Abstract
The 3′-untranslated regions (UTRs) of a group of novel uncapped viral RNAs allow efficient translation initiation at the 5′-proximal AUG. A well-characterized model is the Barley yellow dwarf virus class of cap-independent translation elements (BTE). It facilitates translation by forming kissing stem–loops between the BTE in the 3′-UTR and a BTE-complementary loop in the 5′-UTR. Here we investigate the mechanisms of the long-distance interaction and ribosome entry on the RNA. Upstream AUGs or 5′-extensions of the 5′-UTR inhibit translation, indicating that, unlike internal ribosome entry sites in many viral RNAs, the BTE relies on 5′-end-dependent ribosome scanning. Cap-independent translation occurs when the kissing sites are moved to different regions in either UTR, including outside of the BTE. The BTE can even confer cap-independent translation when fused to the 3′-UTR of a reporter RNA harboring dengue virus sequences that cause base-pairing between the 3′- and 5′-ends. Thus, the BTE serves as a functional sensor to detect sequences capable of long-distance base-pairing. We propose that the kissing interaction is repeatedly disrupted by the scanning ribosome and re-formed in an oscillating process that regulates ribosome entry on the RNA.
Keywords: long-distance RNA interactions, translation initiation, kissing stem–loop; Luteovirus, dengue virus
INTRODUCTION
Initiation of translation for cellular mRNAs can be broken down into three steps (Kozak 2002; Kapp and Lorsch 2004): The 40S ribosomal subunit with the initiation factor complex enters the capped mRNA at the 5′-terminus (step 1) and scans the 5′-untranslated region (UTR) to reach the 5′-proximal AUG (step 2), at which the 60S subunit joins (step 3) and protein synthesis ensues. The recognition of the 5′-cap by the cap-binding protein component (eIF4E) of initiation complex eIF4F enhances ribosome recruitment to the mRNA (von der Haar et al. 2004). The other component of eIF4F, initiation factor 4G (eIF4G), orchestrates the assembly of the scanning machinery (Kapp and Lorsch 2004). eIF4F recruits the RNA helicase eIF4A (Oberer et al. 2005) (not a subunit of eIF4F in plants), which unwinds the 5′-region to favor the attachment and subsequent scanning of the eIF3-bound 40S-ribosomal subunit onto the unstructured RNA (Pestova and Kolupaeva 2002; Siridechadilok et al. 2005; Jivotovskaya et al. 2006).
Many viral RNAs have structures within their 5′- or 3′-UTRs that recruit the host translation machinery in the absence of a 5′-cap and/or a poly(A) tail (Dreher and Miller 2006; Kneller et al. 2006). With the exception of certain plant viral RNAs, cap-independent translation is facilitated by a highly structured internal ribosome entry site (IRES), which varies greatly in size, structure, and mechanism depending on the virus (Hellen and Sarnow 2001). IRESs recruit the 40S ribosomal subunit to an internal region of the RNA in close proximity to the initiation codon, without ribosomal scanning from the 5′-end (Sarnow et al. 2005).
Here we report on a different type of cap-independent translation element discovered in certain uncapped nonpolyadenylated plant viral and satellite RNAs including the Luteoviruses (Wang et al. 1997) and members of the Tombusviridae family (Timmer et al. 1993; Wu and White 1999; Mizumoto et al. 2003; Meulewaeter et al. 2004; Shen and Miller 2004; Batten et al. 2006; Scheets and Redinbaugh 2006). Unlike IRES-mediated translation, the cap-independent translation element is present in the 3′-untranslated region, yet translation initiation occurs at the AUG closest to the 5′-end of the mRNA (Kneller et al. 2006). Through a long-distance kissing stem–loop interaction between the 3′-cap-independent translation element and the 5′-UTR, the 3′-element mediates translation initiation at the 5′-end of the mRNA. Presumably, the long-distance base-pairing facilitates the delivery of the ribosomes and/or initiation factors to the 5′-proximal AUG (Guo et al. 2001; Fabian and White 2006).
At least three different classes of 3′-cap-independent translation element, which show no apparent similarity in sequence or structure to each other, have been identified (for review, see Miller and White 2006). Typical members of each class include Satellite tobacco necrosis virus (STNV, genus Necrovirus) (Timmer et al. 1993), Barley yellow dwarf virus (BYDV, genus Luteovirus) (Wang et al. 1997), and Tomato bushy stunt virus (TBSV, genus Tombusvirus) (Wu and White 1999). Here, we focus on the BYDV (-like) cap-independent translation element (BTE) of BYDV and related viruses.
The genome of BYDV is an uncapped, nonpolyadenylated positive-sense RNA of 5677 nt that encodes six open reading frames (ORFs) (Fig. 1A). The minimal in vitro functional BYDV translation element (BTE) spans nucleotides 4809–4918 and forms a roughly cruciform secondary structure with three major stem–loops (SL-I, SL-II, SL-III) flanked by stem IV (Fig. 1B). Additional sequences in the 3′-UTR, including a domain that functionally replaces a poly(A) tail, are required for full cap-independent and poly(A)-tail-independent translation in plant cells (Wang et al. 1997; Guo et al. 2001).
