S rRNA and was also not dependent on the secondary structure formed for activity. Within this element, two 14-nt segments with an AX6AC motif were chiefly responsible for ribosome recruitment reducing c-Myc IRES activity to these modular units. The IRES found within the APC gene that possibly is responsible for the milder form of adenomatosis polyposis coli is only 84 nt long (Heppner Goss et al. 2002) and may be also representative of a modular unit with IRES activity.
In an acyclovir-resistant strain of herpes simplex virus, very low levels of thymidine kinase are translated by a small IRES that requires only 12 bases and contains a CUG start codon (Griffiths and Coen 2005). The low levels of thymidine kinase prevent its proper activation of acyclovir but are high enough to retain the virus's pathogenicity. As well as being a new IRES modular element, this also shows how very low levels of IRES translation initiation can be physiologically significant.
The above evidence supports the notion that an IRES can be made of modular units that act synergistically with or without trans-acting factors to recruit a ribosome and enhance translation initiation. The overall structure of the UTR for these small units may be somewhat unimportant. A very stable structure like a large hairpin or a tertiary structure that would bury a modular recognition sequence would probably still have an effect on IRES activity. The modulation of the structure by mRNA binding proteins could, therefore, have a regulatory IRES effect by changing the access to these small modules. As these types of short IRES sequences may be available elsewhere on mRNA sequences, they suggest the possibility of a much expanded proteome (Griffiths and Coen 2005).
DATABASES AND IRES
A searchable database of published IRESes exists at http://ifr31w3.toulouse.inserm.fr/IRESdatabase/ (Bonnal et al. 2003a). Within this database, the IRESes are classified in several categories based on the function of the gene in which they are found, how the genes are regulated, and with which factors the IRES elements interact. Links to some of the sequences are directed to NCBI entries. Unfortunately, the database has not been updated since 2002; Table 1 includes all published cellular IRES sequences to date. Many of the IRES publications did not give exact coordinates to specific database entries for the UTR sequences used in their research. For this reason, Table 1 was compiled to include the GenBank Identifier (GI) number and the position on the sequence of the minimal known IRES for all published IRESes to date. Multiple fasta files of all the cellular or viral IRES sequences are available as supplemental data at http://bio.site.uottawa.ca/IRES_rna_supplement/ as either the full-length UTRs or as a collection of minimal IRES sequences or full UTRs where the minimal sequence is not known. Filling the gap left by the previous IRES database, a new online database of IRES sequences has been created at http://www.iresite.org/IRESite_web.php (Mokrejs et al. 2006).
IRES structure data for 16 cellular IRES and five viral IRES sequences are available at Rfam (Griffiths-Jones et al. 2003), a database of noncoding RNA. For the most part, published IRES structures are used initially at Rfam, and a covariance sequence model is built using UTR sequences from different transcript entries and known ortholog sequences. Where no published structure is known, the covariance models for the IRES structures have either been built from energy minimization program prediction (RNAfold) or from a sequence alignment using the PFOLD program. It should be noted that if the alignment sequences did not give any covariance data, the seed structure would dominate the resulting structure model. This could possibly be the case for HCV, which appears to be little different from its seed structure (Lytle et al. 2002) but dissimilar from the HCV NMR structure data (Lukavsky et al. 2003). For tertiary structure information of IRES elements, the data can be found in the structure database at NCBI or RNABase (Murthy and Rose 2003), a specific RNA structure database that may include additional annotations not included in the standard 3D structure files. At present, there are 13 IRES-related elements from GBV-B, enterovirus, and mostly HCV. Tertiary structures can be converted into secondary structure diagrams using RNAVIEW (Yang et al. 2003).
There are other available data sets of IRES-enriched sequences where microarray expression studies have been carried out on cells undergoing a perturbation that would turn down the amount of cap-dependent translation and allow for a greater amount of IRES-mediated translation. Assuming that IRES elements under these conditions would more efficiently recruit ribosomes, experiments that isolated mRNAs bound by multiple ribosomes (polysomes) should show mRNAs with a greater likelihood of containing an IRES element. Johannes et al. (1999) isolated polysomes from poliovirus-infected cells and evaluated any increased amount of bound mRNA compared to noninfected cells using a 10K human cDNA array. They found ∼200 transcripts with a greater than twofold enrichment of polysome-bound mRNA of ∼7000 hits that produced an acceptable signal on the microarray. There are several other studies using microarray examination of polysome-bound mRNA, cells with and without von Hippel-Landu tumor suppressor protein (Galban et al. 2003), rapamycin's effect on translation (Grolleau et al. 2002), resting and mitogenically activated fibroblast (Zong et al. 1999), synchronized cells in the mitotic cycle (Qin and Sarnow 2004), and in yeast during cell cycle arrest (Serikawa et al. 2003) or rapid growth (Arava et al. 2003). Lately some researchers have combined the proteomic approach of measuring both the increase of protein expression using 2D gels and mass spectrometry as well as the mRNA levels (Ideker et al. 2001; Grolleau et al. 2002). This allows one to specifically discover transcripts that are regulated post-transcriptionally. Microarray studies of mRNA bound to mRNA binding proteins implicated as ITAFs will also yield a database of transcripts of which some may contain IRES.
Databases of UTR sequences can be obtained from several sources. Using the Web-based EnsMart in Ensembl (Clamp et al. 2003), UTR data sets from a variety of genomes can be made with a user-defined number of flanking nucleotides. As almost all genes produce several transcripts, one must still consider filtering redundant 5′-UTRs, alternately spliced UTRs, or alternate UTRs produced by different promoters when creating a database. A prefiltered nonredundant database, UTRdb (Pesole et al. 2002), can be found at http://bighost.area.ba.cnr.it/BIG/UTRHome/, in which UTRs with >90% sequence overlap and 95% nucleotide identity within the overlapping region (Grillo et al. 1996) have been removed. UTRdb has annotated the sequences that contain patterns matching possible RNA structure/sequence, which would confer known regulatory elements such as the iron response element (IRE), histone 3′-UTR stem–loop structure (HSL3), or even IRES elements as adapted from the computationally predicted structure of Le and Maizel (1997). In Release 20 of UTRdb, there are ∼34,000 human 5′-UTR sequences and the IRES pattern is found ∼7000 times, ∼20% of all the entries. Although the number of mRNAs containing IRES elements could be quite abundant, there is doubt that the pattern actually detects IRESes. The RefSeq database from NCBI can also be used to produce a data set from the transcript entries but requires the user to write his or her own program/script to pull out the UTR sequences. A redundant version of the RefSeq UTRs is available at http://bighost.area.ba.cnr.it/BIG/UTRHome/.
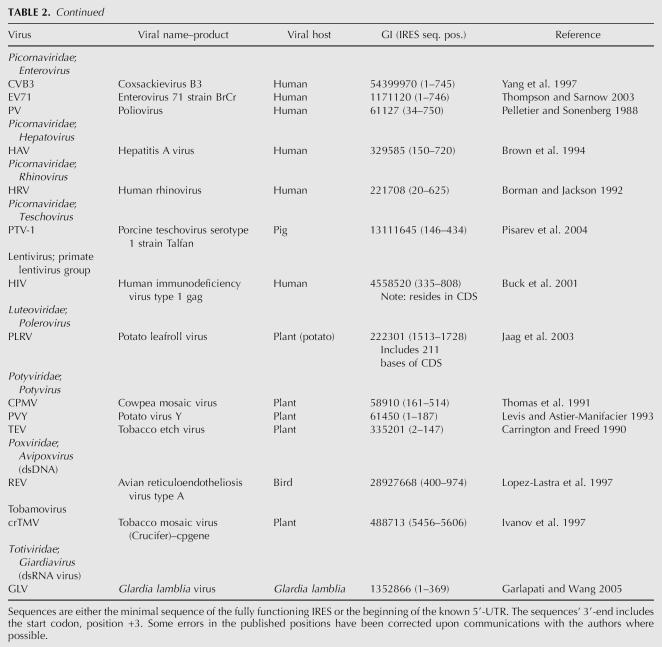
These data sets allow one to compare the overlaps in the UTR sequence characteristics believed to have IRES elements as discovered by different means versus a complete set of a genome's UTRs. For instance, of the ∼23,000 human genes in the human genome, around half of them have AUGs upstream of the defined AUG start codon, as seen in Figure 1, A and B. The number of upstream AUGs does not seem to depend on length as there is a subset of UTRs with more AUGs than expected randomly, as shown in Figure 1B, but the overall frequency is less than expected from random. Although many of the upstream AUGs will most likely be relatively close to the Kozak consensus sequence (Suzuki et al. 2000), this alone does not seem to mark them as proper start codons (Peri and Pandey 2001). Around 10% of the upstream AUGs would be considered strong start codons (RXXAUGG, where R is a purine) as defined by Kozak (2005), whereas 41% to 45% of the coding region start codons would be considered strong start codons. Perhaps the ribosome starts to translate with the first AUG that is in the proper context of the surrounding folded RNA structure, bound proteins, and/or a continuous open reading frame. Possibly the start codon is decided on by a “pioneer” scan (Ishigaki et al. 2001) of the mRNA or an RNA binding protein (McBratney and Sarnow 1996).
FIGURE 1.
(A) The number of AUGs found upstream of the natural start codon in 5′-UTRs is compared in several nonredundant 5′-UTR data sets. Data come from human transcripts from RefSequation (21589, Release 13), RefSequation 5′-UTRs fully reviewed by NCBI staff (1049), and the 66 mammalian UTRs containing published IRES from Table 1. Data bins are represented as the percentage of the total entries in each data set. (B) The frequency of upstream AUGs in several nonredundant data sets. A histogram in several asynchronous bins containing the relative frequency of upstream AUGs as a percentage of each data set compares the 5′-UTR from human transcripts in RefSeq, and fully reviewed RefSeq UTRs. The distribution shows a selection for no upstream AUGs in all the data sets, but approximately half of all transcripts contain an upstream AUG where ∼10% are considered “strong” AUGs (RNNAUGG) by Kozak (2005). Approximately 45% of annotated start codons in these transcripts are “strong” AUGs.
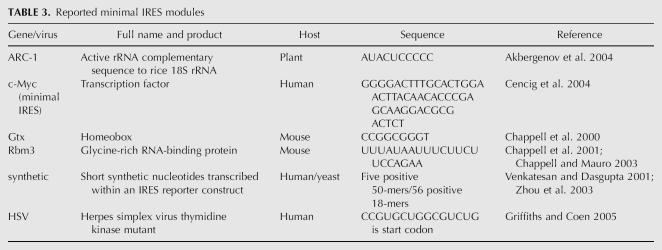
Each data set could be mapped to one common nonredundant set of genes so that ESTs from microarrays as well as genes that are represented by multiple sequence IDs in the public databases could be properly compared. Some basic questions regarding upstream AUGs, polypyrimidine tracks, UTR length, GC content, and putative thermodynamic equilibrium of the structures can be compared as well as comparing the different IRES predictions to further enrich a data set. As an example, we have compared the set of published IRESes from Table 1 with human 5′-UTR sequences from each of the UTRdb and/or RefSeq databases for upstream AUGs as discussed (Fig. 1), UTR length (Fig. 2), and % GC content (Fig. 3). The Ensembl data set gave similar results to RefSeq.
FIGURE 2.
The lengths of 5′-UTRs of human transcripts from nonredundant data sets. The frequency of different lengths of all the UTRs in each data set is placed in increasing bins of 50 nt and plotted as a percentage of the total number of UTRs in each database. A nonredundant data set of 5′-UTRs from UTRdb, RefSeq, fully reviewed transcripts of RefSeq (see Fig. 1 legend), and the mammalian transcripts containing IRES in their 5′-UTR from Table 1 are compared. The legend gives the median, mean, and third quartile values in each data set with the total number of UTRs in each data set in brackets.
FIGURE 3.
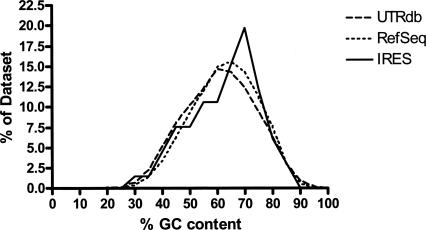
The distribution of the % GC content of human 5′-UTRs. The degree of RNA folding and structure stability can be partially assessed by the percentage of possible G and C base pairing within the UTR sequence and therefore the percent of Gs and Cs within the sequence. The % GC content of mammalian UTRs with published IRES are compared to the human 5′-UTRs from UTRdb and RefSeq and show a similar distribution. Plotted values are grouped in bins of five.
The most notable difference in cellular IRES sequence from the UTR data sets is that IRES-containing UTRs are generally longer (Fig. 2). It should be also noted that the median length of 5′-UTRs is >150 nt, longer than the commonly stated dogma of it being “usually <100 bases” (Lewin 2006). As minimal IRES sequences can be at that length or less, one conclusion from our comparison is that half of all UTRs are long enough to include an IRES. The % GC content of the IRES data set seems quite similar to the overall distribution of UTRs (Fig. 3), suggesting that the degree of RNA secondary structure stability as correlated to the amount of GC pairing possible is no different from that of most UTRs. A similar result is obtained if the secondary structure scanning algorithm of Rivas and Eddy (2000) is used instead of % GC content (data not shown). This does not negate that there could be secondary structure elements within a 5′-UTR that impede translation, but the overall degree of structure in UTRs with IRESes does not appear different from what is found in the distribution of all human 5′-UTRs.
Many more characteristics of the UTR sequences can be examined by comparing and crossing all of these data sets, looking for sequence or structural motifs that may be required for translation initiation in the periods of translation dysregulation.
IRES STRUCTURE
RNA motifs recognizable by RNA binding protein are often made up of structure and/or some primary sequence. Structures can be preserved without preserving the primary sequence, but proteins often bind onto specific sequence motifs, and thus to search for common attributes of known IRES sequences, one would expect a mixture of these two characters, structures, and some small sequence motifs being preserved. Although it is the 5′-UTR sequence that is responsible for recruiting the ribosome, the 3′-UTR may play a similar role (Izquierdo and Cuezva 2000) or synergistically enhance the IRES-mediated translation of an mRNA (Lopez de Quinto et al. 2002; Dobrikova et al. 2003; Koh et al. 2003). It is understood that in 3′-UTRs there is a role played by motifs that control translation (Mazumder et al. 2003). Virtually all the characterized IRESes have been tested and function independently of their 3′-UTRs; for this reason, just the 5′-UTR can be solely considered.
There are several ways to experimentally determine RNA structure; for example, X-ray crystallography, NMR, chemical and enzymatic structural probing, as well as mutational analysis (Kjems and Egebjerg 1998). Structures that have been predicted using computer programs often are functionally tested for validity with small sequence mutations (Kanamori and Nakashima 2001). X-ray crystallography and NMR, while more definitive in determining the structure, have limitations on either the type or length of the RNA molecules that can be investigated. RNA alone tends to form poor crystals that exhibit weak diffraction or heterogeneous samples (Ke and Doudna 2004), possibly because of the dynamic folding of many RNA molecules. The only crystal structures of RNA not complexed with proteins >76 bases in length in the structure databases are either ribonuclease P or group I intron ribozymes. The limitation for NMR is due to the severe spectral overlap of the four different nucleotides. These two methods have been used in examining small, structurally stable motifs extracted from larger RNA molecules. Enzymatic and chemical probing are aids in determining a secondary structure of an RNA molecule but often have some ambiguous results in their experiments caused by the difficulty in the art of doing the experiment and interpreting the results. These wet-laboratory determinations of structure can also be supported with phylogenetic data comparisons in which sequences that have changed have preserved the structure to preserve the function of the structure. The best data from all these approaches are used to support a proposed model, but conflicting secondary structure models do arise in the literature as has been evident in domain II of HCV (Honda et al. 1999; Zhao and Wimmer 2001; Lytle et al. 2002).
We must also consider how much of the proposed mRNA structure is as significant for IRES activity in vivo as it is in a naked state in vitro when it is derived. Messenger RNAs in vivo start folding as they are transcribed and are covered with the protein complexes required for translocation, splicing, stabilization, capping, polyadenylation, cellular localization, and translation. In vivo chemical probing of structures has been performed on relatively abundant RNAs like rRNA, telomerase RNA, and snRNA (Zaug and Cech 1995; Mathews et al. 2004), and could possibly be used to determine whether IRES structures determined in vitro exist in vivo as well.
Viral IRES structures
Several viral IRESes share similar secondary structures, suggesting that similar structures instead of specific sequence recognition sites are used to bind initiation factors used for cap-independent translation. This is a great aid in understanding the IRES mechanism overall, but it must be remembered that ssRNA viruses have a life cycle in the cytoplasm and will not interact with the nucleoprotein complex like cellular mRNA produced in the nucleus. Therefore, we would not expect the noncanonical protein factors to necessarily interact by exactly the same mechanism. Several groups have used characteristics of the viral IRES to separate them into three groups. Type I viral IRESes, which include entero- and rhinoviruses, translate poorly in rabbit reticulocyte lysates (RRL) and require the ribosome to bind and then scan downstream to a start codon 30–150 nt away. Type II viral IRESes include cardio- and apthoviruses, translate very efficiently in RRL, encompass the AUG start codon, and do not require scanning. Type I IRESes are stimulated by the eIF4G cleavage products produced by the viral protease, but Type II are not. Type III IRESes are typified by hepatitis A, do not translate at all in RRL, and encompass the AUG start codon. These classes, which may not be able to apply definitive separation of viral IRES types, serve to show how some viruses use a similar mechanism to initiate translation, while others use distinctly separate mechanisms to arrive at the same end.