FIGURE 1.
(A) Genome organization of BYDV RNA. Open reading frames are numbered. Dashed arrows indicate 5′-ends of subgenomic RNAs (sg1, sg2, sg3). Shown are the positions of stem–loops A, B, C, and D within the 5′-UTR of the genomic RNA; the 5′-proximal stem–loop in sgRNA1 (SL) with potential to base-pair to the BTE; and the position of the BTE. (B) Secondary structures of 5′-UTR stem–loop SL-D and the sgRNA1 5′-stem–loop (in shaded box), which both contain BTE-complementary loops (BCL), and the 3′-BTE. The arrow indicates the GAUC insertion in the BamHI4837 site (BTEBF) that inactivates cap-independent translation activity of the BTE. The italic bases in the BTE are highly conserved in all BTEs. Bases in bold participate in the long-distance kissing interactions (dashed lines).
The 3′-BTE facilitates cap-independent translation initiation at the 5′-proximal AUG via base-pairing of the loop of stem–loop III (SL-III) of the BTE to a complementary loop, SL-D (that we refer to as a BTE-complementary loop, or BCL) located within the 5′-UTR of the viral genomic RNA. A single point mutation within the middle of the five bases of either of these kissing loops disrupts base–pairing and abolishes translation both in cells and in wheat germ extract, while compensatory double mutations that restore base-pairing restore translation (Guo et al. 2001). The BTE also potentially base-pairs to a BCL in the 5′-UTR of BYDV subgenomic RNA1, which serves as the mRNA for the coat protein and other ORFs downstream of ORFs 1 and 2 (Fig. 1B). However, the BCL in the 5′-UTR of sgRNA1 does not facilitate translation in the context of full-length genomic RNA, because no ORFs downstream of the sgRNA1 BCL are translated from genomic RNA (Allen et al. 1999).
Still undetermined are (1) the constraints in sequences, structures, and position of the long-distance kissing-loop interaction to support cap-independent translation; (2) how the ribosomes enter the mRNA; and (3) whether the long-distance kissing loop in the 3′-BTE must be located within the context of the BTE or can be separated from the intrinsic cap-independent translation stimulation activity of the 3′-element.
Here we show that (1) the possible sequences of the long-distance kissing loops that support cap-independent translation are limited, even if complementarity is maintained; (2) ribosomes must enter from the 5′-terminus of the RNA via the BCL–BTE kissing interaction and scan to the 5′-proximal AUG just as on a cellular capped RNA; and (3) the long-distance base-pairing structures can be uncoupled from the BTE structure and even replaced entirely by complementary sequences from an unrelated viral RNA in vitro. Thus, the 3′-BTE can be used as a functional sensor of long-distance interactions of any RNA of interest, even if the RNA is not normally involved in translation. Because the ribosome must scan through the BCL to reach the start codon, the RNA structure must oscillate between kissing stem–loops and disrupted kissing. We propose that this regulates entry of the translation initiation machinery onto the viral RNA.
RESULTS
Sequence specificity required for the BYDV end-to-end interaction
Among different viruses, the kissing loops in the 3′- and 5′-UTRs, which are involved in long-distance interaction, vary in sequence and length with no obvious consensus (Edgil and Harris 2006; Miller and White 2006). The BYDV kissing interaction, required for cap-independent translation, involves a five-base sequence that tolerated an exchange of the middle base of each loop to maintain complementarity (Guo et al. 2001). The five bases involved in base-pairing in each loop are identical, except for the middle base (UGACA:UGUCA). Thus, primary sequence as well as complementarity may be important for this long-distance interaction. This led us to ask whether more sequence variation in the kissing loop could be tolerated. We replaced the natural 3′-BTE loop III and the BTE-complementary loop (BCL) in the 5′-UTR with complementary sequences containing three base changes, UCUGA:UCAGA (altered bases in bold), or 10-base complementary tracts, UCAAUAUGCC:GGCAUAUUGA (Fig. 2A). The 10-base tracts, called cyclization sequences (CS), are located naturally at opposite ends of the 11-kb dengue virus (DEN) genome and are known to base-pair (Khromykh et al. 2001). The mutations were tested in the context of a reporter construct (LUC869) that was used previously to demonstrate the long-distance interaction (Fig. 2; Guo et al. 2001). In LUC869 firefly luciferase, the open reading frame is flanked by the BYDV genomic 5′- and 3′-UTRs (nucleotides 1–142, 4809–5677). The expression efficiency of each mutant was measured both in oat protoplasts (in vivo) and in wheat germ extract (in vitro). Despite retaining complementarity, neither set of altered kissing loop sequences allowed translation in vivo or in vitro (Fig. 2B). In both cases, translation was similar to the level obtained with a four-base insertion in the BamHI site of the BTE that we showed previously renders the BTE completely nonfunctional (BTEBF) (Wang et al. 1997). The BTEBF mutation serves as a negative control throughout this report.
FIGURE 2.