Comparative studies involving covariation analysis as well as enzymatic and chemical probing of structures demonstrated a conservation of structures between members of the Picornaviridae enterovirus family, Poliovirus and coxsackievirus B3, and the human rhinovirus family (Rivera et al. 1988; Pilipenko et al. 1989b). Similar studies found sequence and structural conservation between the Picornaviridae cardiovirus family (EMCV, TMEV) and apthovirus FMDV (Pilipenko et al. 1989a). Further examination of variants and structural probing of PV (Pilipenko et al. 1992), mutational analysis of EMCV (Hoffman and Palmenberg 1995; Kolupaeva et al. 1996), and FMDV (Lopez de Quinto and Martinez-Salas 1997) have further refined the original structure models. The preservation of a stem structure in the base of region 3 of FMDV has been shown to be more important than the sequence that creates the structure (Martinez-Salas et al. 1996; Martinez-Salas et al. 2002). Recently the central domain was analyzed with chemical and enzymatic probing altering slightly the predicted structure (Fernandez-Miragall and Martinez-Salas 2003).
Several Flaviviridae viruses contain IRES sequences and have had their structures determined. In the pestivirus genera of Flaviviridae viruses, there is the bovine viral diarrhea virus (BVDV) and cholera swine fever virus (CSFV); in the hepacivirus genera, there is HCV; and unclassified are the GBV-A, B, and C viruses. Lemon and Honda (1997) reviewed the similarity and structural importance of their IRES structures. The IRES structure that has been studied the most of all IRESes is from the hepatitis C virus (HCV) because of its importance as a human pathogen. The initial secondary structure had been predicted using both enzymatic probing results as constraints in an early version of the MFOLD program (Zuker and Stiegler 1981) along with comparative computer-predicted models with the UTRs of BVDV and CSFV (Brown et al. 1992). There have been numerous studies adding to the refinement of the IRES structure (Lemon and Honda 1997; Lyons et al. 2001; Odreman-Macchioli et al. 2001), arriving at two similar models (Honda et al. 1999; Zhao and Wimmer 2001; Lytle et al. 2002) with some differences in domain II. Smaller stem–loop motifs of the HCV IRES structure have had their tertiary structure determined using both X-ray crystallography (Kieft et al. 2002) and NMR (Klinck et al. 2000; Lukavsky et al. 2000; Collier et al. 2002). Recently, the tertiary structure for domain II has been determined with NMR (Lukavsky et al. 2003) and finally resolves the conflicts of structural models proposed for this domain. It is important to note that the extensive chemical and enzymatic probing of the HCV IRES over 10 years was still not accurate because of the limitations of that approach. A structure model including the NMR and X-ray data is presented in Figure 4. Using cryo-EM, studies of the HCV IRES in the 40S ribosome subunit, domain II, which is necessary for IRES activity, specifically produced conformational changes to the 40S ribosome (Spahn et al. 2001). The segments IIIe and IIIf have recently been shown to interact with ribosomal protein S5 in an IRES-specific manner that does not seem to be important in cap-dependent translation initiation (Ray and Das 2004), suggesting that IRES RNA interaction with the ribosome can be different from that of non-IRES-containing mRNA even without considering nonribosomal protein factors.
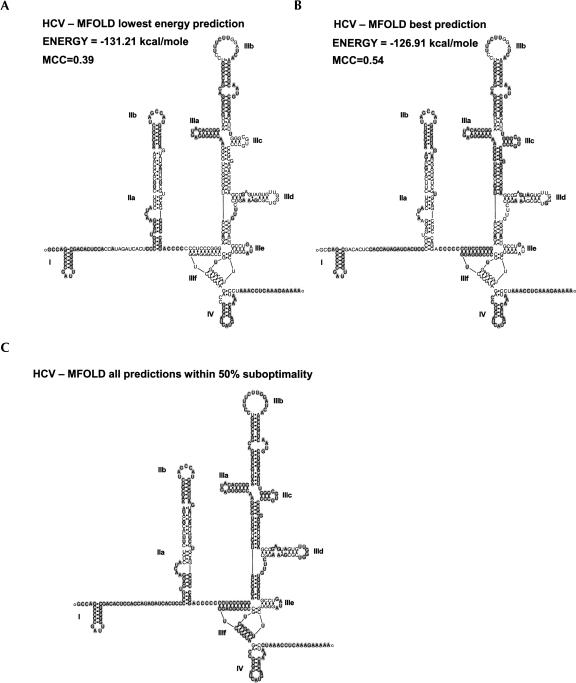
FIGURE 4.
The correctly predicted RNA secondary structure of HCV IRES by MFOLD. (A) The empirically predicted structure of the HCV IRES is shown with correctly predicted basepairs and nonpairing bases with gray background of the lowest energy prediction when using MFOLD with no constraints. (B) The best predicted structure was not the most thermodynamically stable fold, and the predicted base pairs and nonpairing bases shown are in gray. (C) All of the correctly predicted base pairs and nonpairing bases from all of the 36 predicted suboptimal folds using a 50% suboptimal parameter in MFOLD. Structures for figures have been produced with RnaViz2 (De Rijk et al. 2003).
The HCV IRES is an example of how both tertiary and secondary structure is important for IRES function. Conservation of specific stems in domain II preserves IRES function regardless of the sequence used (Honda et al. 1999). Point mutations that alter the tertiary structure of interdomain interactions (Kieft et al. 1999) and mutations to the pseudoknot (Wang et al. 1995) severely affect IRES activity. The importance of these domains has become clearer as domain III has been shown to directly interact with eIF3 and the 40S ribosomal subunit (Kieft et al. 2001) and cryo-EM studies have shown domains II and III wrapped on the 40S subunit, both interacting with the E-site and creating a conformational change of the subunit itself (Spahn et al. 2001). Further studies of this interaction show that the HCV IRES structure sits in the same spot of the 40S normally occupied by initiation factor eIF4G (Siridechadilok et al. 2005), suggesting that the IRES structure has taken the place of eIF4G, which is not needed for IRES function in vitro.
The other known Flaviviridae viral IRES GBV-B, BVDV, and CSFV share similar secondary structures to HCV (Lemon and Honda 1997). BVDV and CSFV structure models (Brown et al. 1992) were originally proposed by aligning four sequences, doing manual sequence covariation analysis, and using the determined base pairs as constraints on the MFOLD program (Zuker and Stiegler 1981). Mutation analysis has substantiated the CSFV pseudoknot (Rijnbrand et al. 1997; Fletcher and Jackson 2002), stem–loop IIIa (Fletcher and Jackson 2002), as well as other sections of the structure with enzymatic probing (Kolupaeva et al. 2000). Only a little supportive work (Grassmann et al. 2005) has been done on the original model of the BVDV UTR structure (Brown et al. 1992). GBV-B was modeled comparing similar sequences to HCV and using those that might base pair as constraints in the MFOLD program, followed by mutational studies on the similar stems and loops in domains II and III (Rijnbrand et al. 2000) as well as NMR on specific domain III stem–loops (Rijnbrand et al. 2004).
RNase P is an endoribonuclease that processes tRNA precursor 5′-ends to the correct length. It has been used to cleave the 5′-UTR IRES-containing regions of HCV, BVDV, CrPV, EMCV, and CSFV (Lyons and Robertson 2003). This suggests that the structures of these IRESes may mimic a portion of a tRNA. Lyons and Robertson (2003) postulate a model in which these viruses would occupy the E-site of the ribosome, positioning it for the AUG start codon downstream. A structural element similar to tRNA spatially situated so that the start codon would be properly placed in the ribosome would greatly enhance the ability to find IRESes in the database, as effective search methods exist that can find these structures within genomic sequences (Lowe and Eddy 1997; Tsui et al. 2003). These putative tRNA-like sequences may not reflect a complete tRNA structure as RNase P cleaves several different RNA structures (Gopalan et al. 2002) including a minimal stem structure of a tRNA molecule as a substrate (Zuleeg et al. 2001). A complete tRNA structure may also not be needed for affinity to the E-site of the ribosome. This proposed structural element used to direct the positioning of mRNA in the ribosome has an even greater role in CrPV IRES-mediated translation, where translation initiation occurs without eIF2 and an activated Met-tRNA in the P-site (Wilson et al. 2000a). The first activated tRNA starts translation on an alanine codon at the A-site. The interactions in CrPV substituting for Met-tRNA would be different from that of just a tRNA, but still suggest structural elements that may be shared with other IRESes. This is a good mechanism by the virus to overcome the antiviral response of the cell to shut off protein synthesis through PKR phosphorylation of eIF2α (Stark et al. 1998). Cellular stress leading to transcription of TNFα and IFN-γ activates PKR as well through conserved RNA motifs in the UTRs of their mRNAs (Kaempfer 2003), requiring cellular proteins to translate in this context in an alternative manner possibly akin to CrPV.
Other Dicistroviridae also seem to be able to initiate translation without a methionine tRNA. A structural model was proposed using the intergenic IRES sequence upstream of the capsid proteins from Plautria stali intestine virus (PSIV), Taura syndrome virus (TSV), and acute bee paralysis virus (ABPV) and used to search nucleotide databases with a pattern searching program (Nishiyama et al. 2003). The key region of the model had experimentally been shown to be a pseudoknot (PK1) 5′ of the capsid-coding region. This structure, even with relaxed search parameters, was not found in extensive database searches, strongly suggesting this functional structure to be distinct to Dicistroviridae.
Coxsackievirus B3, which is in the same family, has a GNRA loop in its IRES that influences the binding of La but not of PTB (Bhattacharyya and Das 2005). La binds to several places on the IRES and not specifically the loop, but the RNA structure of the IRES may change because of mutation of this loop, suggesting a structure requirement for La binding in this case.
There is a definite conservation of RNA structures between groups of viruses within their IRES sequences. The mutation analysis studies have shown that the preservation of these structures in viruses are more important for IRES function than the actual sequence.
Cellular IRES structures
We know that viruses have evolved regulatory mechanisms borrowed from host cells, and therefore it is felt that since some viruses have shared IRES RNA structure, some cellular IRESes most probably have a shared structure/function relationship. At this time, the secondary structure has been derived for several cellular IRESes with enzymatic and chemical probing from the transcripts of c-Myc (Le Quesne et al. 2001), L-Myc (Jopling et al. 2004), Apaf-1 (Mitchell et al. 2003), FGF-2 (Bonnal et al. 2003b), FGF1 (Martineau et al. 2004), Kv1.4 (Jang et al. 2004), Bag-1 (Pickering et al. 2004), Igf2 (Pedersen et al. 2002), cat-1 (Yaman et al. 2003), Mnt, and MTG8a (Mitchell et al. 2005). Their structure/function relationships are described below.
As has been mentioned previously, a common Y structure (Le and Maizel 1997) had been predicted for cellular IRESes based on the computational comparison of several orthologs of Bip and FGF2 UTRs. This pattern had been adapted for the PATSEARCH program (Grillo et al. 2003) to annotate the UTRdb entries as putative IRES motifs and is used by the UTRscan Web server. Of the 33,677 entries in Release 20 of UTRdb, 7122 entries have been annotated as IRESes, which is ∼21% of all the entries. Using the same pattern to search the cellular IRESes from Table 1 with RNAMotif finds the pattern in ∼21% of the entries, showing that this pattern is no more common in known IRES-containing UTRs than in all UTRs.
Some studies have proposed that mRNA binding proteins open up the natural RNA structure of the IRES and present single-stranded RNA for other ITAFs or the small ribosomal subunit to bind to. Therefore, the structure would not be the “landing pad” for the ribosome but the “attenuater” that is controlled by the ability of some proteins to change it. For example, the poly rC binding protein 1 (PCBP1) appears to open the Bag-1 IRES structure allowing PTB-1 to bind. Mutations that open up the binding region of the PCBP1 on the Bag-1 IRES seem to remove any requirement for this factor, even enhancing IRES activity after PTB-1 is added (Pickering et al. 2004). Although PCBP1’s role seems to be to open up the structure for PTB-1, it is not clear whether the structure is necessary for its binding. A similar mechanism was proposed for the ITAFs Unr and PTB in the APAF-1 IRES (Mitchell et al. 2003). In that case, Unr opens up the secondary structure allowing PTB to bind by possibly exposing recognition sequences that were previously buried or double stranded. IRES activity regulated by structure is also described by the “zipper model” of the CAT-1 (cationic amino acid transporter) transcript. This IRES structure only attains its active form upon amino acid starvation and the translation of an upstream ORF (Yaman et al. 2003). Structural probing has shown RNA–RNA interactions that inhibit IRES activity are opened up by the scanning ribosome and perhaps unknown ITAFs during the translation of the uORF.
The relevance of the c-Myc IRES structure is still outstanding. Initially, full activity was assigned to a 394-base region upstream of the AUG whose structure was derived using chemical and enzymatic probing (Le Quesne et al. 2001). Mapping of the ribosome landing site by engineering several AUGs along the 5′-UTR revealed that start codons that were at least 220 bases upstream of the standard AUG had no effect on the downstream reporter ORF. Interestingly, domain 1 (−380 to −221) upstream of the proposed ribosome binding site could have a negative effect on IRES-initiated translation if a couple of the dsRNA regions were disrupted, suggesting some structural requirements. Mutations that disrupt the predicted pseudoknots in position −185 to −347 only seemed to enhance IRES activity, perhaps showing in the case of this IRES that the pseudoknots attenuate IRES activity and proteins that would open them up would allow for IRES activity. More recent work has shown that the major portion of c-Myc IRES activity can be minimized to a 50-base region from −143 to −94 (Cencig et al. 2004). Analysis revealed that the sequence and not the proposed structure was important for IRES activity (Cencig et al. 2004). In vivo, the overall structure of the UTR may provide a role for IRES regulation, as increased levels of PCBP1 and PCBP2 enhance c-Myc IRES activity (Evans et al. 2003), possibly opening up the structure for other ITAFs and the ribosome as with Bag-1.
An interesting structural study has been performed on a segment of the Kv1.4 IRES where a series of deletion mutants have had their structures derived from enzymatic probing (Jang et al. 2004). A 0.2-kb fragment on the 3′-end of the 1.2-kb Kv1.4 UTR has only ∼25% of the activity of the full UTR, but specific internal deletions raised the activity to that of the full UTR. In contrast, any deletions of the polypyrimidine tract seemed to remove all activity even if some of the original structural elements were mostly preserved. As the 1-kb region upstream of this segment also retains activity on its own without the polypyrimidine tract, the conclusion seems to be that several modular elements of sequence and structure contribute to overall IRES activity. Unfortunately, the data are not clear on what defines the contributing elements other than the polypyrimidine tract.
FGF2 has a UTR with a unique G-quartet structure within its IRES (Bonnal et al. 2003b). Although deletion of this element cuts the relative IRES activity in half, deletion analysis has shown that stem–loop II has the greatest effect on overall activity. Deletion or changes of the loop alone can achieve this effect. Not all of the stem is required for activity, and it is not clear whether only the sequence of the loop needs to be available as a single-stranded region to preserve the effect of stem–loop II.
The minimal IRES sequence of FGF1A seems to be conserved over several mammalian species. Computationally predicted structures from the mouse, rat, cow, and two primate sequences seem very similar to the empirically derived human sequence structure (Martineau et al. 2004). The most important element DII, as shown by deletion analysis, retained more activity when mutations were made to it that preserved the structure rather than ones that changed the structure. This suggests the need for some structural elements for complete activity.
In all these studies on cellular IRES, only some show a preference for structure preservation to retain full activity, suggesting that there is no need for an overall structure as seen in some viral IRES.
RNA STRUCTURE PREDICTION AND SEARCH SOFTWARE
RNA secondary structure is usually predicted computationally by biologists in two ways. The first would use thermodynamic values for the base-paring, base-stacking, and near-neighbor forces to calculate the most thermodynamically stable structure in the “minimum free energy” method. The second method is a comparative method that examines the covariation of several homologous sequences to predict sequence positions that would consistently form base pairs even if specific base pairs change from one homologous sequence to another. Covariation is seen as the most accurate method of structure prediction but requires many sequences with similar structures to do this properly. The most extensive structure prediction using this method has been done by Guttell and colleagues for more than 25 years on ribosomal RNA, where crystallographic data of the ribosome have shown their structure models to be 97% accurate (Gutell et al. 2002). Covariation structure models have been predicted for viral IRESes, but unfortunately the relatively small number of similar sequences for cellular IRESes does not allow this method to be used to fully define a structure. Covariation models can also produce evolutionarily preserved structures or specific base pairing that may rely on cofactors such as ions or proteins. These structural elements would not be apparent when empirically examining naked RNA structures without these factors present. Variations and combinations of these two methodologies, thermodynamics and covariation, combined with other computational algorithms have increased the predictive ability of computer programs. Several programs have not been considered in this review if the limit on the sequence length is too short or if there is not an available implementation of the program for the typical user. For a review on the development of secondary structure prediction programs, see Zuker (2000), Higgs (2000), Mathews (2006) , and Mathews and Turner (2006). There have been a few studies (Perriquet et al. 2003; Doshi et al. 2004; Dowell and Eddy 2004; Ji et al. 2004) that have evaluated and compared the accuracy of some prediction software, some of which are described in the following section.
SINGLE SEQUENCE STRUCTURE PREDICTION
Thermodynamics
It has been hard to predict noncoding regions of transcripts within genomic sequence, leaving sequence annotation of UTR regions of messenger RNAs to empirical evidence of cDNA, RNA–RACE methods, and cap trapping methods (Carninci et al. 1996). Although distinct ordered structures may exist, the overall degree of secondary structure as measured by the lowest free energy of possible folds of noncoding regions of mRNA was not seen to be different from random sequence or from coding sequence (Workman and Krogh 1999; Rivas and Eddy 2000; Clote et al. 2005). There could be problems in assessing the actual energy status of the structure because tertiary interactions that would affect the overall energy calculations are not calculated or the global secondary structure's effects on the actual folding are not considered with each prediction (Workman and Krogh 1999). There has been some evidence to show that if one examines simpler putative hairpin structures within a nematode genomic sequence below a certain level of free energy, there are less of these sequences in UTR regions than intron and intergenic regions (Pervouchine et al. 2003), suggesting lower overall secondary structure in UTRs. We examined a few commonly used computer approaches to RNA structure or pattern predictions in more detail as it applies to IRESes.