Effect of covarying mutations in the kissing loop sequence on BTE-mediated translation. (A) Mutants of the LUC869 luciferase reporter construct that consists of the BYDV genomic RNA 5′- and 3′-UTRs flanking the firefly luciferase ORF. Mutations within the kissing bases (boxed) of 5′-BCL (LD-3, LD-CS) and the BTE LIII (LIII-3, LIII-CS) are indicated. (CS) Cyclization sequences from dengue virus RNA. Mutated bases are shown in bold. (B) Relative luciferase activity in oat protoplasts (in vivo) and in wheat germ extract (in vitro) produced by uncapped mRNAs containing the indicated mutations. (C) Relative luciferase activity produced in wheat germ extract by the mRNAs containing the mutant BTE-SLIII in the 5′-UTR of the luciferase reporter (5′-BTE-LUC) with 15 nt of nonviral sequence derived from the pGEM3Zf(+) plasmid (Promega), as the 3′-UTR.
The 3′-BTE must perform at least two functions: mediate the recruitment of the translational machinery, and communicate with the 5′-end where translation initiates. The need for the latter function can be eliminated by placing the BTE in the 5′-UTR, from which it can mediate cap-independent translation in vitro (Wang et al. 1997; Guo et al. 2000). We refer to any BTE mutant capable of mediating cap-independent translation from the 5′-UTR as having the core cap-independent translation activity. These mutants may or may not have the long-distance base-pairing capability. To determine whether the above alternative sequences of the kissing interaction disrupted the core cap-independent translation activity of the BTE, the BTE mutants (LIII-3; LIII-CS) were tested in the 5′-UTR context of the luciferase reporter gene with nonviral sequences in the 3′-UTR (5′-BTE-LUC) (Fig. 2C). Translation of these uncapped BTE-LUC RNAs was at least as efficient as the “wild-type” 5′-BTE-LUC RNA (Fig. 2C), indicating that the mutations within loop III of the BTE did not interfere with the cap-independent translation activity of the BTE. Thus, the LIII-3 and LIII-CS mutants lost the ability to mediate translation from the 3′-UTR due to an inability to interact productively with the 5′-UTR or because the compensating mutants in the 5′-BCL (absent in the constructs with the BTE in the 5′-UTR) disrupted translation.
Structural flexibility in the 5′-UTR but not in the BTE
Previously, we showed that the stem portions of the kissing stem–loops can be exchanged between the SL-III and the 5′-BCL, SL-D (Guo et al. 2001). Here, we test whether the stem is necessary at all. Mutations were introduced to disrupt the base-pairing in the stem portion of the BTE SL-III (SIII-m1, SIII-m2) (Fig. 3A). Although the BTE loop III bases were predicted to remain single-stranded and available for base-pairing to the 5′-end, no translation activity above background level (obtained with the BTEBF construct) was observed in cells or in wheat germ extract (Fig. 3B). The BTE mutants also failed to promote cap-independent translation above background level when placed in the 5′-UTR context of the luciferase gene (Fig. 3C), revealing that disruption of the stem III structure abolished the core cap-independent translation function of the BTE, even in the 5′-UTR context that lacks a requirement for the long-distance kissing interaction. The double compensatory mutation, predicted to restore the BTE stem III structure (SIII-r), rescued the translation of the LUC869 RNA fully in vitro and to one-third of the wild-type level in vivo (Fig. 3B). These levels of translation were about fivefold and 200-fold, respectively, greater than the expression obtained for the mutants with the disrupted stem III. As expected, double mutant SIII-r facilitated full translation in the 5′-BTE-LUC context (SIII-r) (Fig. 3C). Thus, the stem portion of SL-III tolerates changes in the primary sequence as long as the stem remains intact. Disruption of the stem causes structural changes sufficient to abolish cap-independent translation in any context.
FIGURE 3.
Effect of disruption and restoration of the stem portions of the kissing stem–loops on the BTE-mediated translation. (A) LUC869 luciferase reporter constructs with the mutations within the stem portion of the BTE stem–loop III. The mutated bases are shown in bold with the effect on base-pairing indicated in the boxed regions. (B) Relative luciferase activity measured in vivo (gray bars) and in vitro (white bars) from the LUC869 RNAs containing the mutations in the stem of the BTE SL-III in the natural 3′-UTR context. (C) Relative luciferase activity in vitro measured from the mRNAs containing the mutant BTE stem III in the 5′-UTR context of the 5′-BTE-LUC luciferase reporter construct. (D) LUC869 construct with mutations within the stem portion of the 5′-SL-D structure. Mutated bases are shown in bold, with the effect on base-pairing indicated in the boxed regions. Relative luciferase activity of LUC869 constructs in vivo with the mutations within the stem portion of the 5′-BCL.
Unlike SL-III in the BTE, disruption of the stem flanking the BCL in the 5′-UTR (SD-m1, -m2) (Fig. 3D) still allowed significant translation of the uncapped reporter transcript in vivo (Fig. 3E), while the double compensatory mutant that restored the stem-D (SD-r) translated as efficiently as wild-type LUC869 RNA. Thus, regardless of the flanking structure, the BCL bases are sufficient to support the long-distance RNA–RNA interaction with the 3′-BTE.