ED_SCAN (Le et al. 2003)
ED_SCAN looks for local energy differences within a sequence from a sliding window. The energy difference (E diff) is represented by the lowest energy for a possible fold within the window subtracted by the lowest energy possible for a fold that has been constrained to prohibit any base pairing that has been used in the lowest energy fold. The greater the E diff of a window, the greater the order of the structure. In order to compare the E diff across the sequence, a Zscr is used that incorporates the E diff mean of all the windows analyzed and the standard deviation. Plotting the Zscr across a sequence has shown peaks for the iron response elements (IRE) in ferritin RNA, Rev elements in HIV, and let-7 RNAs in nematode and fruitfly (Le et al. 2003). The source code for the program can be downloaded for use; or if you do not have a compatible Fortran compiler, a Web server can be accessed through http://protein3d.ncifcrf.gov/shuyun/edscan.html.
We used ED_SCAN to predict small structures within the UTR of the known IRES structures of HCV, APAF-1, CSFV, or c-Myc. No known stem–loops correlate with any peak in the plots of Zscr against the sequence position (data not shown). The peaks within the UTR segments were not significantly higher than would be expected with random sequence with Zscr < 3. Therefore, in these four IRES structures, a notable local IRES structure could not be detected by ED_SCAN.
MFOLD (Zuker 2003), RNASTRUCTURE (Mathews 2004; Mathews et al. 2004), and RNAFOLD (VIENNA PACKAGE) (Hofacker 2003)
MFOLD is a powerful “minimum free energy” program and probably the most commonly used software for predicting RNA secondary structures based on the thermodynamics of the possible folds using a well-defined empirically determined energy table (Mathews et al. 1999). It has been evaluated in studies (Mathews et al. 1999; Doshi et al. 2004) and most recently has shown to average 41% accuracy of predicted base pairs for 16S and 23S RNA (Doshi et al. 2004). Studies on other data sets have shown an MFOLD accuracy of 56% (Dowell and Eddy 2004). Users of this software tend to assume that RNA folds will undertake their most thermodynamically stable form, the structure with the minimal free energy, but unfortunately in nature, if one examines functional RNAs like tRNA and RNase P RNA, this is not always true. Correct structures are believed to usually lie within 10% of the minimum energy value predicted (Shapiro et al. 2001), and therefore MFOLD and similar programs will also produce suboptimal folds to increase the likelihood of an accurate prediction. MFOLD has some common limitations in that it cannot predict interloop base pairing or pseudoknots and, like all other programs, does not show noncanonical base pairing with the exception of the G-U wobble, although noncanonical pairing energy values are calculated from the energy tables. PKNOT (Rivas and Eddy 1999) takes the MFOLD algorithm a step further to allow pseudoknots, but it does not allow user-defined constraints on the predicted folds. The computational expense of inclusion of pseudoknots restricts the sequences examined by PKNOT to ∼140 bases. HotKnots uses a heuristic method of solving the most stable structure with pseudoknots and takes larger sequences (Ren et al. 2005). Pseudoknots were formerly considered rare, but several viral and cellular IRESes contain them. A recent analysis of RNA structure topologies estimates pseudoknots in 90% of the sequences over 120 bases in length (Kim et al. 2004). One of MFOLD's great advantages is that experimental or phylogenetic comparative data can be used as constraints in the structure prediction, greatly increasing the accuracy of the prediction. Since MFOLD has often been used by researchers for de novo prediction of IRES structures (Negulescu et al. 1998; Schiavi et al. 1999; Oumard et al. 2000; Sehgal et al. 2000; Xiao et al. 2003; Sherrill et al. 2004; Van Eden et al. 2004), we tested MFOLD with the most accurately defined IRES structure from HCV as well as the APAF1 IRES.
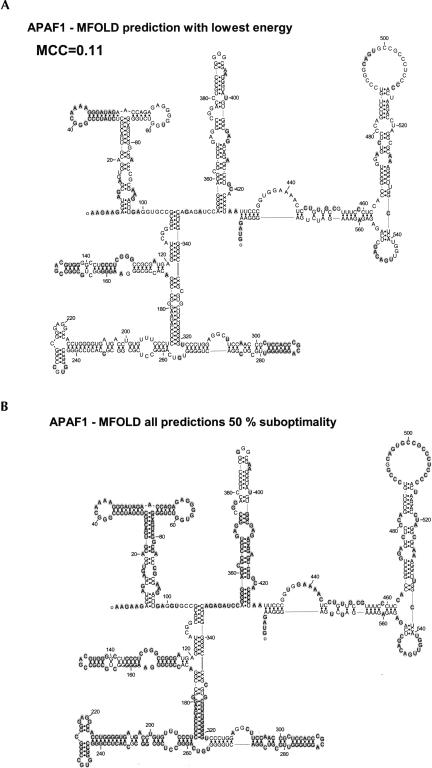
In Figures 4 and 5, the empirically determined structures of the 5′-UTRs of HCV and APAF-1 have been presented with the base pairs and single-stranded residues correctly predicted by MFOLD denoted with gray circles. Using efn2 from MFOLD, the energy states of the experimental and predicted structures were calculated. The RNA structure version of the Matthews correlation coefficient (MCC) as presented by Stormo's group (Gorodkin et al. 2001; Ji et al. 2004) was used as a comparative metric between the actual and predicted sequences, as it takes into account both the sensitivity and accuracy of the prediction by including both true- and false-positive predicted base pairing as well as true- and false-negative base pairings. Of a possible MCC from 0 to 1, the best MFOLD prediction for HCV is 0.59 and for the prediction with the lowest energy is 0.39. All of the base pairs and single-stranded regions that were correctly predicted in the 36 structures produced by MFOLD with the suboptimality parameter set at 50% are shown in Figure 4C. No more structures were produced at 50% suboptimality than at 10%. This is a cursory examination of the folding space considered by this program and is overall quite good with some exceptions. The NMR results of domain II (Lukavsky et al. 2003) and of domain IIId (Klinck et al. 2000) indicate several noncanonical base pairings, suggesting that these regions cannot be expected to be properly predicted by any secondary structure prediction program. In Figure 4 these base pairs are aligned together but are not connected as a base pair. Several base pairs in domains IIIe, IIId, and IIa are not predicted in the set of low free energy secondary structures predicted by MFOLD. The predictions of the larger UTR of Apaf-1 in Figure 5 fare more poorly compared to the experimentally predicted structure. This experimental structure has also not been as rigorously examined by as many studies as the HCV structure. Much of the experimental structure is not predicted correctly with the 50% suboptimality parameter as can be seen in Figure 5B. The stems that are missed in the suboptimal structures in APAF-1 and HCV would suggest that MFOLD is not considering all possible stems in its search space and, therefore, would never be able to predict a correct structure for these molecules. The examples shown in Figures 4 and 5 suggest that predictions should not be attempted until there is at least some wet-laboratory experimental data of the structure. Virtually all the IRES secondary structures that have been empirically derived using chemical or enzymatic probing have used MFOLD or a similar program using the probing results as constraints to determine the final structures (Le Quesne et al. 2001; Bonnal et al. 2003b; Mitchell et al. 2003; Jang et al. 2004; Jopling et al. 2004; Pickering et al. 2004).
FIGURE 5.
The correctly predicted RNA secondary structure of APAF1 IRES by MFOLD. (A) The empirically predicted IRES structure of Apaf-1 (Mitchell et al. 2003) is shown with the MFOLD correctly predicted base pairs or nonpairing bases shadowed in gray for the most thermodynamically stable structure. (B) All of the nonpairing bases or base pairs that were correctly predicted from all of the suboptimal folds predicted using a 50% suboptimal parameter in MFOLD are presented as gray shadows behind each base. This stresses the need for some empirically derived constraints for large RNA fold predictions when using MFOLD. Structures for figures have been produced with RnaViz2 (De Rijk et al. 2003).
Mathews (2004) has updated the energy calculations in his Windows program RNAstructure, a derivative of MFOLD. It also can incorporate chemical modification probing of the structure as constraints, showing an increase in accuracy by 26%–86% over in vivo structure prediction (Mathews 2004). The latest version also includes a partition function calculation that predicts the base-pairing probabilities for all possible base pairs. These probabilities can then be used to estimate the confidence in predicted base pairs in low free energy structures. Knowing stems with a strong probability of forming would be advantageous in the design of further experiments. A comparative survey of computationally predicted RNA structures of Flaviviridae (Thurner et al. 2004) reported their prediction of the HCV IRES being similar to empirically derived published structures when using McCaskill's partition algorithm (McCaskill 1990). The Vienna package also offers a minimal free energy program, RNAfold, similar to MFOLD. It is available from a Web server (Hofacker 2003) or as a stand-alone Unix package containing a variety of algorithms for the prediction and analysis of RNA secondary structure. Although these three programs use the same energy data table, they use different energy rules to calculate the structures. While RNAfold gives very similar results to MFOLD for the minimal free energy structure of a given sequence, RNAstructure produces a different lowest energy structure and a variety of different suboptimal folds.
Genetic algorithm
The genetic algorithm (GA) is based on the concept of biological evolution and is used throughout science and engineering. Whereas dynamic programming as demonstrated by MFOLD finds the mathematically optimal path through a matrix of all the possible base pairings formed by an RNA molecule, genetic algorithms select structures from a large, changing population of structures. From a large population of arbitrarily generated structures, the foldings with the highest degree of fitness, which usually have the lowest free energy, are selected for the next generation of structures. With each iteration of a generation, random mutations of the base pairings are formed and crossovers between structures are followed by a selection of the fittest for the next generation. Eventually there is a convergence with subsequent iterations upon one structure. GA programs can also consider pseudoknots in their structures.
A massively parallel implementation of this algorithm for several supercomputers has predicted 80%–90% of the correct base pairing in the poliovirus 5′-UTR (Shapiro et al. 2001). The accuracy is dependent on the initial population size. Accuracy improves with larger sizes of the starting population to a certain point, depending on which structure is being analyzed. Formally limited to specific high-performance machines, it has been adapted to other parrellel computers like Linux clusters (http://wwwlecb.ncifcrf.gov/~bshapiro/mpgaFold/mpgaFold.html).
Structure prediction from sequence alignments
X2S (Juan and Wilson 1999)
Although it has not been possible to get a large number of homologous cellular IRES sequences, more will become available with the completion of vertebrate genome and EST sequencing projects. With a minimum of five sequences, X2S can take an alignment in a clustal style and calculate covariation statistics and stem and loop energy scores and provide an overall score from each region to build a secondary structure. This is similar to giving phylogenetic or structure probing constraints to MFOLD to improve its accuracy (Mathews et al. 2004).
Conserved structures in unaligned sequences
Although the overall structure is important for IRESes like that in HCV, several cellular IRESes show reduced activity when nonoverlapping pieces of the sequences are tested for cap-independent translation, suggesting smaller modules that may be structural motifs for RNA binding proteins. The cellular IRES secondary structures published so far do not share an overall similarity, and if classes of IRESes exhibit activity in different cellular contexts, they may only share a similar motif. If this is the case, software capable of finding specific structural motifs in unaligned sequences may be more useful than finding global similarities.
Dynalign (Mathews and Turner 2002; Mathews 2005)
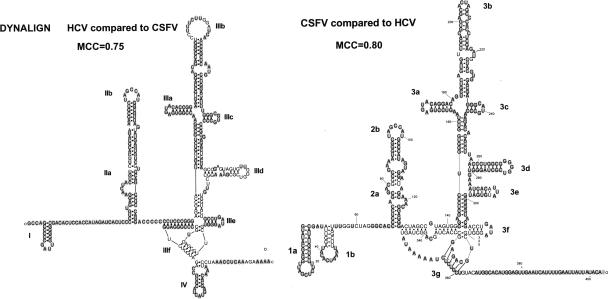
This program is an adaptation of the Sankoff dynamic programming algorithm (Sankoff 1985), which seeks similar structures in between two RNA sequences using free energy minimization and not sequence identity. In contrast to the original Sankoff algorithm, Dynalign limits the comparison between only two sequences within user-defined parameters, limiting the distance between aligned nucleotides to make the computations more manageable. As the program looks for a global structure alignment, the two sequences would be expected to be of similar length and overall structure. If one of the sequences has a known structure, it could possibly help to assess the accuracy of the prediction of an unknown sequence. In the example in Figure 6, CSFV IRES is known to have a similar structure as HCV IRES even though the sequence identity is very low. HCV is predicted quite accurately with an MCC of 0.75, which could allow us to assume that the prediction of CSFV is also accurate. In this case, we know the structure of CSFV, and it was predicted with an MCC of 0.8. It has not been shown whether Dynalign could predict a shared motif if several sequences were used in a pairwise comparison that did not share a global structure.
FIGURE 6.
Simultaneous prediction of HCV and CSFV IRES secondary structure with Dynalign. Two phylogenetically related sequences like HCV and CSFV are well predicted by Dynalign. The correctly predicted base pairs and nonpairing bases are shown with gray background. The Matthews Correlation Coefficient (MCC) (Gorodkin et al. 2001) is used to measure the accuracy of the prediction when compared to the empirically derived structures. Structures for figures have been produced with RnaViz2 (De Rijk et al. 2003).
Dynalign is very computer intensive and is available for Unix/Linux machines or for windows within the RNAStructure package. It has been updated to include experimental and comparative data as constraints as well as updated energy values (Mathews 2005).
CARNAC (Perriquet et al. 2003)
CARNAC (computer alignment of RNA by Cofolding) takes two or more homologous sequences and using energy minimization, sequence similarity, and covariation, devises a shared secondary structure model. It is available as a Web server at http://bioinfo.lifl.fr/carnac/carnac.html. Using HCV, CSFV, and BVDV as an example data set, the predicted folds of HCV and CSFV were compared to the known structures. The MCC of the CARNAC predicted structure for HCV was 0.282 when compared to the known structure (as seen in Fig. 4) and 0.587 for the predicted structure of CSFV.
PFOLD (Knudsen and Hein 1999, 2003)
PFOLD uses a comparative method using stochastic context-free grammars (SCFG) to predict a common structure. SCFG are sets of rules determined by probability from examining a set of sequences. PFOLD assumes that all the sequences are related through evolution and therefore can be aligned and share a common secondary structure. Input requires a structurally correct alignment of the sequences for accurate results. Since this is not possible without some structural data, a multiple alignment from a program like CLUSTALW with as many sequences as possible that are close in evolutionary distance will improve accuracy. PFOLD has been used to predict the IRES structures of FGF2, MNT, BIP, HSP-70, and BAG-1 in the Rfam database using UTRs from several transcripts or orthologs. If we compare the published structures of FGF2, MNT, and BAG-1 determined by enzymatic/chemical probing to the PFOLD/Rfam structures, we find a poor correlation in structure models with an MCC of 0.11 for FGF2, 0.25 for MNT, and 0.00 for BAG-1.
RNAGA (Chen et al. 2000)
In this program, a genetic algorithm is used to predict an overall structure from a group of structurally related sequences. A population of structures is generated from each of the related sequences. At each iteration, the population is regenerated by creating altered structures from the selected structures of the preceding generation and crossing over sections of the structures. At the first set of iterations, the fittest of the populations are selected with free energy until an overall stability is attained. Selection is then attained by giving a higher value to conserved structures within all of the populations. The top scoring structures are taken from the final population. A Web interface of RNAGA is available from the Pasteur Institute (http://bioweb.pasteur.fr/seqanal/interfaces/rnaga.html).
RNAProfile (Pavesi et al. 2004)
As the cellular IRES structures may not share an overall global similarity but may share smaller motifs, this program may be more appropriate for the task. The inputs for this program are a group of sequences that “mostly” share a conserved motif and the number of hairpins being sought. By creating a number of possible regions in each sequence and comparing the regions to each of the regions of the other sequences, the alignments with the best profile scores are kept and built on (greedily) in further iterations. Initial energy values of the first iteration are quite relaxed, as any fold with a lower free energy value than the unfolded state is considered. Tests on sequences of widely varying lengths gave good results for test cases of the iron response element, SECIS motifs, as well as hairpins with RNase P (Pavesi et al. 2004).
ComRNA (Ji et al. 2004)
This program takes unaligned sequences as an input, and like RNAProfile in the first step, produces all the stems that exist in each sequence. The stems are compared pairwise between each sequence, and those sharing a similarity above a set score are connected together in a graph theoretical approach. If a critical number of sequences share a subset of similar stems, the assembled compatible stems are used to construct a secondary structure profile, and with refinement the best structures are reported. Importantly for IRES structures, pseudoknots are also considered. This is a command line program written in C++, available as source code, and the output is single lines of sequence with a single line stem pattern description underneath. Ji et al. (2004) believe that the upper limit input scenario would restrict a limit of 20 sequences of <300 nt each. Using only the HCV, CSFV, and BVDV UTRs as input sequences, which are from 372 to 403 bases in length, the computations were too great to produce useful results after 24 h of processing time on a Sun Fire V20z computer with an AMD Opteron 248 (2.2 GHz) processor and 9 GB of memory.
GPRM (Hu 2003)
GPRM (genetic programming for RNA motifs) searches for elements that are common in a large set of coregulated RNA molecules. A second “negative” data set of random sequences is also created using the alphabet of the “positive” data set. The larger the positive data set, the better; only 10 members would be too small. Pseudoknots are not a problem.
Database searching
RSEARCH (Klein and Eddy 2003)
Whereas Dynalign is able to search all the possible folds in comparing two sequences in order to find a structural similarity between the two sequences, this method would be overly taxing in terms of computational time and space to use it to search a complete database. One comparison takes ∼15 min on a machine of moderate speed; therefore, a database search of 20,000 entries would take 6 mo. RSEARCH is designed to search a database with a sequence that has a known secondary structure. Scoring matrices analogous to the BLOSUM (Henikoff and Henikoff 1993) protein matrices used by BLAST and FASTA were built with alignments of the small subunit rRNA and are called RIBOSUM. Comparison of the query sequence to the unknown sequence is done using an algorithm termed “profile stochastic context-free-grammar” (SCFG). A profile SCFG also termed “covariance model” is produced from the query sequence, and each state of the comparison to an unknown sequence is taken care of in the model whether it is a nucleotide match, a base-pair match, a gap on either side, or an end of a stem–loop. This program seems closest to what a molecular biologist would use to find new sequences sharing a similar structure/function relationship with a query sequence. The computational expense is very high for this type of search, and it would be prohibitive to use a complete IRES-containing UTR like HCV, APAF1, or EMCV to search a UTR database without a multiprocessor system.