Ribosome scanning from the 5′-end is required for cap-independent translation
Previously we reported that addition of a highly stable stem–loop at the extreme 5′-end of the gRNA abolished both BTE- and cap-mediated translation (Guo et al. 2001), presumably by blocking access of the 5′-end for ribosome binding (Kozak 1989). Such a major change to the 5′-UTR could prevent BTE–BCL interactions or affect translation in unpredicted ways. Thus, we further examined the possibility of 5′-end-dependency of the BTE-mediated cap-independent translation with different alterations of the 5′-UTR. An out-of-frame AUG (uAUG) was inserted just upstream of the 5′-BCL in the LUC869 reporter construct context, creating a small upstream ORF (uORF) of 29 codons (Fig. 4A). If scanning occurred, the ribosomes would encounter the uORF before reaching the start codon of the luciferase gene, which would greatly reduce initiation at the LUC AUG.
FIGURE 4.
Effect of upstream AUG (uAUG) on BTE-mediated translation. (A) Out-of-frame AUG (uAUG) was inserted upstream of the 5′-BCL. The arrows indicate the position of the uAUG and LUC AUG within nucleotides 74–177 of BYDV 5′-UTR. The underlined bases indicate the complementary bases forming the stem portion of the 5′-BCL, SL-D. The kissing bases are in the open box. The gray shading shows the sequence context of the upstream AUGs and the start codon of the luciferase gene. The uAUG construct has the same sequence context as the LUC AUG. In the Kozak uAUG construct, the uAUG is in the optimal context for initiation. The presence of the uAUG creates a small ORF (uORF) of 29 codons, which overlaps the 5′-end of the firefly luciferase coding region (fLUC). The amino acid residues encoded by the uORF and the 5′-end of the luciferase ORF are shown. (B) Relative luciferase activity of translation products of mRNAs containing the uAUG. (C) 35S-methionine-labeled in vitro translation products of indicated transcripts. Due to the presence of only one methionine residue in the uORF, the bottom panel was exposed at higher sensitivity using ImageQuant software to allow visualization of the uORF-encoded protein.
Luciferase expression in vivo was compared to that of the wild-type RNA and a capped BTEBF RNA construct, in which translation is cap-mediated and, thus, scanning-dependent (Fig. 4B). The presence of the uAUG reduced cap- and BTE-mediated translation of luciferase to 13% and 27%, respectively, of that obtained from the wild-type LUC869 RNA (capped uAUG-BTEBF, uAUG). The reduction of the luciferase expression correlated with the accumulation of a 2.9-kDa peptide encoded by the uORF, except for the uAUG-BTEBF construct in which no translation above background of either ORF was expected (Fig. 4C). In this construct, both the uAUG and the LUC AUG had the same sequence context (GCGCAUGG). We next placed the uAUG in the optimal Kozak context for efficient initiation (GCCACCAUGG) (Kozak 1991). This construct abolished luciferase expression (Kozak uAUG) (Fig. 4B). These data support the notion that cap-independent translation mediated by the 3′-BTE relies on a ribosome scanning mechanism that obeys conventional Kozak rules, starting from a region upstream of the 5′-kissing site (BCL), most likely the 5′-end itself (Guo et al. 2001), just as on a cellular capped mRNA.
Increasing distance of the BTE-complementary loop from the 5′-end reduces translation efficiency
In BYDV gRNA, the BCL is located at base positions 103–107 of the 5′-UTR and is the fourth stem–loop from the 5′-end (Guo et al. 2000). In contrast, in all other 3′-TE-bearing RNAs including BYDV sgRNA1 and other luteoviruses, the BCL is located within 10–30 nt of the 5′-UTR in the 5′-proximal stem–loop (Fig. 5A). This raises the question of the importance of the position of the BCL relative to the 5′-end for the efficiency of the BTE-mediated cap-independent translation. We propose that the 3′–5′ interaction is necessary for the delivery of either the ribosome or the initiation factors to the 5′-end of the RNA, from where the ribosome enters and scans to the first AUG via conventional mechanisms (above). This model predicts that (1) if the BCL is farther away from the 5′-end, the BTE-bound ribosome and/or initiation factors may be less likely to find the 5′-terminus of the mRNA, and translation efficiency would be reduced; and (2) if the BCL is brought closer to the 5′-end, the delivery of the translation complex may be more efficient.
FIGURE 5.
(Legend on next page).