Pattern searching
Pattern matching programs serve the user by searching databases for structural patterns that they have defined or already know exist. Several programs have been developed with their own search mechanism and pattern description syntax: RNAMOT (Laferriere et al. 1994), PALINGOL (Billoud et al. 1996), PATSEARCH (Grillo et al. 2003), and RNAMotif (Macke et al. 2001).
RNAMotif (Macke et al. 2001)
This program is a successor to the secondary structure pattern searching programs RNABOB (http://www.genetics.wustl.edu/eddy/software/) and RNAMOT. It is the most powerful pattern-searching program now available because of its extensive and flexible descriptor scripting. It can find pseudoknots as well as triplexes and G-quartets that are found in FGF2 IRES (Bonnal et al. 2003b). A descriptor of an RNA structure is made using the program's scripting language and is then used to search primary sequence databases. It has been used successfully to predict Rev response elements in lentivirus (Lesnik et al. 2002), small RNA molecules in the Escherichia coli genome (Chen et al. 2002), and tRNA genes (Tsui et al. 2003). It has also been coupled with an “evolutionary algorithm” starting with a loose structure descriptor with no sequence constraints and successfully picked up iron response elements and signal recognition particles in a wide variety of organisms (Fogel et al. 2002).
As RNAMotif has been used successfully to find tRNA sequences (Tsui et al. 2003), we examined if this program can pick out those tRNA-like regions believed to be in IRESes as exhibited by viral IRESes from CrPV, HCV, EMCV, and BVDV. Using the original descriptor of Tsui et al. (2003) for eukaryotic tRNA but with no introns, RNAMotif finds only one putative tRNA structure in the 78 published eukaryotic sequences and only 23 sequences in the 33,677 sequences in UTRdb. By relaxing the descriptor so that only two of the eight specific nucleotides of the script are required, >60% of the IRES data shows a pattern match to this “loose” descriptor, as seen in Figure 7. As many of these matches gave structures with positive free energy values, a third tRNA script was made with cutoff matches >−10 kcal/mol. This value is calculated using the energy tables of Mathews et al. (1999) with the efn2 program included in the RNAMotif package. It must be noted that the loose tRNA descriptor did not find a match in CrPV, which is known to interact with the ribosome without need of an initiator Met-tRNA. The pattern for the capsid IRES of Discistroviridae built from empirical data of CrPV and PSIV did not find any matches in eukaryotic mRNAs using another pattern searching program, “Scan for Matches” (Dsouza et al. 1997; Hatakeyama et al. 2004). This suggests a structure/function mechanism for these Discistroviridae viral IRESes distinct from eukaryotic IRES and other viral IRESes in their interaction with ribosomes.
FIGURE 7.
RNAMotif pattern matches to tRNA descriptors of different levels of stringency within 5′-UTR human sequences. Using the eukaryote tRNA descriptor of Tsui et al. (2003) without introns, the number of pattern matches is shown as a percentage of the total number of UTRs in either UTRdb, RefSeq, fully reviewed UTRs in RefSeq, or published IRES from eukaryote transcripts in Table 1 (see data sets in Fig. 1). The first pattern is strict because of the specific sequences required in the pattern matches as well as the overall tRNA structure. The second pattern only requires two of the eight specific base sequences that are standard in tRNAs as well as a maximal −10.0 kCal/mol free energy cut-off for the putative matching RNA structure as calculated by the efn2 program included with RNAMotif software. The last pattern is the same as the second pattern but does not have a free energy cutoff.
ERPIN server (Lambert et al. 2004)
ERPIN (easy RNA profile identification) takes pattern searching to a higher level of complexity by adding sequence alignments to the structure annotation, creating a profile for each residue position. As the difficulty in preparing a correct RNA sequence alignment and parameter tuning requires expertise and time to fine tune, only preprepared patterns are available on the ERPIN Web server for searching user-submitted sequences. There is a limit of 5 Mb per search; larger amounts require the data to be split or the program to be installed locally. An expect value (e-value) analogous to the value produced by BLAST is calculated to estimate finding the pattern by chance with that score or higher in the same data set (Lambert et al. 2005). The IRES from type 1 poliovirus is one of ∼50 RNA genes or elements available as search patterns. This pattern was not found within the IRES database created from Table 1 and also was not found in 10 of 25 complete type 1 poliovirus genome sequences from GenBank. This shows that this particular IRES pattern is quite restrictive and not useful for finding unknown IRES sequences. Functionally significant or not, the pattern for tRNA type 1 was found in VEGF (mouse) at ∼−1200 upstream of the start codon with an e-value of 10−6.
CONCLUDING REMARKS
The standard translation initiation mechanism is inhibited or impeded during mitosis, cell stress, or viral infection. During these times, the efficiency of translation of a subset of mRNAs is elevated relative to the total population of transcripts. One mechanism by which this is achieved is through the recruitment of ribosomes by IRES elements in the 5′-UTR. Viral IRES sequences have shown some RNA structural conservation between different viral families necessary for translation initiation. Recent viral IRES/ribosome studies suggest that these structures allow the bypassing of other cofactors involved in standard translation initiation by direct interactions with the ribosome as seen by HCV or the tRNA structure of CrPV. It has been postulated that there are shared sequence/structural motifs between cellular IRES as well, allowing subsets of mRNAs to be more efficiently translated. Perhaps these shared motifs will not be required to hijack the translation process overall as viruses will, but just to retain enough translation efficiency to be biologically relevant in that cellular context. The noncanonical protein factors, the ITAFs, which have been shown to be involved in IRES function, are all involved in normal mRNA processing, and their binding may not be so specific as to require a particular stem–loop for binding, but the IRES structure may dictate whether an ITAF is able to bind. So far only PTB has shown some structural preference with a binding sequence motif in a double-stranded context (Mitchell et al. 2005). Other ITAFs like PCBP open up the structure for PTB to bind (Mitchell et al. 2003). Do the cellular IRESes have structures that prevent the ribosome binding to it unless specific ITAFs are present? Some mRNA binding proteins are needed to specifically recruit the small ribosomal subunit to the transcript, while others change or open up the structure to allow ITAFs and the ribosome to bind.
Some generalized points related to IRESes in UTRs can be made from what is known so far. IRESes tend to be found in longer UTRs but with half of all UTRs from human transcripts being at least 150 bases long, a large portion of UTRs are long enough to contain IRES elements. Upstream AUGs are common in UTRs, and although UTRs with IRESes possibly have more upstream AUGs than found in the normal distribution, UTRs with IRESes containing no upstream AUGs are common as well. It is often stated that UTRs with IRESes tend to have greater levels of secondary structure. It is true that upstream open reading frames and specific inhibitory stable RNA structures can impede the cap-dependent scanning mechanism for translation, but it is not clear that all UTRs with IRESes contain these elements. The overall % GC content appears no different for UTRs containing IRES elements from that found in the normal distribution of % GC content with 5′-UTRs of human transcripts.
The structure prediction examples given are not qualitative in the assessment of the predictive powers of these programs and although only anecdotal, they give a frame of reference to the study of IRES sequences and the search for possible shared structural elements. Although RNA secondary structure prediction has benefited from a depth of intellect, as is exhibited by the algorithms that exist today, accuracy of de novo secondary structure prediction of large RNA molecules depends somewhat on good fortune. Protein binding along with the dynamics of RNA folding as the mRNA is transcribed may produce a somewhat alternative structure than the in vitro defined structure. However, some in vitro transcribed IRES sequences still function in cell-free translation experiments. It is important to have either empirically derived data of the structure or covariance data from truly related structures as additional inputs for structure prediction. In the case of cellular IRES, the diversity of regulatory contexts may be the biggest hindrance to a global understanding of the IRES mechanisms held within the primary sequence of UTRs containing IRES elements. If there was a wish list of program functions to help define IRES structure/sequence/function from primary sequence, one might be the ability to take all the sequences and do a “cluster” motif search similar to microarray clustering, assuming that IRESes will separate into groups. There is also quite a bit of deletion and directed mutation analysis data that alter the activity of IRESes. These data seem underutilized within current prediction software. Could these data be used similarly to structure probing data so that a high score could be placed on sequences that were crucial to activity and a low score on sequences that showed no effect when altered? The apparent modular elements within some IRESes as exhibited by partial activity by nonoverlapping fragments of the UTRs suggest no need for an overall similar structure in every cellular IRES sequence.
The possibility of different classes of IRESes and of cellular IRES diversity may exist for different changes to the translational machinery, different forms of initiation factors, different factors, and different ribosome assemblies. Even in the studies of IRES, there is a need to avoid becoming dogmatic on only one mechanism of translation initiation, one universal ITAF, one structure, or one way to recruit a ribosome. Diversity of regulation seems to work well for the cell and even better for whole organisms. The design may not seem intelligent, but it is good enough.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.157806.
REFERENCES
- Akbergenov, R., Zhanybekova, S., Kryldakov, R.V., Zhigailov, A., Polimbetova, N.S., Hohn, T., Iskakov, B.K. ARC-1, a sequence element complementary to an internal 18S rRNA segment, enhances translation efficiency in plants when present in the leader or intercistronic region of mRNAs. Nucleic Acids Res. 2004;32:239–247. doi: 10.1093/nar/gkh176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, I.K., McKendrick, L., Morley, S.J., Jackson, R.J. Truncated initiation factor eIF4G lacking an eIF4E binding site can support capped mRNA translation. EMBO J. 2001;20:4233–4242. doi: 10.1093/emboj/20.15.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar, A., Ali, N., Tanveer, R., Siddiqui, A. Demonstration of functional requirement of polypyrimidine tract-binding protein by SELEX RNA during hepatitis C virus internal ribosome entry site-mediated translation initiation. J. Biol. Chem. 2000;275:34231–34235. doi: 10.1074/jbc.M006343200. [DOI] [PubMed] [Google Scholar]
- Arava, Y., Wang, Y., Storey, J.D., Liu, C.L., Brown, P.O., Herschlag, D. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attal, J., Theron, M.C., Taboit, F., Cajero-Juarez, M., Kann, G., Bolifraud, P., Houdebine, L.M. The RU5 (‘R’) region from human leukaemia viruses (HTLV-1) contains an internal ribosome entry site (IRES)-like sequence. FEBS Lett. 1996;392:220–224. doi: 10.1016/0014-5793(96)00815-0. [DOI] [PubMed] [Google Scholar]
- Back, S.H., Kim, Y.K., Kim, W.J., Cho, S., Oh, H.R., Kim, J.E., Jang, S.K. Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-binding proteins executed by polioviral 3C(pro) J. Virol. 2002a;76:2529–2542. doi: 10.1128/jvi.76.5.2529-2542.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back, S.H., Shin, S., Jang, S.K. Polypyrimidine tract-binding proteins are cleaved by caspase-3 during apoptosis. J. Biol. Chem. 2002b;277:27200–27209. doi: 10.1074/jbc.M203887200. [DOI] [PubMed] [Google Scholar]
- Barraille, P., Chinestra, P., Bayard, F., Faye, J.C. Alternative initiation of translation accounts for a 67/45 kDa dimorphism of the human estrogen receptor ERα. Biochem. Biophys. Res. Commun. 1999;257:84–88. doi: 10.1006/bbrc.1999.0334. [DOI] [PubMed] [Google Scholar]
- Belsham, G.J., Sonenberg, N. RNA–protein interactions in regulation of picornavirus RNA translation. Microbiol. Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlioz, C., Darlix, J.L. An internal ribosomal entry mechanism promotes translation of murine leukemia virus gag polyprotein precursors. J. Virol. 1995;69:2214–2222. doi: 10.1128/jvi.69.4.2214-2222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlioz, C., Torrent, C., Darlix, J.L. An internal ribosomal entry signal in the rat VL30 region of the Harvey murine sarcoma virus leader and its use in dicistronic retroviral vectors. J. Virol. 1995;69:6400–6407. doi: 10.1128/jvi.69.10.6400-6407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, J., Sella, O., Le, S.Y., Elroy-Stein, O. PDGF2/c-sis mRNA leader contains a differentiation-linked internal ribosomal entry site (D-IRES) J. Biol. Chem. 1997;272:9356–9362. doi: 10.1074/jbc.272.14.9356. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya, S., Das, S. Mapping of secondary structure of the spacer region within the 5′-untranslated region of the coxsackievirus B3 RNA: Possible role of an apical GAGA loop in binding La protein and influencing internal initiation of translation. Virus Res. 2005;108:89–100. doi: 10.1016/j.virusres.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Bieleski, L., Talbot, S.J. Kaposi's sarcoma-associated herpesvirus vCyclin open reading frame contains an internal ribosome entry site. J. Virol. 2001;75:1864–1869. doi: 10.1128/JVI.75.4.1864-1869.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieleski, L., Hindley, C., Talbot, S.J. A polypyrimidine tract facilitates the expression of Kaposi's sarcoma-associated herpesvirus vFLIP through an internal ribosome entry site. J. Gen. Virol. 2004;85:615–620. doi: 10.1099/vir.0.19733-0. [DOI] [PubMed] [Google Scholar]
- Billoud, B., Kontic, M., Viari, A. Palingol: A declarative programming language to describe nucleic acids’ secondary structures and to scan sequence database. Nucleic Acids Res. 1996;24:1395–1403. doi: 10.1093/nar/24.8.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnal, S., Boutonnet, C., Prado-Lourenco, L., Vagner, S. IRESdb: The Internal Ribosome Entry Site database. Nucleic Acids Res. 2003a;31:427–428. doi: 10.1093/nar/gkg003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnal, S., Schaeffer, C., Creancier, L., Clamens, S., Moine, H., Prats, A.C., Vagner, S. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J. Biol. Chem. 2003b;278:39330–39336. doi: 10.1074/jbc.M305580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnal, S., Pileur, F., Orsini, C., Parker, F., Pujol, F., Prats, A.C., Vagner, S. Heterogeneous nuclear ribonucleoprotein A1 is a novel internal ribosome entry site trans-acting factor that modulates alternative initiation of translation of the fibroblast growth factor 2 mRNA. J. Biol. Chem. 2005;280:4144–4153. doi: 10.1074/jbc.M411492200. [DOI] [PubMed] [Google Scholar]
- Borman, A., Jackson, R.J. Initiation of translation of human rhinovirus RNA: Mapping the internal ribosome entry site. Virology. 1992;188:685–696. doi: 10.1016/0042-6822(92)90523-r. [DOI] [PubMed] [Google Scholar]
- Brown, E.C., Jackson, R.J. All five cold-shock domains of unr (upstream of N-ras) are required for stimulation of human rhinovirus RNA translation. J. Gen. Virol. 2004;85:2279–2287. doi: 10.1099/vir.0.80045-0. [DOI] [PubMed] [Google Scholar]
- Brown, E.A., Zhang, H., Ping, L.H., Lemon, S.M. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, E.A., Zajac, A.J., Lemon, S.M. In vitro characterization of an internal ribosomal entry site (IRES) present within the 5′ nontranslated region of hepatitis A virus RNA: Comparison with the IRES of encephalomyocarditis virus. J. Virol. 1994;68:1066–1074. doi: 10.1128/jvi.68.2.1066-1074.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck, C.B., Shen, X., Egan, M.A., Pierson, T.C., Walker, C.M., Siliciano, R.F. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 2001;75:181–191. doi: 10.1128/JVI.75.1.181-191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci, P., Kvam, C., Kitamura, A., Ohsumi, T., Okazaki, Y., Itoh, M., Kamiya, M., Shibata, K., Sasaki, N., Izawa, M., et al. High-efficiency full-length cDNA cloning by biotinylated CAP trapper. Genomics. 1996;37:327–336. doi: 10.1006/geno.1996.0567. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C., Freed, D.D. Cap-independent enhancement of translation by a plant potyvirus 5′ nontranslated region. J. Virol. 1990;64:1590–1597. doi: 10.1128/jvi.64.4.1590-1597.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, M.S., Sarnow, P. Distinct mRNAs that encode La autoantigen are differentially expressed and contain internal ribosome entry sites. J. Biol. Chem. 2000;275:28301–28307. doi: 10.1074/jbc.M004657200. [DOI] [PubMed] [Google Scholar]