To test this hypothesis, we first added an extra sequence to the 5′-UTR of the LUC869 reporter construct. To rule out potential deleterious effects of structure in the added sequence on ribosome scanning, we used a fragment of a sequence with little secondary structure that is known not to inhibit translation (Fig. 5B). We used portions of the Tobacco mosaic virus (TMV) Ω sequence, a translation enhancer that allows significant translation of uncapped mRNA but is strongly stimulated by the presence of a 5′-cap (Gallie et al. 1987). Moreover, Ω sequence has no stimulatory effect on BTE-mediated cap-independent translation (Wang et al. 1997). When the 5′-BCL was positioned 30–63 bases farther downstream of the 5′-end, by inserting portions of the Ω sequence at the extreme 5′-end of the 5′-UTR, cap-independent translation in vivo dropped by 56% and 77%, respectively (+5′-30 nt, +5′-63 nt) (Fig. 5B). This reduction in expression correlated roughly with the distance of the BCL from the 5′-end. To determine whether the insertions could affect the stability of the mRNA, the functional half-life of the mutant RNA with the extended 5′-UTR (+5′-63 nt) was estimated by monitoring the rate of luciferase accumulation in cells (Fig. 5C; Meulewaeter et al. 1998b). While the half-life of the mutant was reduced, this did not account for the fourfold drop in luciferase activity relative to the reporter with the wild-type 5′-UTR. Thus, the 63-nt extension of the 5′-UTR significantly reduced translation efficiency.
When repeats of the same sequence were inserted at the 3′-end of the 5′-UTR, downstream of the BCL, RNA translated similarly to the wild-type construct (+3′-63 nt; +3′-200 nt) (Fig. 5B). These insertions positioned the BCL ∼100–230 bases upstream of the AUG, instead of the natural 30 bases upstream, while the BCL remained at the wild-type distance from the 5′-end of the RNA. Thus, as expected, the sequences of the Ω fragments were not inhibitory to translation. Moreover, they did not provide any cap-independent translation activity, as no translation was observed in the context of the uncapped BTEBF mutant RNAs bearing the 3′-extension of the 5′-UTR (+3′-63 nt BTEBF; +3′-200 nt BTEBF).
All of the above data support a model in which a longer distance between the BCL and the 5′-end reduces the likelihood of the BCL–BTE kissing to deliver components required for cap-independent translation in the competitive environment of the cell. Furthermore, the data show that a longer 5′-UTR per se does not affect translatability, because insertion of extra sequence between the BCL and the start codon did not interfere with cap-independent translation.
We next tested the effect of relocating the BCL to a site closer to the 5′-end of the gRNA, similar to the positions of the BCLs of other 3′-TE-bearing RNAs (Fig. 5A). The loop sequence of the 5′-proximal stem–loop structure (SL-A) at bases 12–16 in the gRNA 5′-UTR was converted from UGACC to a BCL by changing one base (C16 to A; C16A) (Fig. 5D). First, we observed that the presence of this additional BCL in the 5′-UTR of the genomic reporter construct allowed translation in vivo (construct 2-BCL) (Fig. 5D). By mutating the SL-D loop (GAC104–106 to CUG) and thereby preventing its interaction with the BTE, the 3′-BTE was able to promote translation, presumably via the surrogate kissing interaction with the artificial BCL (construct A-BCL). The translation efficiency of the artificial BCL reporter constructs was somewhat lower than the wild-type level. In contrast, when the ability of the 5′-SL-D loop to base-pair to the BTE was disrupted by an A105-to-U mutation that alone abolished translation (construct no-BCL) (Fig. 5D; Guo et al. 2001), the alternative SL-A C16A–BTE interaction was unable to rescue the translation of the reporter RNA (construct A*-BCL) (Fig. 5D). Presumably, due to sequence complementarity between the SL-A C16A (UGACA) and SL-D A105U (UGUCA), nonproductive base-pairing between these two nearby stem–loops out-competed the long-distance interaction between the loop III of the BTE, which has the same sequence as the SL-D A105U, and SL-A C16A.
Taken together, these results support our hypothesis that an essential portion of the translation machinery is recruited to the 5′-end via a 5′-BCL–3′-BTE base-pairing and that the translation complex is transferred via the 3′-BTE to the 5′-terminus of the RNA. The translational machinery can find the 5′-end with similar efficiency when the BCL is located within a range of 10 nt to ∼110 nt from the 5′-end of the mRNA. The lack of translation by the A*-BCL construct supports the notion that A105U acts as a competitive decoy by base-pairing to the complementary sequence in SL-A C16A.
An alternative long-distance kissing interaction supports BYDV RNA replication
To determine the biological relevance of moving the BCL to SL-A, this time in the natural viral context, the SL-A C16A and SL-D GAC104–106 to CUG mutations (vA-BCL) and others were introduced into the full-length BYDV viral RNA, and the effects of these changes on RNA translatability and viral replication were analyzed (Fig. 6). The translation efficiency of the mutant RNA in vitro was determined by monitoring the incorporation of 35S-labeled methionine into the translation products of ORF 1 (39 kDa) and ORFs 12 + 22 (99 kDa), the latter resulting from −1 ribosomal frameshifting (Fig. 6A). As expected, the GAC-to-CUG mutation in SL-D that disrupted the 3′–5′-end interaction abolished gRNA translation (vno-BCL). In contrast, the viral transcript bearing two BTE-complementary loops (v2-BCL) and the transcript with the surrogate kissing interaction (vA-BCL) translated at near-wild-type levels (Fig. 6A). Thus, the effect of the mutations on translation of the full-length viral RNA correlated with their effects observed in the reporter gene translation (above).