- Cecconi, F., Alvarez-Bolado, G., Meyer, B.I., Roth, K.A., Gruss, P. Apaf1 (CED-4 homolog) regulates programmed cell death in mammalian development. Cell. 1998;94:727–737. doi: 10.1016/s0092-8674(00)81732-8. [DOI] [PubMed] [Google Scholar]
- Cencig, S., Nanbru, C., Le, S.Y., Gueydan, C., Huez, G., Kruys, V. Mapping and characterization of the minimal internal ribosome entry segment in the human c-Myc mRNA 5′ untranslated region. Oncogene. 2004;23:267–277. doi: 10.1038/sj.onc.1207017. [DOI] [PubMed] [Google Scholar]
- Chappell, S.A., Mauro, V.P. The internal ribosome entry site (IRES) contained within the RNA-binding motif protein 3 (Rbm3) mRNA is composed of functionally distinct elements. J. Biol. Chem. 2003;278:33793–33800. doi: 10.1074/jbc.M303495200. [DOI] [PubMed] [Google Scholar]
- Chappell, S.A., Edelman, G.M., Mauro, V.P. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc. Natl. Acad. Sci. 2000;97:1536–1541. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell, S.A., Owens, G.C., Mauro, V.P. A 5′ leader of Rbm3, a cold stress-induced mRNA, mediates internal initiation of translation with increased efficiency under conditions of mild hypothermia. J. Biol. Chem. 2001;276:36917–36922. doi: 10.1074/jbc.M106008200. [DOI] [PubMed] [Google Scholar]
- Chen, J.H., Le, S.Y., Maizel, J.V. Prediction of common secondary structures of RNAs: A genetic algorithm approach. Nucleic Acids Res. 2000;28:991–999. doi: 10.1093/nar/28.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., Lesnik, E.A., Hall, T.A., Sampath, R., Griffey, R.H., Ecker, D.J., Blyn, L.B. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. Biosystems. 2002;65:157–177. doi: 10.1016/s0303-2647(02)00013-8. [DOI] [PubMed] [Google Scholar]
- Cherry, S., Doukas, T., Armknecht, S., Whelan, S., Wang, H., Sarnow, P., Perrimon, N. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes & Dev. 2005;19:445–452. doi: 10.1101/gad.1267905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, P.W., Carpenter, L.E., Hagerman, P.J. The 5′-untranslated region of the FMR1 message facilitates translation by internal ribosome entry. J. Biol. Chem. 2001;276:37916–37921. doi: 10.1074/jbc.M105584200. [DOI] [PubMed] [Google Scholar]
- Cho, S., Kim, J.H., Back, S.H., Jang, S.K. Polypyrimidine tract-binding protein enhances the internal ribosomal entry site-dependent translation of p27Kip1mRNA and modulates transition from G1to S phase. Mol. Cell. Biol. 2005;25:1283–1297. doi: 10.1128/MCB.25.4.1283-1297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp, M., Andrews, D., Barker, D., Bevan, P., Cameron, G., Chen, Y., Clark, L., Cox, T., Cuff, J., Curwen, V., et al. Ensembl 2002: Accommodating comparative genomics. Nucleic Acids Res. 2003;31:38–42. doi: 10.1093/nar/gkg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens, M.J. Translational regulation in cell stress and apoptosis. Roles of the eIF4E binding proteins. J. Cell. Mol. Med. 2001;5:221–239. doi: 10.1111/j.1582-4934.2001.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens, M.J., Bushell, M., Jeffrey, I.W., Pain, V.M., Morley, S.J. Translation initiation factor modifications and the regulation of protein synthesis in apoptotic cells. Cell Death Differ. 2000;7:603–615. doi: 10.1038/sj.cdd.4400695. [DOI] [PubMed] [Google Scholar]
- Clote, P., Ferre, F., Kranakis, E., Krizanc, D. Structural RNA has lower folding energy than random RNA of the same dinucleotide frequency. RNA. 2005;11:578–591. doi: 10.1261/rna.7220505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coldwell, M.J., Mitchell, S.A., Stoneley, M., MacFarlane, M., Willis, A.E. Initiation of Apaf-1 translation by internal ribosome entry. Oncogene. 2000;19:899–905. doi: 10.1038/sj.onc.1203407. [DOI] [PubMed] [Google Scholar]
- Coldwell, M.J., deSchoolmeester, M.L., Fraser, G.A., Pickering, B.M., Packham, G., Willis, A.E. The p36 isoform of BAG-1 is translated by internal ribosome entry following heat shock. Oncogene. 2001;20:4095–4100. doi: 10.1038/sj.onc.1204547. [DOI] [PubMed] [Google Scholar]
- Collier, A.J., Gallego, J., Klinck, R., Cole, P.T., Harris, S.J., Harrison, G.P., Aboul-Ela, F., Varani, G., Walker, S. A conserved RNA structure within the HCV IRES eIF3-binding site. Nat. Struct. Biol. 2002;9:375–380. doi: 10.1038/nsb785. [DOI] [PubMed] [Google Scholar]
- Cornelis, S., Bruynooghe, Y., Denecker, G., Van Huffel, S., Tinton, S., Beyaert, R. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol. Cell. 2000;5:597–605. doi: 10.1016/s1097-2765(00)80239-7. [DOI] [PubMed] [Google Scholar]
- Cornelis, S., Tinton, S.A., Schepens, B., Bruynooghe, Y., Beyaert, R. UNR translation can be driven by an IRES element that is negatively regulated by polypyrimidine tract binding protein. Nucleic Acids Res. 2005;33:3095–3108. doi: 10.1093/nar/gki611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli, M., Svitkin, Y., Sonenberg, N. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol. Cell. Biol. 2004;24:6861–6870. doi: 10.1128/MCB.24.15.6861-6870.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta, R., Laroia, G., Schneider, R.J. Chaperone hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap-initiation complexes. Genes & Dev. 2000;14:1460–1470. [PMC free article] [PubMed] [Google Scholar]
- Czibener, C., Alvarez, D., Scodeller, E., Gamarnik, A.V. Characterization of internal ribosomal entry sites of Triatoma virus. J. Gen. Virol. 2005;86:2275–2280. doi: 10.1099/vir.0.80842-0. [DOI] [PubMed] [Google Scholar]
- Deffaud, C., Darlix, J.L. Characterization of an internal ribosomal entry segment in the 5′ leader of murine leukemia virus env RNA. J. Virol. 2000a;74:846–850. doi: 10.1128/jvi.74.2.846-850.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deffaud, C., Darlix, J.L. Rous sarcoma virus translation revisited: Characterization of an internal ribosome entry segment in the 5′ leader of the genomic RNA. J. Virol. 2000b;74:11581–11588. doi: 10.1128/jvi.74.24.11581-11588.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pietri Tonelli, D., Mihailovich, M., Schnurbus, R., Pesole, G., Grohovaz, F., Zacchetti, D. Translational control of Scamper expression via a cell-specific internal ribosome entry site. Nucleic Acids Res. 2003;31:2508–2513. doi: 10.1093/nar/gkg357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rijk, P., Wuyts, J., De Wachter, R. RnaViz 2: An improved representation of RNA secondary structure. Bioinformatics. 2003;19:299–300. doi: 10.1093/bioinformatics/19.2.299. [DOI] [PubMed] [Google Scholar]
- Dever, T.E. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- Dinkova, T.D., Zepeda, H., Martinez-Salas, E., Martinez, L.M., Nieto-Sotelo, J., de Jimenez, E.S. Cap-independent translation of maize Hsp101. Plant J. 2005;41:722–731. doi: 10.1111/j.1365-313X.2005.02333.x. [DOI] [PubMed] [Google Scholar]
- Dobrikova, E., Florez, P., Bradrick, S., Gromeier, M. Activity of a type 1 picornavirus internal ribosomal entry site is determined by sequences within the 3′ nontranslated region. Proc. Natl. Acad. Sci. 2003;100:15125–15130. doi: 10.1073/pnas.2436464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson, T., Minic, A., Nielsen, K., Amiott, E., Krushel, L. Internal initiation of translation of the TrkB mRNA is mediated by multiple regions within the 5′ leader. Nucleic Acids Res. 2005;33:2929–2941. doi: 10.1093/nar/gki605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domier, L.L., McCoppin, N.K., D'Arcy, C.J. Sequence requirements for translation initiation of Rhopalosiphum padi virus ORF2. Virology. 2000;268:264–271. doi: 10.1006/viro.2000.0189. [DOI] [PubMed] [Google Scholar]
- Doshi, K.J., Cannone, J.J., Cobaugh, C.W., Gutell, R.R. Evaluation of the suitability of free-energy minimization using nearest-neighbor energy parameters for RNA secondary structure prediction. BMC Bioinformatics. 2004;5 doi: 10.1186/1471-2105-5-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell, R.D., Eddy, S.R. Evaluation of several lightweight stochastic context-free grammars for RNA secondary structure prediction. BMC Bioinformatics. 2004;5 doi: 10.1186/1471-2105-5-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss, G., Kim, V.N., Kataoka, N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Dsouza, M., Larsen, N., Overbeek, R. Searching for patterns in genomic data. Trends Genet. 1997;13:497–498. doi: 10.1016/s0168-9525(97)01347-4. [DOI] [PubMed] [Google Scholar]
- Evans, J.R., Mitchell, S.A., Spriggs, K.A., Ostrowski, J., Bomsztyk, K., Ostarek, D., Willis, A.E. Members of the poly (rC) binding protein family stimulate the activity of the c-Myc internal ribosome entry segment in vitro and in vivo. Oncogene. 2003;22:8012–8020. doi: 10.1038/sj.onc.1206645. [DOI] [PubMed] [Google Scholar]
- Fernandez, J., Yaman, I., Mishra, R., Merrick, W.C., Snider, M.D., Lamers, W.H., Hatzoglou, M. Internal ribosome entry site-mediated translation of a mammalian mRNA is regulated by amino acid availability. J. Biol. Chem. 2001;276:12285–12291. doi: 10.1074/jbc.M009714200. [DOI] [PubMed] [Google Scholar]
- Fernandez-Miragall, O., Martinez-Salas, E. Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA. 2003;9:1333–1344. doi: 10.1261/rna.5950603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher, S.P., Jackson, R.J. Pestivirus internal ribosome entry site (IRES) structure and function: Elements in the 5′ untranslated region important for IRES function. J. Virol. 2002;76:5024–5033. doi: 10.1128/JVI.76.10.5024-5033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel, G.B., Porto, V.W., Weekes, D.G., Fogel, D.B., Griffey, R.H., McNeil, J.A., Lesnik, E., Ecker, D.J., Sampath, R. Discovery of RNA structural elements using evolutionary computation. Nucleic Acids Res. 2002;30:5310–5317. doi: 10.1093/nar/gkf653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galban, S., Fan, J., Martindale, J.L., Cheadle, C., Hoffman, B., Woods, M.P., Temeles, G., Brieger, J., Decker, J., Gorospe, M. von Hippel-Lindau protein-mediated repression of tumor necrosis factor alpha translation revealed through use of cDNA arrays. Mol. CellBiol. 2003;23:2316–2328. doi: 10.1128/MCB.23.7.2316-2328.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego, J. Internal initiation of translation by viral and cellular IRESs—A new avenue for specific inhibition of protein synthesis? Curr. Opin. Drug Discov. Devel. 2002;5:777–784. [PubMed] [Google Scholar]
- Garlapati, S., Wang, C.C. Structural elements in the 5′-untranslated region of giardiavirus transcript essential for internal ribosome entry site-mediated translation initiation. Eukaryot. Cell. 2005;4:742–754. doi: 10.1128/EC.4.4.742-754.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras, A.C., Raught, B., Sonenberg, N. eIF4 initiation factorsEffectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Giraud, S., Greco, A., Brink, M., Diaz, J.J., Delafontaine, P. Translation initiation of the insulin-like growth factor I receptor mRNA is mediated by an internal ribosome entry site. J. Biol. Chem. 2001;276:5668–5675. doi: 10.1074/jbc.M005928200. [DOI] [PubMed] [Google Scholar]
- Gooding, C., Kemp, P., Smith, C.W. A novel polypyrimidine tract-binding protein paralog expressed in smooth muscle cells. J. Biol. Chem. 2003;278:15201–15207. doi: 10.1074/jbc.M210131200. [DOI] [PubMed] [Google Scholar]
- Gopalan, V., Vioque, A., Altman, S. RNase P: Variations and uses. J. Biol. Chem. 2002;277:6759–6762. doi: 10.1074/jbc.R100067200. [DOI] [PubMed] [Google Scholar]
- Gorodkin, J., Stricklin, S.L., Stormo, G.D. Discovering common stem–loop motifs in unaligned RNA sequences. Nucleic Acids Res. 2001;29:2135–2144. doi: 10.1093/nar/29.10.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grace, K., Gartland, M., Karayiannis, P., McGarvey, M.J., Clarke, B. The 5′ untranslated region of GB virus B shows functional similarity to the internal ribosome entry site of hepatitis C virus. J. Gen. Virol. 1999;80:2337–2341. doi: 10.1099/0022-1317-80-9-2337. [DOI] [PubMed] [Google Scholar]
- Gradi, A., Imataka, H., Svitkin, Y.V., Rom, E., Raught, B., Morino, S., Sonenberg, N. A novel functional human eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 1998a;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradi, A., Svitkin, Y.V., Imataka, H., Sonenberg, N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. 1998b;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassmann, C.W., Yu, H., Isken, O., Behrens, S.E. Hepatitis C virus and the related bovine viral diarrhea virus considerably differ in the functional organization of the 5′ non-translated region: Implications for the viral life cycle. Virology. 2005;333:349–366. doi: 10.1016/j.virol.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Griffiths, A., Coen, D.M. An unusual internal ribosome entry site in the herpes simplex virus thymidine kinase gene. Proc. Natl. Acad. Sci. 2005;102:9667–9672. doi: 10.1073/pnas.0504132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones, S., Bateman, A., Marshall, M., Khanna, A., Eddy, S.R. Rfam: An RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo, G., Attimonelli, M., Liuni, S., Pesole, G. CLEANUP: A fast computer program for removing redundancies from nucleotide sequence databases. Comput. Appl. Biosci. 1996;12:1–8. doi: 10.1093/bioinformatics/12.1.1. [DOI] [PubMed] [Google Scholar]
- Grillo, G., Licciulli, F., Liuni, S., Sbisa, E., Pesole, G. PatSearch: A program for the detection of patterns and structural motifs in nucleotide sequences. Nucleic Acids Res. 2003;31:3608–3612. doi: 10.1093/nar/gkg548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grolleau, A., Bowman, J., Pradet-Balade, B., Puravs, E., Hanash, S., Garcia-Sanz, J.A., Beretta, L. Global and specific translational control by rapamycin in T cells uncovered by microarrays and proteomics. J. Biol. Chem. 2002;277:22175–22184. doi: 10.1074/jbc.M202014200. [DOI] [PubMed] [Google Scholar]
- Grundhoff, A., Ganem, D. Mechanisms governing expression of the v-FLIP gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 2001;75:1857–1863. doi: 10.1128/JVI.75.4.1857-1863.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell, R.R., Lee, J.C., Cannone, J.J. The accuracy of ribosomal RNA comparative structure models. Curr. Opin. Struct. Biol. 2002;12:301–310. doi: 10.1016/s0959-440x(02)00339-1. [DOI] [PubMed] [Google Scholar]
- Han, B., Dong, Z., Zhang, J.T. Tight control of platelet-derived growth factor B/c-sis expression by interplay between the 5′-untranslated region sequence and the major upstream promoter. J. Biol. Chem. 2003;278:46983–46993. doi: 10.1074/jbc.M304976200. [DOI] [PubMed] [Google Scholar]
- Hart, K., Bienz, M. A test for cell autonomy, based on di-cistronic messenger translation. Development. 1996;122:747–751. doi: 10.1242/dev.122.3.747. [DOI] [PubMed] [Google Scholar]
- Hatakeyama, Y., Shibuya, N., Nishiyama, T., Nakashima, N. Structural variant of the intergenic internal ribosome entry site elements in dicistroviruses and computational search for their counterparts. RNA. 2004;10:779–786. doi: 10.1261/rna.5208104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen, C.U., Sarnow, P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes & Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., Henikoff, J.G. Performance evaluation of amino acid substitution matrices. Proteins. 1993;17:49–61. doi: 10.1002/prot.340170108. [DOI] [PubMed] [Google Scholar]
- Henis-Korenblit, S., Strumpf, N.L., Goldstaub, D., Kimchi, A. A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol. Cell. Biol. 2000;20:496–506. doi: 10.1128/mcb.20.2.496-506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henis-Korenblit, S., Shani, G., Sines, T., Marash, L., Shohat, G., Kimchi, A. The caspase-cleaved DAP5 protein supports internal ribosome entry site-mediated translation of death proteins. Proc. Natl. Acad. Sci. 2002;99:5400–5405. doi: 10.1073/pnas.082102499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennecke, M., Kwissa, M., Metzger, K., Oumard, A., Kroger, A., Schirmbeck, R., Reimann, J., Hauser, H. Composition and arrangement of genes define the strength of IRES-driven translation in bicistronic mRNAs. Nucleic Acids Res. 2001;29:3327–3334. doi: 10.1093/nar/29.16.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppner Goss, K., Trzepacz, C., Tuohy, T.M., Groden, J. Attenuated APC alleles produce functional protein from internal translation initiation. Proc. Natl. Acad. Sci. 2002;99:8161–8166. doi: 10.1073/pnas.112072199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, G., Vazquez-Pianzola, P., Sierra, J.M., Rivera-Pomar, R. Internal ribosome entry site drives cap-independent translation of reaper and heat shock protein 70 mRNAs in Drosophila embryos. RNA. 2004;10:1783–1797. doi: 10.1261/rna.7154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs, P.G. RNA secondary structure: Physical and computational aspects. Q. Rev. Biophys. 2000;33:199–253. doi: 10.1017/s0033583500003620. [DOI] [PubMed] [Google Scholar]
- Hofacker, I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, M.A., Palmenberg, A.C. Mutational analysis of the J-K stem–loop region of the encephalomyocarditis virus IRES. J. Virol. 1995;69:4399–4406. doi: 10.1128/jvi.69.7.4399-4406.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik, M. Targeting translation for treatment of cancer—A novel role for IRES? Curr. Cancer Drug Targets. 2004;4:299–311. doi: 10.2174/1568009043333005. [DOI] [PubMed] [Google Scholar]
- Holcik, M., Korneluk, R.G. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: Role of La autoantigen in XIAP translation. Mol. Cell. Biol. 2000;20:4648–4657. doi: 10.1128/mcb.20.13.4648-4657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik, M., Sonenberg, N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- Holcik, M., Lefebvre, C., Yeh, C., Chow, T., Korneluk, R.G. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat. Cell Biol. 1999;1:190–192. doi: 10.1038/11109. [DOI] [PubMed] [Google Scholar]
- Holcik, M., Sonenberg, N., Korneluk, R.G. Internal ribosome initiation of translation and the control of cell death. Trends Genet. 2000;16:469–473. doi: 10.1016/s0168-9525(00)02106-5. [DOI] [PubMed] [Google Scholar]
- Holcik, M., Gordon, B.W., Korneluk, R.G. The internal ribosome entry site-mediated translation of antiapoptotic protein XIAP is modulated by the heterogeneous nuclear ribonucleoproteins C1 and C2. Mol. Cell. Biol. 2003;23:280–288. doi: 10.1128/MCB.23.1.280-288.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik, M., Graber, T., Lewis, S.M., Lefebvre, C.A., Lacasse, E., Baird, S. Spurious splicing within the XIAP 5′ UTR occurs in the Rluc/Fluc but not the βgal/CAT bicistronic reporter system. RNA. 2005;11:1605–1609. doi: 10.1261/rna.2158605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, M., Beard, M.R., Ping, L.H., Lemon, S.M. A phylogenetically conserved stem–loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 1999;73:1165–1174. doi: 10.1128/jvi.73.2.1165-1174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y.J. GPRM: A genetic programming approach to finding common RNA secondary structure elements. Nucleic Acids Res. 2003;31:3446–3449. doi: 10.1093/nar/gkg521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, M.C., Tranque, P., Edelman, G.M., Mauro, V.P. rRNA-complementarity in the 5′ untranslated region of mRNA specifying the Gtx homeodomain protein: Evidence that base-pairing to 18S rRNA affects translational efficiency. Proc. Natl. Acad. Sci. 1999;96:1339–1344. doi: 10.1073/pnas.96.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudder, A., Werner, R. Analysis of a Charcot-Marie-Tooth disease mutation reveals an essential internal ribosome entry site element in the connexin-32 gene. J. Biol. Chem. 2000;275:34586–34591. doi: 10.1074/jbc.M005199200. [DOI] [PubMed] [Google Scholar]
- Huez, I., Bornes, S., Bresson, D., Creancier, L., Prats, H. New vascular endothelial growth factor isoform generated by internal ribosome entry site-driven CUG translation initiation. Mol. Endocrinol. 2001;15:2197–2210. doi: 10.1210/mend.15.12.0738. [DOI] [PubMed] [Google Scholar]
- Ideker, T., Thorsson, V., Ranish, J.A., Christmas, R., Buhler, J., Eng, J.K., Bumgarner, R., Goodlett, D.R., Aebersold, R., Hood, L. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- Iizuka, N., Najita, L., Franzusoff, A., Sarnow, P. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae . Mol. Cell. Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki, Y., Li, X., Serin, G., Maquat, L.E. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- Ivanov, P.A., Karpova, O.V., Skulachev, M.V., Tomashevskaya, O.L., Rodionova, N.P., Dorokhov Yu, L., Atabekov, J.G. A tobamovirus genome that contains an internal ribosome entry site functional in vitro. Virology. 1997;232:32–43. doi: 10.1006/viro.1997.8525. [DOI] [PubMed] [Google Scholar]
- Izquierdo, J.M., Cuezva, J.M. Internal-ribosome-entry-site functional activity of the 3′-untranslated region of the mRNA for the β subunit of mitochondrial H+-ATP synthase. Biochem. J. 2000;346:849–855. [PMC free article] [PubMed] [Google Scholar]
- Jaag, H.M., Kawchuk, L., Rohde, W., Fischer, R., Emans, N., Prufer, D. An unusual internal ribosomal entry site of inverted symmetry directs expression of a potato leafroll polerovirus replication-associated protein. Proc. Natl. Acad. Sci. 2003;100:8939–8944. doi: 10.1073/pnas.1332697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, R.J., Hunt, S.L., Reynolds, J.E., Kaminski, A. Cap-dependent and cap-independent translation: Operational distinctions and mechanistic interpretations. Curr. Top. Microbiol. Immunol. 1995;203:1–29. doi: 10.1007/978-3-642-79663-0_1. [DOI] [PubMed] [Google Scholar]
- Jacobs, J.L., Dinman, J.D. Systematic analysis of bicistronic reporter assay data. Nucleic Acids Res. 2004;32 doi: 10.1093/nar/gnh157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, S.K., Krausslich, H.G., Nicklin, M.J., Duke, G.M., Palmenberg, A.C., Wimmer, E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, G.M., Leong, L.E., Hoang, L.T., Wang, P.H., Gutman, G.A., Semler, B.L. Structurally distinct elements mediate internal ribosome entry within the 5′-noncoding region of a voltage-gated potassium channel mRNA. J. Biol. Chem. 2004;279:47419–47430. doi: 10.1074/jbc.M405885200. [DOI] [PubMed] [Google Scholar]
- Ji, Y., Xu, X., Stormo, G.D. A graph theoretical approach for predicting common RNA secondary structure motifs including pseudoknots in unaligned sequences. Bioinformatics. 2004;20:1591–1602. doi: 10.1093/bioinformatics/bth131. [DOI] [PubMed] [Google Scholar]
- Jimenez, J., Jang, G.M., Semler, B.L., Waterman, M.L. An internal ribosome entry site mediates translation of lymphoid enhancer factor-1. RNA. 2005;11:1385–1399. doi: 10.1261/rna.7226105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes, G., Sarnow, P. Cap-independent polysomal association of natural mRNAs encoding c-Myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA. 1998;4:1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes, G., Carter, M.S., Eisen, M.B., Brown, P.O., Sarnow, P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl. Acad. Sci. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, K.N., Christian, P.D. The novel genome organization of the insect picorna-like virus Drosophila C virus suggests this virus belongs to a previously undescribed virus family. J. Gen. Virol. 1998;79:191–203. doi: 10.1099/0022-1317-79-1-191. [DOI] [PubMed] [Google Scholar]
- Jopling, C.L., Willis, A.E. N-Myc translation is initiated via an internal ribosome entry segment that displays enhanced activity in neuronal cells. Oncogene. 2001;20:2664–2670. doi: 10.1038/sj.onc.1204404. [DOI] [PubMed] [Google Scholar]
- Jopling, C.L., Spriggs, K.A., Mitchell, S.A., Stoneley, M., Willis, A.E. L-Myc protein synthesis is initiated by internal ribosome entry. RNA. 2004;10:287–298. doi: 10.1261/rna.5138804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan, V., Wilson, C. RNA secondary structure prediction based on free energy and phylogenetic analysis. J. Mol. Biol. 1999;289:935–947. doi: 10.1006/jmbi.1999.2801. [DOI] [PubMed] [Google Scholar]
- Kaempfer, R. RNA sensors: Novel regulators of gene expression. EMBO Rep. 2003;4:1043–1047. doi: 10.1038/sj.embor.7400005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski, A., Belsham, G.J., Jackson, R.J. Translation of encephalomyocarditis virus RNA: Parameters influencing the selection of the internal initiation site. EMBO J. 1994;13:1673–1681. doi: 10.1002/j.1460-2075.1994.tb06431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori, Y., Nakashima, N. A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA. 2001;7:266–274. doi: 10.1017/s1355838201001741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke, A., Doudna, J.A. Crystallization of RNA and RNA–protein complexes. Methods. 2004;34:408–414. doi: 10.1016/j.ymeth.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Keene, J.D., Tenenbaum, S.A. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell. 2002;9:1161–1167. doi: 10.1016/s1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
- Kieft, J.S., Zhou, K., Jubin, R., Murray, M.G., Lau, J.Y., Doudna, J.A. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- Kieft, J.S., Zhou, K., Jubin, R., Doudna, J.A. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft, J.S., Zhou, K., Grech, A., Jubin, R., Doudna, J.A. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat. Struct. Biol. 2002;9:370–374. doi: 10.1038/nsb781. [DOI] [PubMed] [Google Scholar]
- Kim, J.G., Armstrong, R.C., Berndt, J.A., Kim, N.W., Hudson, L.D. A secreted DNA-binding protein that is translated through an internal ribosome entry site (IRES) and distributed in a discrete pattern in the central nervous system. Mol. Cell. Neurosci. 1998;12:119–140. doi: 10.1006/mcne.1998.0701. [DOI] [PubMed] [Google Scholar]
- Kim, Y.K., Hahm, B., Jang, S.K. Polypyrimidine tract-binding protein inhibits translation of bip mRNA. J. Mol. Biol. 2000;304:119–133. doi: 10.1006/jmbi.2000.4179. [DOI] [PubMed] [Google Scholar]
- Kim, N., Shiffeldrim, N., Gan, H.H., Schlick, T. Candidates for novel RNA topologies. J. Mol. Biol. 2004;341:1129–1144. doi: 10.1016/j.jmb.2004.06.054. [DOI] [PubMed] [Google Scholar]
- Kjems, J., Egebjerg, J. Modern methods for probing RNA structure. Curr. Opin. Biotechnol. 1998;9:59–65. doi: 10.1016/s0958-1669(98)80085-2. [DOI] [PubMed] [Google Scholar]
- Klein, R.J., Eddy, S.R. RSEARCH: Finding homologs of single structured RNA sequences. BMC Bioinformatics. 2003;4 doi: 10.1186/1471-2105-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinck, R., Westhof, E., Walker, S., Afshar, M., Collier, A., Aboul-Ela, F. A potential RNA drug target in the hepatitis C virus internal ribosomal entry site. RNA. 2000;6:1423–1431. doi: 10.1017/s1355838200000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen, B., Hein, J. RNA secondary structure prediction using stochastic context-free grammars and evolutionary history. Bioinformatics. 1999;15:446–454. doi: 10.1093/bioinformatics/15.6.446. [DOI] [PubMed] [Google Scholar]
- Knudsen, B., Hein, J. Pfold: RNA secondary structure prediction using stochastic context-free grammars. Nucleic Acids Res. 2003;31:3423–3428. doi: 10.1093/nar/gkg614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, D.C., Liu, D.X., Wong, S.M. A six-nucleotide segment within the 3′ untranslated region of hibiscus chlorotic ringspot virus plays an essential role in translational enhancement. J. Virol. 2002;76:1144–1153. doi: 10.1128/JVI.76.3.1144-1153.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, D.C., Wong, S.M., Liu, D.X. Synergism of the 3′-untranslated region and an internal ribosome entry site differentially enhances the translation of a plant virus coat protein. J. Biol. Chem. 2003;278:20565–20573. doi: 10.1074/jbc.M210212200. [DOI] [PubMed] [Google Scholar]
- Kolupaeva, V.G., Hellen, C.U., Shatsky, I.N. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA. 1996;2:1199–1212. [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva, V.G., Pestova, T.V., Hellen, C.U. Ribosomal binding to the internal ribosomal entry site of classical swine fever virus. RNA. 2000;6:1791–1807. doi: 10.1017/s1355838200000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar, A.A., Hatzoglou, M. Internal ribosome entry sites in cellular mRNAs: Mystery of their existence. J. Biol. Chem. 2005;280:23425–23428. doi: 10.1074/jbc.R400041200. [DOI] [PubMed] [Google Scholar]
- Kozak, M. New ways of initiating translation in eukaryotes? Mol. Cell. Biol. 2001;21:1899–1907. doi: 10.1128/MCB.21.6.1899-1907.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak, M. Alternative ways to think about mRNA sequences and proteins that appear to promote internal initiation of translation. Gene. 2003;318:1–23. doi: 10.1016/s0378-1119(03)00774-1. [DOI] [PubMed] [Google Scholar]
- Kozak, M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37. doi: 10.1016/j.gene.2005.06.037. [DOI] [PubMed] [Google Scholar]
- Kuhn, R., Luz, N., Beck, E. Functional analysis of the internal translation initiation site of foot-and-mouth disease virus. J. Virol. 1990;64:4625–4631. doi: 10.1128/jvi.64.10.4625-4631.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laferriere, A., Gautheret, D., Cedergren, R. An RNA pattern matching program with enhanced performance and portability. Comput. Appl. Biosci. 1994;10:211–212. doi: 10.1093/bioinformatics/10.2.211. [DOI] [PubMed] [Google Scholar]
- Lahlou, H., Fanjul, M., Pradayrol, L., Susini, C., Pyronnet, S. Restoration of functional gap junctions through internal ribosome entry site-dependent synthesis of endogenous connexins in density-inhibited cancer cells. Mol. Cell. Biol. 2005;25:4034–4045. doi: 10.1128/MCB.25.10.4034-4045.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert, A., Fontaine, J.F., Legendre, M., Leclerc, F., Permal, E., Major, F., Putzer, H., Delfour, O., Michot, B., Gautheret, D. The ERPIN server: An interface to profile-based RNA motif identification. Nucleic Acids Res. 2004;32:W160–W165. doi: 10.1093/nar/gkh418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert, A., Legendre, M., Fontaine, J.F., Gautheret, D. Computing expectation values for RNA motifs using discrete convolutions. BMC Bioinformatics. 2005;6 doi: 10.1186/1471-2105-6-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, K.J., Kappel, A., Goodall, G.J. Hypoxia-inducible factor-1α mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol. Biol. Cell. 2002;13:1792–1801. doi: 10.1091/mbc.02-02-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring, A.S., Overbaugh, J. Evidence that an IRES within the Notch2 coding region can direct expression of a nuclear form of the protein. Mol. Cell. 2000;6:939–945. doi: 10.1016/s1097-2765(05)00084-5. [DOI] [PubMed] [Google Scholar]
- Le, S.Y., Maizel, J.V., Jr. A common RNA structural motif involved in the internal initiation of translation of cellular mRNAs. Nucleic Acids Res. 1997;25:362–369. doi: 10.1093/nar/25.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, S.Y., Chen, J.H., Konings, D., Maizel, J.V., Jr. Discovering well-ordered folding patterns in nucleotide sequences. Bioinformatics. 2003;19:354–361. doi: 10.1093/bioinformatics/btf826. [DOI] [PubMed] [Google Scholar]
- Lemon, S.M., Honda, M. Internal ribosome entry sites within the RNA genomes of hepatitis C virus and other flaviviruses. Semin. Virol. 1997;8:274–288. [Google Scholar]
- Le Quesne, J.P., Stoneley, M., Fraser, G.A., Willis, A.E. Derivation of a structural model for the c-Myc IRES. J. Mol. Biol. 2001;310:111–126. doi: 10.1006/jmbi.2001.4745. [DOI] [PubMed] [Google Scholar]
- Lesnik, E.A., Sampath, R., Ecker, D.J. Rev response elements (RRE) in lentiviruses: An RNAMotif algorithm-based strategy for RRE prediction. Med. Res. Rev. 2002;22:617–636. doi: 10.1002/med.10027. [DOI] [PubMed] [Google Scholar]
- Levis, C., Astier-Manifacier, S. The 5′ untranslated region of PVY RNA, even located in an internal position, enables initiation of translation. Virus Genes. 1993;7:367–379. doi: 10.1007/BF01703392. [DOI] [PubMed] [Google Scholar]
- Lewin, B. Essential genes, Pearson Education Inc., Prentice Hall; Upper Saddle River, NJ: 2006. [Google Scholar]
- Lewis, S.M., Holcik, M. IRES in distress: Translational regulation of the inhibitor of apoptosis proteins XIAP and HIAP2 during cell stress. Cell Death Differ. 2005;12:547–553. doi: 10.1038/sj.cdd.4401602. [DOI] [PubMed] [Google Scholar]
- Ling, J., Morley, S.J., Traugh, J.A. Inhibition of cap-dependent translation via phosphorylation of eIF4G by protein kinase Pak2. EMBO J. 2005;24:4094–4105. doi: 10.1038/sj.emboj.7600868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., Dong, Z., Han, B., Yang, Y., Liu, Y., Zhang, J.T. Regulation of expression by promoters versus internal ribosome entry site in the 5′-untranslated sequence of the human cyclin-dependent kinase inhibitor p27kip1 . Nucleic Acids Res. 2005;33:3763–3771. doi: 10.1093/nar/gki680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Quinto, S., Martinez-Salas, E. Conserved structural motifs located in distal loops of aphthovirus internal ribosome entry site domain 3 are required for internal initiation of translation. J. Virol. 1997;71:4171–4175. doi: 10.1128/jvi.71.5.4171-4175.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Quinto, S., Saiz, M., de la Morena, D., Sobrino, F., Martinez-Salas, E. IRES-driven translation is stimulated separately by the FMDV 3′-NCR and poly(A) sequences. Nucleic Acids Res. 2002;30:4398–4405. doi: 10.1093/nar/gkf569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Lastra, M., Gabus, C., Darlix, J.L. Characterization of an internal ribosomal entry segment within the 5′ leader of avian reticuloendotheliosis virus type A RNA and development of novel MLV-REV-based retroviral vectors. Hum. Gene Ther. 1997;8:1855–1865. doi: 10.1089/hum.1997.8.16-1855. [DOI] [PubMed] [Google Scholar]