FIGURE 6.
Effect of the BCL mutations on the full-length infectious BYDV transcripts. (A) (Top) BYDV genomic RNA map showing locations of mutations in SL-A and/or SL-D. Full-length genomic transcripts are named as in the reporter constructs (Fig. 5) preceded by a “v.” (Bottom) 35S-methionine-labeled in vitro translation products of indicated full-length viral transcripts. The translation products of the full-length RNA are ORF 1 (39 kDa) and ORF 12 + 22 (99 kDa) resulting from −1 ribosomal frameshifting. (B) Northern blot analysis revealing the accumulation of wild-type and mutant viral RNAs in oat protoplasts 48 h post-inoculation. Mobility of sgRNAs accumulated during replication is indicated where visible.
The effect of mutations in the BCL on viral replication was assessed by observing accumulation of viral RNAs in transfected protoplasts 48 h after inoculation (Fig. 6B). As expected, no viral replication was observed in cells transfected with the translationally incompetent RNA (vno-BCL). The construct in which both SL-A and SL-D serve as BCLs replicated (v2-BCL). Most importantly, the vA-BCL RNA, in which the role of BCL was moved to SL-A, replicated to levels close to that of wild-type viral RNA (Fig. 6B). These results reveal that (1) the BTE–SL-A interaction sustains cap-independent translation in the context of replicating viral RNA in vivo, and (2) either the SL-A loop plays no role in viral RNA replication, or viral RNA replication tolerates the C-to-A point mutation in SL-A and the unnatural interaction of the BTE with SL-A.
Sequences outside of the BTE can mediate the long-distance interaction to facilitate translation
We wondered whether the 3′–5′-end interaction can be maintained by sequences outside of the BTE with the BTE still retaining its core activity as a recruiter of key translation components. To test this, a stem–loop downstream of the BTE, called the long-distance frameshift element (LDFE), was made complementary to the 5′-BCL by changing the LDFE loop from UCUGU to the BTE loop III sequence UGUCA (cLDFE) (Fig. 7A). The LDFE normally base-pairs to a sequence near the frameshift site 4 kb upstream (Barry and Miller 2002). The LDFE is located at bases 5057–5075 in the BYDV genome and is not required for cap-independent translation (Paul et al. 2001). In the luciferase reporter construct, no sequence in the wild-type BYDV UTRs or LUC ORF is predicted to base-pair to the LDFE, indicating that the LDFE is unlikely to pair to other sites in the wild-type reporter. The BTE loop III was altered (LIII-3) to disrupt base-pairing to the 5′-BCL without affecting the core cap-independent translation activity of the BTE (Fig. 2C). Despite the sequence complementarity of the cLDFE loop with SL-D, the LIII-3 + 2cLDFE construct failed to support cap-independent translation (Fig. 7A). To determine whether the location of the cLDFE rendered the loop inaccessible to the 5′-BCL, the cLDFE stem–loop was relocated upstream of the BTE at base position 4809 (uLDFE) (Fig. 7A). Translation was still not recovered, suggesting that either the BTE itself must mediate the long-distance interaction and/or in both constructs, the mutant RNAs fold in unpredicted ways that disrupt the BTE structure and/or cLDFE–BCL base-pairing.
FIGURE 7.
Effect of moving the 5′-UTR-complementary stem–loop outside of the BTE. (A) The long-distance frameshift element (LDFE) loop, located downstream from the BTE, was made complementary to the 5′-BCL (cLDFE). The BTE was altered (LIII-3) to prevent base-pairing to it by the 5′-BCL. The mutated bases are indicated in bold in the boxed regions. The cLDFE was also moved upstream of the BTE at nucleotide 4809 (uLDFE). The solid and the dashed arrows show the alternative and the natural long-distance interactions, respectively. The table shows relative luciferase activity in vivo measured for the RNAs containing the indicated mutations. The loops that are predicted to base-pair to the 5′-BCL are indicated in italics. (B) Relative luciferase activity in vivo of the mRNA with altered SL-D loop. The long-distance interaction is mediated by the BCL mutated to be complementary to the LDFE (cSLD). The mutated bases are shown in bold in boxed regions. In the table, the loops that are predicted to mediate the long-distance interaction of the RNA are shown in italics.
To answer this question, we investigated an alternative interaction in which the 3′-UTR remained unaltered. The 5′-BCL was made complementary to the loop portion of the wild-type LDFE and rendered unable to base-pair to the wild-type BTE loop III, by changing the BCL sequence from UGACA to ACAGA (cSL-D) (Fig. 7B). Indeed, this construct promoted cap-independent translation of the mutant LUC869 RNA in protoplasts to about one-third of the wild-type translation efficiency level (cSL-D) (Fig. 7B). This was a 28-fold increase over the expression of the single mutation in which the BCL could base-pair to neither the BTE nor the LDFE loop (A105U) (Fig. 7B). Thus, the long-distance kissing interaction can be mediated by sequence outside of the BTE as long as the 3′-UTR or (more likely) the BTE structure remains unaltered. This suggests some additional constraints in sequence and/or structure of the core-cap-independent translation function of the BTE in the natural 3′-UTR context.