- Low, W., Harries, M., Ye, H., Du, M.Q., Boshoff, C., Collins, M. Internal ribosome entry site regulates translation of Kaposi's sarcoma-associated herpesvirus FLICE inhibitory protein. J. Virol. 2001;75:2938–2945. doi: 10.1128/JVI.75.6.2938-2945.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, T.M., Eddy, S.R. tRNAscan-SE:A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky, P.J., Otto, G.A., Lancaster, A.M., Sarnow, P., Puglisi, J.D. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat. Struct. Biol. 2000;7:1105–1110. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- Lukavsky, P.J., Kim, I., Otto, G.A., Puglisi, J.D. Structure of HCV IRES domain II determined by NMR. Nat. Struct. Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- Lyons, A.J., Robertson, H.D. Detection of tRNA-like structure through RNase P cleavage of viral internal ribosome entry site RNAs near the AUG start triplet. J. Biol. Chem. 2003;278:26844–26850. doi: 10.1074/jbc.M304052200. [DOI] [PubMed] [Google Scholar]
- Lyons, A.J., Lytle, J.R., Gomez, J., Robertson, H.D. Hepatitis C virus internal ribosome entry site RNA contains a tertiary structural element in a functional domain of stem–loop II. Nucleic Acids Res. 2001;29:2535–2541. doi: 10.1093/nar/29.12.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle, J.R., Wu, L., Robertson, H.D. Domains on the hepatitis C virus internal ribosome entry site for 40s subunit binding. RNA. 2002;8:1045–1055. doi: 10.1017/s1355838202029965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macejak, D.G., Sarnow, P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- Macke, T.J., Ecker, D.J., Gutell, R.R., Gautheret, D., Case, D.A., Sampath, R. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 2001;29:4724–4735. doi: 10.1093/nar/29.22.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, D., Nagel, A.C., Preiss, A. Two isoforms of the Notch antagonist Hairless are produced by differential translation initiation. Proc. Natl. Acad. Sci. 2002;99:15480–15485. doi: 10.1073/pnas.242596699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marash, L., Kimchi, A. DAP5 and IRES-mediated translation during programmed cell death. Cell Death Differ. 2005;12:554–562. doi: 10.1038/sj.cdd.4401609. [DOI] [PubMed] [Google Scholar]
- Martin, M.M., Garcia, J.A., McFarland, J.D., Duffy, A.A., Gregson, J.P., Elton, T.S. Translation of the human angiotensin II type 1 receptor mRNA is mediated by a highly efficient internal ribosome entry site. Mol. Cell. Endocrinol. 2003;212:51–61. doi: 10.1016/j.mce.2003.09.017. [DOI] [PubMed] [Google Scholar]
- Martineau, Y., Le Bec, C., Monbrun, L., Allo, V., Chiu, I.M., Danos, O., Moine, H., Prats, H., Prats, A.C. Internal ribosome entry site structural motifs conserved among mammalian fibroblast growth factor 1 alternatively spliced mRNAs. Mol. Cell. Biol. 2004;24:7622–7635. doi: 10.1128/MCB.24.17.7622-7635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Salas, E., Regalado, M.P., Domingo, E. Identification of an essential region for internal initiation of translation in the aphthovirus internal ribosome entry site and implications for viral evolution. J. Virol. 1996;70:992–998. doi: 10.1128/jvi.70.2.992-998.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Salas, E., Quinto, S.L., Ramos, R., Fernandez-Miragall, O. IRES elements: Features of the RNA structure contributing to their activity. Biochimie. 2002;84:755–763. doi: 10.1016/s0300-9084(02)01408-6. [DOI] [PubMed] [Google Scholar]
- Maser, R.S., Zinkel, R., Petrini, J.H. An alternative mode of translation permits production of a variant NBS1 protein from the common Nijmegen breakage syndrome allele. Nat. Genet. 2001;27:417–421. doi: 10.1038/86920. [DOI] [PubMed] [Google Scholar]
- Mathews, D.H. Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA. 2004;10:1178–1190. doi: 10.1261/rna.7650904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews, D.H. Predicting a set of minimal free energy RNA secondary structures common to two sequences. Bioinformatics. 2005;21:2246–2253. doi: 10.1093/bioinformatics/bti349. [DOI] [PubMed] [Google Scholar]
- Mathews, D.H. Revolutions in RNA secondary structure prediction. J. Mol. Biol. 2006;359:526–532. doi: 10.1016/j.jmb.2006.01.067. [DOI] [PubMed] [Google Scholar]
- Mathews, D.H., Turner, D.H. Dynalign: An algorithm for finding the secondary structure common to two RNA sequences. J. Mol. Biol. 2002;317:191–203. doi: 10.1006/jmbi.2001.5351. [DOI] [PubMed] [Google Scholar]
- Mathews, D.H., Turner, D.H. Prediction of RNA secondary structure by free energy minimization. Curr. Opin. Struct. Biol. 2006;16:270–278. doi: 10.1016/j.sbi.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Mathews, D.H., Sabina, J., Zuker, M., Turner, D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Mathews, D.H., Disney, M.D., Childs, J.L., Schroeder, S.J., Zuker, M., Turner, D.H. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro, V.P., Edelman, G.M. The ribosome filter hypothesis. Proc. Natl. Acad. Sci. 2002;99:12031–12036. doi: 10.1073/pnas.192442499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder, B., Seshadri, V., Fox, P.L. Translational control by the 3′-UTR: The ends specify the means. Trends Biochem. Sci. 2003;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- McBratney, S., Sarnow, P. Evidence for involvement of trans-acting factors in selection of the AUG start codon during eukaryotic translational initiation. Mol. Cell. Biol. 1996;16:3523–3534. doi: 10.1128/mcb.16.7.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaskill, J.S. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990;29:1105–1119. doi: 10.1002/bip.360290621. [DOI] [PubMed] [Google Scholar]
- Merrill, M.K., Dobrikova, E.Y., Gromeier, M. Cell-type-specific repression of internal ribosome entry site activity by double-stranded RNA-binding protein 76. J. Virol. 2006;80:3147–3156. doi: 10.1128/JVI.80.7.3147-3156.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard, S.S., Vidal, A., Markus, M., Koff, A. A U-rich element in the 5′ untranslated region is necessary for the translation of p27 mRNA. Mol. Cell. Biol. 2000;20:5947–5959. doi: 10.1128/mcb.20.16.5947-5959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miskimins, W.K., Wang, G., Hawkinson, M., Miskimins, R. Control of cyclin-dependent kinase inhibitor p27 expression by cap-independent translation. Mol. Cell. Biol. 2001;21:4960–4967. doi: 10.1128/MCB.21.15.4960-4967.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, S.A., Brown, E.C., Coldwell, M.J., Jackson, R.J., Willis, A.E. Protein factor requirements of the Apaf-1 internal ribosome entry segment: Roles of polypyrimidine tract binding protein and upstream of N-ras. Mol. Cell. Biol. 2001;21:3364–3374. doi: 10.1128/MCB.21.10.3364-3374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, S.A., Spriggs, K.A., Coldwell, M.J., Jackson, R.J., Willis, A.E. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell. 2003;11:757–771. doi: 10.1016/s1097-2765(03)00093-5. [DOI] [PubMed] [Google Scholar]
- Mitchell, S.A., Spriggs, K.A., Bushell, M., Evans, J.R., Stoneley, M., Le Quesne, J.P., Spriggs, R.V., Willis, A.E. Identification of a motif that mediates polypyrimidine tract-binding protein-dependent internal ribosome entry. Genes & Dev. 2005;19:1556–1571. doi: 10.1101/gad.339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, P., Thompson, J., Chakkalakal, J.V., Holcik, M., Jasmin, B.J. The utrophin A 5′ untranslated region confers internal ribosome entry site-mediated translation control during regeneration of skeletal muscle fibers. J. Biol. Chem. 2005;280:32997–33005. doi: 10.1074/jbc.M503994200. [DOI] [PubMed] [Google Scholar]
- Mokrejs, M., Vopalensky, V., Kolenaty, O., Masek, T., Feketova, Z., Sekyrova, P., Skaloudova, B., Kriz, V., Pospisek, M. IRESite: The database of experimentally verified IRES structures ( www.iresite.org) Nucleic Acids Res. 2006;34:D125–D130. doi: 10.1093/nar/gkj081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish, B.C., Rumsby, M.G. The 5′ untranslated region of protein kinase Cδ directs translation by an internal ribosome entry segment that is most active in densely growing cells and during apoptosis. Mol. Cell. Biol. 2002;22:6089–6099. doi: 10.1128/MCB.22.17.6089-6099.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy, V.L., Rose, G.D. RNABase: An annotated database of RNA structures. Nucleic Acids Res. 2003;31:502–504. doi: 10.1093/nar/gkg012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanbru, C., Prats, A.C., Droogmans, L., Defrance, P., Huez, G., Kruys, V. Translation of the human c-Myc P0 tricistronic mRNA involves two independent internal ribosome entry sites. Oncogene. 2001;20:4270–4280. doi: 10.1038/sj.onc.1204548. [DOI] [PubMed] [Google Scholar]
- Negulescu, D., Leong, L.E., Chandy, K.G., Semler, B.L., Gutman, G.A. Translation initiation of a cardiac voltage-gated potassium channel by internal ribosome entry. J. Biol. Chem. 1998;273:20109–20113. doi: 10.1074/jbc.273.32.20109. [DOI] [PubMed] [Google Scholar]
- Nevins, T.A., Harder, Z.M., Korneluk, R.G., Holcik, M. Distinct regulation of internal ribosome entry site-mediated translation following cellular stress is mediated by apoptotic fragments of eIF4G translation initiation factor family members eIF4GI and p97/DAP5/NAT1. J. Biol. Chem. 2003;278:3572–3579. doi: 10.1074/jbc.M206781200. [DOI] [PubMed] [Google Scholar]
- Nishiyama, T., Yamamoto, H., Shibuya, N., Hatakeyama, Y., Hachimori, A., Uchiumi, T., Nakashima, N. Structural elements in the internal ribosome entry site of Plautia stali intestine virus responsible for binding with ribosomes. Nucleic Acids Res. 2003;31:2434–2442. doi: 10.1093/nar/gkg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizuka, S., Charboneau, L., Young, L., Major, S., Reinhold, W.C., Waltham, M., Kouros-Mehr, H., Bussey, K.J., Lee, J.K., Espina, V., et al. Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc. Natl. Acad. Sci. 2003;100:14229–14234. doi: 10.1073/pnas.2331323100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odreman-Macchioli, F., Baralle, F.E., Buratti, E. Mutational analysis of the different bulge regions of hepatitis C virus domain II and their influence on internal ribosome entry site translational ability. J. Biol. Chem. 2001;276:41648–41655. doi: 10.1074/jbc.M104128200. [DOI] [PubMed] [Google Scholar]
- Oh, S.K., Scott, M.P., Sarnow, P. Homeotic gene Antennapedia mRNA contains 5′-noncoding sequences that confer translational initiation by internal ribosome binding. Genes & Dev. 1992;6:1643–1653. doi: 10.1101/gad.6.9.1643. [DOI] [PubMed] [Google Scholar]
- Ohlmann, T., Lopez-Lastra, M., Darlix, J.L. An internal ribosome entry segment promotes translation of the simian immunodeficiency virus genomic RNA. J. Biol. Chem. 2000;275:11899–11906. doi: 10.1074/jbc.275.16.11899. [DOI] [PubMed] [Google Scholar]
- Oltean, S., Banerjee, R. A B12-responsive internal ribosome entry site (IRES) element in human methionine synthase. J. Biol. Chem. 2005;280:32662–32668. doi: 10.1074/jbc.M501964200. [DOI] [PubMed] [Google Scholar]
- Oumard, A., Hennecke, M., Hauser, H., Nourbakhsh, M. Translation of NRF mRNA is mediated by highly efficient internal ribosome entry. Mol. Cell. Biol. 2000;20:2755–2759. doi: 10.1128/mcb.20.8.2755-2759.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens, G.C., Chappell, S.A., Mauro, V.P., Edelman, G.M. Identification of two short internal ribosome entry sites selected from libraries of random oligonucleotides. Proc. Natl. Acad. Sci. 2001;98:1471–1476. doi: 10.1073/pnas.98.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, E., Lee, J.M., Blais, J.D., Bell, J.C., Pelletier, J. Internal translation initiation mediated by the angiogenic factor Tie2. J. Biol. Chem. 2005;280:20945–20953. doi: 10.1074/jbc.M412744200. [DOI] [PubMed] [Google Scholar]
- Pavesi, G., Mauri, G., Stefani, M., Pesole, G. RNAProfile: An algorithm for finding conserved secondary structure motifs in unaligned RNA sequences. Nucleic Acids Res. 2004;32:3258–3269. doi: 10.1093/nar/gkh650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen, S.K., Christiansen, J., Hansen, T.O., Larsen, M.R., Nielsen, F.C. Human insulin-like growth factor II leader 2 mediates internal initiation of translation. Biochem. J. 2002;363:37–44. doi: 10.1042/0264-6021:3630037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier, J., Sonenberg, N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- Peri, S., Pandey, A. A reassessment of the translation initiation codon in vertebrates. Trends Genet. 2001;17:685–687. doi: 10.1016/s0168-9525(01)02493-3. [DOI] [PubMed] [Google Scholar]
- Perriquet, O., Touzet, H., Dauchet, M. Finding the common structure shared by two homologous RNAs. Bioinformatics. 2003;19:108–116. doi: 10.1093/bioinformatics/19.1.108. [DOI] [PubMed] [Google Scholar]
- Pervouchine, D.D., Graber, J.H., Kasif, S. On the normalization of RNA equilibrium free energy to the length of the sequence. Nucleic Acids Res. 2003;31 doi: 10.1093/nar/gng049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesole, G., Liuni, S., Grillo, G., Licciulli, F., Mignone, F., Gissi, C., Saccone, C. UTRdb and UTRsite: Specialized databases of sequences and functional elements of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Update 2002. Nucleic Acids Res. 2002;30:335–340. doi: 10.1093/nar/30.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering, B.M., Mitchell, S.A., Evans, J.R., Willis, A.E. Polypyrimidine tract binding protein and poly r(C) binding protein 1 interact with the BAG-1 IRES and stimulate its activity in vitro and in vivo. Nucleic Acids Res. 2003;31:639–646. doi: 10.1093/nar/gkg146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering, B.M., Mitchell, S.A., Spriggs, K.A., Stoneley, M., Willis, A.E. Bag-1 internal ribosome entry segment activity is promoted by structural changes mediated by poly(rC) binding protein 1 and recruitment of polypyrimidine tract binding protein 1. Mol. Cell. Biol. 2004;24:5595–5605. doi: 10.1128/MCB.24.12.5595-5605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko, E.V., Blinov, V.M., Chernov, B.K., Dmitrieva, T.M., Agol, V.I. Conservation of the secondary structure elements of the 5′-untranslated region of cardio- and aphthovirus RNAs. Nucleic Acids Res. 1989a;17:5701–5711. doi: 10.1093/nar/17.14.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko, E.V., Blinov, V.M., Romanova, L.I., Sinyakov, A.N., Maslova, S.V., Agol, V.I. Conserved structural domains in the 5′-untranslated region of picornaviral genomes: An analysis of the segment controlling translation and neurovirulence. Virology. 1989b;168:201–209. doi: 10.1016/0042-6822(89)90259-6. [DOI] [PubMed] [Google Scholar]
- Pilipenko, E.V., Gmyl, A.P., Maslova, S.V., Svitkin, Y.V., Sinyakov, A.N., Agol, V.I. Prokaryotic-like cis elements in the cap-independent internal initiation of translation on picornavirus RNA. Cell. 1992;68:119–131. doi: 10.1016/0092-8674(92)90211-t. [DOI] [PubMed] [Google Scholar]
- Pilipenko, E.V., Gmyl, A.P., Maslova, S.V., Belov, G.A., Sinyakov, A.N., Huang, M., Brown, T.D., Agol, V.I. Starting window, a distinct element in the cap-independent internal initiation of translation on picornaviral RNA. J. Mol. Biol. 1994;241:398–414. doi: 10.1006/jmbi.1994.1516. [DOI] [PubMed] [Google Scholar]
- Pilipenko, E.V., Viktorova, E.G., Guest, S.T., Agol, V.I., Roos, R.P. Cell-specific proteins regulate viral RNA translation and virus-induced disease. EMBO J. 2001;20:6899–6908. doi: 10.1093/emboj/20.23.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkstaff, J.K., Chappell, S.A., Mauro, V.P., Edelman, G.M., Krushel, L.A. Internal initiation of translation of five dendritically localized neuronal mRNAs. Proc. Natl. Acad. Sci. 2001;98:2770–2775. doi: 10.1073/pnas.051623398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev, A.V., Chard, L.S., Kaku, Y., Johns, H.L., Shatsky, I.N., Belsham, G.J. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J. Virol. 2004;78:4487–4497. doi: 10.1128/JVI.78.9.4487-4497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole, T.L., Wang, C., Popp, R.A., Potgieter, L.N., Siddiqui, A., Collett, M.S. Pestivirus translation initiation occurs by internal ribosome entry. Virology. 1995;206:750–754. doi: 10.1016/s0042-6822(95)80003-4. [DOI] [PubMed] [Google Scholar]
- Pozner, A., Goldenberg, D., Negreanu, V., Le, S.Y., Elroy-Stein, O., Levanon, D., Groner, Y. Transcription-coupled translation control of AML1/RUNX1 is mediated by cap- and internal ribosome entry site-dependent mechanisms. Mol. Cell. Biol. 2000;20:2297–2307. doi: 10.1128/mcb.20.7.2297-2307.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud, C.G. eIF2 and the control of cell physiology. Semin. Cell Dev. Biol. 2005;16:3–12. doi: 10.1016/j.semcdb.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Pyronnet, S., Pradayrol, L., Sonenberg, N. A cell cycle-dependent internal ribosome entry site. Mol. Cell. 2000;5:607–616. doi: 10.1016/s1097-2765(00)80240-3. [DOI] [PubMed] [Google Scholar]
- Pyronnet, S., Dostie, J., Sonenberg, N. Suppression of cap-dependent translation in mitosis. Genes & Dev. 2001;15:2083–2093. doi: 10.1101/gad.889201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, X., Sarnow, P. Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J. Biol. Chem. 2004;279:13721–13728. doi: 10.1074/jbc.M312854200. [DOI] [PubMed] [Google Scholar]
- Rabadan-Diehl, C., Volpi, S., Nikodemova, M., Aguilera, G. Translational regulation of the vasopressin v1b receptor involves an internal ribosome entry site. Mol. Endocrinol. 2003;17:1959–1971. doi: 10.1210/me.2001-0336. [DOI] [PubMed] [Google Scholar]