The 3′-BTE promotes cap-independent translation of an mRNA containing dengue virus UTRs
Next, we set out to investigate the ability of the BTE to mediate cap-independent translation from the 3′-end of a normally capped mRNA that harbors complementary sequences in each end that are known to base-pair. We used a luciferase reporter RNA representing the dengue virus 2 genomic RNA (DEN) (Fig. 8). The luciferase ORF is flanked by the DEN 5′-UTR followed by the first 72 bases of the viral capsid protein-coding region (nucleotides 1–168), and by the DEN 3′-UTR (nucleotides 10269–10723) (Holden and Harris 2004). The naturally capped DEN RNA contains conserved complementary 10-base tracts (UCAAUAUGCC:GGCAUAUUGA) within the UTRs, constituting the cyclization sequences (CS). CS base-pairing is necessary for replication (You and Padmanabhan 1999; Alvarez et al. 2005b) but not for translation of DEN RNA (Alvarez et al. 2005a; Chiu et al. 2005; Edgil and Harris 2006). We fused the BTE, along with flanking bases from the BYDV genome (nucleotides 4809–5045), to the extreme 3′-end of the DEN 3′-UTR on this reporter construct (Fig. 8A). We predicted that, via the 5′–3′ CS interaction, the 3′-BTE would be brought into close proximity to the 5′-end of the RNA to mediate cap-independent translation, by the same mechanism as for BYDV RNA translation.
FIGURE 8.
The 3′-BTE mediates cap-independent translation of a reporter with dengue viral UTRs. (A) In the dengue viral reporter construct, the luciferase reporter gene is flanked by the dengue 5′-UTR and the first 72 nt of the 5′-coding region of the capsid protein, and by the dengue 3′-UTR. The cyclization sequences (CS) and the mutated versions (mut 3′CS) are shown in gray. The 3′-BTE with flanking bases from the BYDV genome (nucleotides 4809–5045) was inserted at the 3′-end of the dengue viral reporter construct. (B) Relative luciferase activity in wheat germ extract of the dengue viral reporter mRNAs containing a 5′-cap and/or the 3′-BTE where indicated. The DEN-BTEBF is the same as DEN-3′BTE construct except that it harbors the BamHI-fill-in mutation (BTEBF). DEN-LIII-3 bears the three-base change within loop III of the BTE as shown in Figure 2.
The translation efficiency of the chimeric DEN-BTE RNA was measured in wheat germ extract and compared to that of capped and uncapped DEN RNAs (Fig. 8B). Uncapped DEN-3′BTE RNA translated as efficiently as capped DEN RNA lacking the 3′-BTE. In contrast, uncapped DEN RNA and the negative control, DEN-3′BTEBF RNA, translated about one-fourth as efficiently as did DEN RNAs with a cap or a BTE. This apparently high background level of cap and BTE-independent translation in wheat germ extract is because capped DEN RNA, defined as 100% here, translates only about one-third as efficiently as LUC869 RNA (data not shown). Therefore, the background translation level of uncapped DEN-3′BTEBF RNA is about one-twelfth that of LUC869 RNA. It is noteworthy that the BTE bearing the three base changes on its kissing loop (LIII-3) (see Fig. 2) facilitated full cap-independent translation of the DEN reporter RNA (DEN-LIII-3), supporting the notion that loop III of the BTE did not interfere with the core cap-independent translation activity of the BTE and that wild-type loop III did not base-pair fortuitously to a sequence in the 5′-UTR of the DEN reporter.
To determine whether the 3′-BTE-mediated translation required CS base-pairing as predicted, the potential base-pairing of the CS elements was disrupted by mutating each CS (mut 5′CS, CUCUCUCUG; mut 3′CS, CAGAGAGAG). Alteration of either CS sequence reduced cap-independent translation significantly (mut 5′CS-DEN-3′BTE, DEN-3′BTE-mut 3′CS) but had no effect on cap-mediated translation (capped DEN-mut 3′CS). The translation of the DEN-3′BTE-mut 3′CS RNA could be rescued either by the presence of a 5′-cap (capped DEN-3′BTE-mut 3′CS) or, most importantly, by combining the two CS mutants to restore base-pairing (mut 5′CS-DEN-3′BTE-mut 3′CS). In summary, the BTE, incapable of base-pairing to the 5′-UTR, facilitates cap-independent translation of DEN reporter RNA from a 3′-position as long as the CS sequences, or their mutated version, were able to base-pair to each other.
DISCUSSION
The results presented here support our previous observation that translation activation by a 3′-cap-independent translation element involves at least two major steps (Guo et al. 2001). One is recruitment of translation initiation machinery onto the uncapped RNA, which may itself involve multiple steps, such as binding of translation factors to the 3′-element (Wang et al. 1997; Gazo et al. 2004). Second is the long-distance interaction between the 3′-element and the 5′-UTR, to facilitate the delivery of the translation machinery to the 5′-proximal AUG. Here we find that the translation machinery recruitment and long-distance kissing interaction are two independent events that together regulate translation via 5′-end-dependent ribosome scanning.