- Ray, P.S., Das, S. Inhibition of hepatitis C virus IRES-mediated translation by small RNAs analogous to stem–loop structures of the 5′-untranslated region. Nucleic Acids Res. 2004;32:1678–1687. doi: 10.1093/nar/gkh328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, J., Rastegari, B., Condon, A., Hoos, H.H. HotKnots: Heuristic prediction of RNA secondary structures including pseudoknots. RNA. 2005;11:1494–1504. doi: 10.1261/rna.7284905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee, S., Yang, S.J., Lee, S.J., Park, D. βPix-b(L), a novel isoform of βPix, is generated by alternative translation. Biochem. Biophys. Res. Commun. 2004;318:415–421. doi: 10.1016/j.bbrc.2004.04.039. [DOI] [PubMed] [Google Scholar]
- Rijnbrand, R., van der Straaten, T., van Rijn, P.A., Spaan, W.J., Bredenbeek, P.J. Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initiation codon. J. Virol. 1997;71:451–457. doi: 10.1128/jvi.71.1.451-457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijnbrand, R., Abell, G., Lemon, S.M. Mutational analysis of the GB virus B internal ribosome entry site. J. Virol. 2000;74:773–783. doi: 10.1128/jvi.74.2.773-783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijnbrand, R., Thiviyanathan, V., Kaluarachchi, K., Lemon, S.M., Gorenstein, D.G. Mutational and structural analysis of stem–loop IIIC of the hepatitis C virus and GB virus B internal ribosome entry sites. J. Mol. Biol. 2004;343:805–817. doi: 10.1016/j.jmb.2004.08.095. [DOI] [PubMed] [Google Scholar]
- Rivas, E., Eddy, S.R. A dynamic programming algorithm for RNA structure prediction including pseudoknots. J. Mol. Biol. 1999;285:2053–2068. doi: 10.1006/jmbi.1998.2436. [DOI] [PubMed] [Google Scholar]
- Rivas, E., Eddy, S.R. Secondary structure alone is generally not statistically significant for the detection of noncoding RNAs. Bioinformatics. 2000;16:583–605. doi: 10.1093/bioinformatics/16.7.583. [DOI] [PubMed] [Google Scholar]
- Rivera, V.M., Welsh, J.D., Maizel, J.V., Jr. Comparative sequence analysis of the 5′ noncoding region of the enteroviruses and rhinoviruses. Virology. 1988;165:42–50. doi: 10.1016/0042-6822(88)90656-3. [DOI] [PubMed] [Google Scholar]
- Rubtsova, M.P., Sizova, D.V., Dmitriev, S.E., Ivanov, D.S., Prassolov, V.S., Shatsky, I.N. Distinctive properties of the 5′-untranslated region of human hsp70 mRNA. J. Biol. Chem. 2003;278:22350–22356. doi: 10.1074/jbc.M303213200. [DOI] [PubMed] [Google Scholar]
- Sanderbrand, S.A., Tautz, N., Thiel, H.J., Ochs, K., Beck, E., Niepmann, M. Translation from the internal ribosome entry site of bovine viral diarrhea virus is independent of the interaction with polypyrimidine tract-binding protein. Vet. Microbiol. 2000;77:215–227. doi: 10.1016/s0378-1135(00)00278-9. [DOI] [PubMed] [Google Scholar]
- Sankoff, D. Simultaneous solution of the RNA folding alignment and protosequence problems. SIAM J. Appl. Math. 1985;45:810–825. [Google Scholar]
- Sasaki, J., Nakashima, N. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J. Virol. 1999;73:1219–1226. doi: 10.1128/jvi.73.2.1219-1226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavi, A., Hudder, A., Werner, R. Connexin43 mRNA contains a functional internal ribosome entry site. FEBS Lett. 1999;464:118–122. doi: 10.1016/s0014-5793(99)01699-3. [DOI] [PubMed] [Google Scholar]
- Sehgal, A., Briggs, J., Rinehart-Kim, J., Basso, J., Bos, T.J. The chicken c-Jun 5′ untranslated region directs translation by internal initiation. Oncogene. 2000;19:2836–2845. doi: 10.1038/sj.onc.1203601. [DOI] [PubMed] [Google Scholar]
- Sella, O., Gerlitz, G., Le, S.Y., Elroy-Stein, O. Differentiation-induced internal translation of c-sis mRNA: Analysis of the cis elements and their differentiation-linked binding to the hnRNP C protein. Mol. Cell. Biol. 1999;19:5429–5440. doi: 10.1128/mcb.19.8.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serikawa, K.A., Xu, X.L., MacKay, V.L., Law, G.L., Zong, Q., Zhao, L.P., Bumgarner, R., Morris, D.R. The transcriptome and its translation during recovery from cell cycle arrest in Saccharomyces cerevisiae . Mol. Cell. Proteomics. 2003;2:191–204. doi: 10.1074/mcp.D200002-MCP200. [DOI] [PubMed] [Google Scholar]
- Seroussi, E., Shani, N., Ben-Meir, D., Chajut, A., Divinski, I., Faier, S., Gery, S., Karby, S., Kariv-Inbal, Z., Sella, O., et al. Uniquely conserved non-translated regions are involved in generation of the two major transcripts of protein phosphatase 2Cβ. J. Mol. Biol. 2001;312:439–451. doi: 10.1006/jmbi.2001.4967. [DOI] [PubMed] [Google Scholar]
- Shapiro, B.A., Bengali, D., Kasprzak, W., Wu, J.C. RNA folding pathway functional intermediates: Their prediction and analysis. J. Mol. Biol. 2001;312:27–44. doi: 10.1006/jmbi.2001.4931. [DOI] [PubMed] [Google Scholar]
- Sherrill, K.W., Byrd, M.P., Van Eden, M.E., Lloyd, R.E. BCL-2 translation is mediated via internal ribosome entry during cell stress. J. Biol. Chem. 2004;279:29066–29074. doi: 10.1074/jbc.M402727200. [DOI] [PubMed] [Google Scholar]
- Shi, Y., Sharma, A., Wu, H., Lichtenstein, A., Gera, J. Cyclin D1 and c-Myc internal ribosome entry site (IRES)-dependent translation is regulated by AKT activity and enhanced by rapamycin through a p38 MAPK- and ERK-dependent pathway. J. Biol. Chem. 2005;280:10964–10973. doi: 10.1074/jbc.M407874200. [DOI] [PubMed] [Google Scholar]
- Shimazaki, T., Honda, M., Kaneko, S., Kobayashi, K. Inhibition of internal ribosomal entry site-directed translation of HCV by recombinant IFN-α correlates with a reduced La protein. Hepatology. 2002;35:199–208. doi: 10.1053/jhep.2002.30202. [DOI] [PubMed] [Google Scholar]
- Shiroki, K., Ohsawa, C., Sugi, N., Wakiyama, M., Miura, K., Watanabe, M., Suzuki, Y., Sugano, S. Internal ribosome entry site-mediated translation of Smad5 in vivo: requirement for a nuclear event. Nucleic Acids Res. 2002;30:2851–2861. doi: 10.1093/nar/gkf408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siridechadilok, B., Fraser, C.S., Hall, R.J., Doudna, J.A., Nogales, E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- Sonenberg, N., Dever, T.E. Eukaryotic translation initiation factors and regulators. Curr. Opin. Struct. Biol. 2003;13:56–63. doi: 10.1016/s0959-440x(03)00009-5. [DOI] [PubMed] [Google Scholar]
- Spahn, C.M., Kieft, J.S., Grassucci, R.A., Penczek, P.A., Zhou, K., Doudna, J.A., Frank, J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- Stark, G.R., Kerr, I.M., Williams, B.R., Silveran, R.H., Schreiber, R.D. How cells respond to interferons. Annu. Rev. Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Stein, I., Itin, A., Einat, P., Skaliter, R., Grossman, Z., Keshet, E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: Implications for translation under hypoxia. Mol. Cell. Biol. 1998;18:3112–3119. doi: 10.1128/mcb.18.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneley, M., Willis, A.E. Cellular internal ribosome entry segments: Structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- Stoneley, M., Paulin, F.E., Le Quesne, J.P., Chappell, S.A., Willis, A.E. c-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- Stoneley, M., Subkhankulova, T., Le Quesne, J.P., Coldwell, M.J., Jopling, C.L., Belsham, G.J., Willis, A.E. Analysis of the c-Myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 2000;28:687–694. doi: 10.1093/nar/28.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneley, M., Spencer, J.P., Wright, S.C. An internal ribosome entry segment in the 5′ untranslated region of the mnt gene. Oncogene. 2001;20:893–897. doi: 10.1038/sj.onc.1204157. [DOI] [PubMed] [Google Scholar]
- Sudarsan, N., Barrick, J.E., Breaker, R.R. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9:644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, Y., Ishihara, D., Sasaki, M., Nakagawa, H., Hata, H., Tsunoda, T., Watanabe, M., Komatsu, T., Ota, T., Isogai, T., et al. Statistical analysis of the 5′ untranslated region of human mRNA using “Oligo-Capped” cDNA libraries. Genomics. 2000;64:286–297. doi: 10.1006/geno.2000.6076. [DOI] [PubMed] [Google Scholar]
- Svitkin, Y.V., Gradi, A., Imataka, H., Morino, S., Sonenberg, N. Eukaryotic initiation factor 4GII (eIF4GII), but not eIF4GI, cleavage correlates with inhibition of host cell protein synthesis after human rhinovirus infection. J. Virol. 1999;73:3467–3472. doi: 10.1128/jvi.73.4.3467-3472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teerink, H., Voorma, H.O., Thomas, A.A. The human insulin-like growth factor II leader 1 contains an internal ribosomal entry site. Biochim. Biophys. Acta. 1995;1264:403–408. doi: 10.1016/0167-4781(95)00185-9. [DOI] [PubMed] [Google Scholar]
- Tenenbaum, S.A., Lager, P.J., Carson, C.C., Keene, J.D. Ribonomics: Identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 2002;26:191–198. doi: 10.1016/S1046-2023(02)00022-1. [DOI] [PubMed] [Google Scholar]
- Tenenbaum, S.A., Carson, C.C., Atasoy, U., Keene, J.D. Genome-wide regulatory analysis using en masse nuclear run-ons and ribonomic profiling with autoimmune sera. Gene. 2003;317:79–87. doi: 10.1016/s0378-1119(03)00661-9. [DOI] [PubMed] [Google Scholar]
- Thomas, A.A., ter Haar, E., Wellink, J., Voorma, H.O. Cowpea mosaic virus middle component RNA contains a sequence that allows internal binding of ribosomes and that requires eukaryotic initiation factor 4F for optimal translation. J. Virol. 1991;65:2953–2959. doi: 10.1128/jvi.65.6.2953-2959.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, S.R., Sarnow, P. Enterovirus 71 contains a type I IRES element that functions when eukaryotic initiation factor eIF4G is cleaved. Virology. 2003;315:259–266. doi: 10.1016/s0042-6822(03)00544-0. [DOI] [PubMed] [Google Scholar]
- Thurner, C., Witwer, C., Hofacker, I.L., Stadler, P.F. Conserved RNA secondary structures in Flaviviridae genomes. J. Gen. Virol. 2004;85:1113–1124. doi: 10.1099/vir.0.19462-0. [DOI] [PubMed] [Google Scholar]
- Tinton, S.A., Schepens, B., Bruynooghe, Y., Beyaert, R., Cornelis, S. Regulation of the cell-cycle-dependent internal ribosome entry site of the PITSLRE protein kinase: Roles of Unr (upstream of N-ras) protein and phosphorylated translation initiation factor eIF-2α. Biochem.J. 2005;385:155–163. doi: 10.1042/BJ20040963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui, V., Macke, T., Case, D.A. A novel method for finding tRNA genes. RNA. 2003;9:507–517. doi: 10.1261/rna.2193703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama-Kohara, K., Iizuka, N., Kohara, M., Nomoto, A. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner, S., Gensac, M.C., Maret, A., Bayard, F., Amalric, F., Prats, H., Prats, A.C. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol. Cell. Biol. 1995a;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner, S., Waysbort, A., Marenda, M., Gensac, M.C., Amalric, F., Prats, A.C. Alternative translation initiation of the Moloney murine leukemia virus mRNA controlled by internal ribosome entry involving the p57/PTB splicing factor. J. Biol. Chem. 1995b;270:20376–20383. doi: 10.1074/jbc.270.35.20376. [DOI] [PubMed] [Google Scholar]
- Vagner, S., Touriol, C., Galy, B., Audigier, S., Gensac, M.C., Amalric, F., Bayard, F., Prats, H., Prats, A.C. Translation of CUG- but not AUG-initiated forms of human fibroblast growth factor 2 is activated in transformed and stressed cells. J. Cell Biol. 1996;135:1391–1402. doi: 10.1083/jcb.135.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eden, M.E., Byrd, M.P., Sherrill, K.W., Lloyd, R.E. Translation of cellular inhibitor of apoptosis protein 1 (c-IAP1) mRNA is IRES mediated and regulated during cell stress. RNA. 2004;10:469–481. doi: 10.1261/rna.5156804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan, A., Dasgupta, A. Novel fluorescence-based screen to identify small synthetic internal ribosome entry site elements. Mol. Cell. Biol. 2001;21:2826–2837. doi: 10.1128/MCB.21.8.2826-2837.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier, S.B., Jean-Jean, O. Complementarity between the mRNA 5′ untranslated region and 18S ribosomal RNA can inhibit translation. RNA. 2000;6:584–597. doi: 10.1017/s1355838200992239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., Le, S.Y., Ali, N., Siddiqui, A. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA. 1995;1:526–537. [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Weaver, M., Magnuson, N.S. Cryptic promoter activity in the DNA sequence corresponding to the pim-1 5′-UTR. Nucleic Acids Res. 2005;33:2248–2258. doi: 10.1093/nar/gki523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnakulasuriyarachchi, D., Cerquozzi, S., Cheung, H.H., Holcik, M. Translational induction of the inhibitor of apoptosis protein HIAP2 during endoplasmic reticulum stress attenuates cell death and is mediated via an inducible internal ribosome entry site element. J. Biol. Chem. 2004;279:17148–17157. doi: 10.1074/jbc.M308737200. [DOI] [PubMed] [Google Scholar]
- Watada, H., Mirmira, R.G., Leung, J., German, M.S. Transcriptional and translational regulation of β-cell differentiation factor Nkx6.1. J. Biol. Chem. 2000;275:34224–34230. doi: 10.1074/jbc.M004981200. [DOI] [PubMed] [Google Scholar]
- Wilson, J.E., Pestova, T.V., Hellen, C.U., Sarnow, P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000a;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- Wilson, J.E., Powell, M.J., Hoover, S.E., Sarnow, P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell. Biol. 2000b;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese, C.R. Translation: In retrospect and prospect. RNA. 2001;7:1055–1067. doi: 10.1017/s1355838201010615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollerton, M.C., Gooding, C., Robinson, F., Brown, E.C., Jackson, R.J., Smith, C.W. Differential alternative splicing activity of isoforms of polypyrimidine tract binding protein (PTB) RNA. 2001;7:819–832. doi: 10.1017/s1355838201010214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman, C., Krogh, A. No evidence that mRNAs have lower folding free energies than random sequences with the same dinucleotide distribution. Nucleic Acids Res. 1999;27:4816–4822. doi: 10.1093/nar/27.24.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Z.S., Simpson, L.G., Quarles, L.D. IRES-dependent translational control of Cbfa1/Runx2 expression. J. Cell. Biochem. 2003;88:493–505. doi: 10.1002/jcb.10375. [DOI] [PubMed] [Google Scholar]
- Yaman, I., Fernandez, J., Liu, H., Caprara, M., Komar, A.A., Koromilas, A.E., Zhou, L., Snider, M.D., Scheuner, D., Kaufman, R.J., et al. The zipper model of translational control: A small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell. 2003;113:519–531. doi: 10.1016/s0092-8674(03)00345-3. [DOI] [PubMed] [Google Scholar]
- Yang, D., Wilson, J.E., Anderson, D.R., Bohunek, L., Cordeiro, C., Kandolf, R., McManus, B.M. In vitro mutational and inhibitory analysis of the cis-acting translational elements within the 5′ untranslated region of coxsackievirus B3: Potential targets for antiviral action of antisense oligomers. Virology. 1997;228:63–73. doi: 10.1006/viro.1996.8366. [DOI] [PubMed] [Google Scholar]
- Yang, H., Jossinet, F., Leontis, N., Chen, L., Westbrook, J., Berman, H., Westhof, E. Tools for the automatic identification and classification of RNA base pairs. Nucleic Acids Res. 2003;31:3450–3460. doi: 10.1093/nar/gkg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yueh, A., Schneider, R.J. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes & Dev. 2000;14:414–421. [PMC free article] [PubMed] [Google Scholar]
- Zaug, A.J., Cech, T.R. Analysis of the structure of Tetrahymena nuclear RNAs in vivo: Telomerase RNA, the self-splicing rRNA intron, and U2 snRNA. RNA. 1995;1:363–374. [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Richie, C., Legerski, R.J. Translation of hSNM1 is mediated by an internal ribosome entry site that upregulates expression during mitosis. DNA Repair (Amst.) 2002;1:379–390. [PubMed] [Google Scholar]
- Zhao, W.D., Wimmer, E. Genetic analysis of a poliovirus/hepatitis C virus chimera: New structure for domain II of the internal ribosomal entry site of hepatitis C virus. J. Virol. 2001;75:3719–3730. doi: 10.1128/JVI.75.8.3719-3730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, W., Edelman, G.M., Mauro, V.P. Transcript leader regions of two Saccharomyces cerevisiae mRNAs contain internal ribosome entry sites that function in living cells. Proc. Natl. Acad. Sci. 2001;98:1531–1536. doi: 10.1073/pnas.98.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, W., Edelman, G.M., Mauro, V.P. Isolation and identification of short nucleotide sequences that affect translation initiation in Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. 2003;100:4457–4462. doi: 10.1073/pnas.0437993100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong, Q., Schummer, M., Hood, L., Morris, D.R. Messenger RNA translation state: The second dimension of high-throughput expression screening. Proc. Natl. Acad. Sci. 1999;96:10632–10636. doi: 10.1073/pnas.96.19.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. Calculating nucleic acid secondary structure. Curr. Opin. Struct. Biol. 2000;10:303–310. doi: 10.1016/s0959-440x(00)00088-9. [DOI] [PubMed] [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M., Stiegler, P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuleeg, T., Hansen, A., Pfeiffer, T., Schubel, H., Kreutzer, R., Hartmann, R.K., Limmer, S. Correlation between processing efficiency for ribonuclease P minimal substrates and conformation of the nucleotide −1 at the cleavage position. Biochemistry. 2001;40:3363–3369. doi: 10.1021/bi0016974. [DOI] [PubMed] [Google Scholar]