Structural constraints and mechanism of the long-distance kissing interaction
The primary sequence of the kissing loops appears in part to determine the efficiency of the long-distance interaction. Some complementary sequences, UCUGA:UCAGA and the dengue virus cyclization sequences UCAAUAUGCC:GGCAUAUUGA, within the kissing loops were unable to support 3′-BTE-mediated cap-independent translation, even though MFOLD (Zuker 2003) predicted that these kissing loops should remain intact (Fig. 2). Neither of these mutations disrupted the core cap-independent translation activity of the BTE, as indicated by the ability of the BTE to function from the 5′-UTR (LIII-3, LIII-CS) (Fig. 2C).
We postulate that either non-Watson-Crick interactions affect the stability of the kissing interaction, or base-pairing is enhanced in a sequence-specific fashion by a host protein. Interactions among bases flanking the Watson-Crick-paired bases influence annealing of kissing stem–loops in HIV RNA dimerization (Mujeeb et al. 1998). RNA chaperone proteins are known to enhance base-pairing of certain complementary RNAs (Moller et al. 2002; Wilusz and Wilusz 2005). However, structure probing data reveal that at least some measure of base-pairing between BYDV kissing stem–loops can take place in the absence of protein (Guo et al. 2001). In contrast to the BTE–BCL interaction, the unrelated TBSV 3′-cap-independent translation enhancer sustained translation with extensive base changes as long as the sequence complementarity between 3′- and 5′-UTRs was retained (Fabian and White 2004, 2006).
We show here and previously (Guo et al. 2000) that the core cap-independent translation activity of the 3′-element does not require long-distance base-pairing because the BTE functions in the 5′-UTR (Fig. 2). This rules out any model in which the kissing interaction must form in order to recruit the ribosome. However, it was not known whether the long-distance interaction requires that the sequence complementary to the 5′-UTR must be located within the context of the BTE. The functional construct in which the BCL was altered to base-pair to the LDFE—which is not part of the BTE—in the 3′-UTR (Fig. 7B), and the ability of DEN UTRs to replace BYDV sequences entirely for the inter-UTR interactions (Fig. 8), demonstrated clearly that the long-distance base-pairing interaction can be uncoupled from BTE structure and function. While the 5′-complementary portion of the BTE, loop III, is not part of the core cap-independent translation function sequence (Figs. 2C, 8), integrity of the sequence and/or structure of the BTE appears necessary for its function in the natural 3′-UTR context both in vivo and in vitro (Figs. 2B, 7). In the context of DEN UTRs, the 5′–3′ interaction that supported the BTE-mediated translation required base-pairing between the cyclization sequences (Fig. 8). The above-background level of translation from 5′-CS mutant RNA may result from fortuitous base-pairing between the mutant 5′-CS sequence (CUCUCUCUG) to a contiguous six-base complementary sequence downstream of the 3′-CS (CAGAGA) (nucleotides 10622–10627). Note that the DEN UTRs facilitate circularization in the absence of mammalian proteins in a wheat germ extract, consistent with observations that DEN RNA circularizes in the absence of any proteins (Alvarez et al. 2005b).
The 5′-proximal stem–loop permits replication when serving as the BTE-complementary loop
It is significant that mutations in the infectious BYDV genome that force the 5′-proximal stem–loop, SL-A, to replace SL-D as the BCL still allow viral RNA replication (Fig. 6B). The 5′-proximal stem–loop plays a key role in replication of many positive-strand viral RNAs (Barton et al. 2001; Vlot and Bol 2003), including members of the Tombusviridae (Ray et al. 2004), which resemble BYDV in many aspects (Miller et al. 2002). Either BYDV is exceptional in that its 5′-proximal stem–loop is not involved in replication, or (more likely) it tolerates the single base change that converted SL-A to a BCL and kissing by the BTE, and presence of associated translational machinery does not inhibit the role of SL-A in replication.
In many other viruses that have 3′-cap-independent translation elements including BYDV-like luteoviruses, the 3′-element is predicted to base-pair to the 5′-proximal loop on the viral RNA (Fig. 5A; Miller and White 2006). Why, then, has BYDV evolved a 5′-BCL farther downstream from the 5′-end? Recent data suggest that the distal position of the 5′-BCL in BYDV genomic RNA allows selective regulation of translation in trans by BYDV subgenomic RNA2 (Shen et al., in prep.). In the presence of subgenomic RNA2, translation of genomic RNA but not subgenomic RNA1 is inhibited (Wang et al. 1999), suggesting that subgenomic RNA2 provides a switch from early to late gene expression. The position of the BCL, which is naturally in the 5′-proximal stem–loop of sgRNA1 but at the 5′-distal stem–loop of gRNA, determines the selectivity of this trans-inhibition (Shen et al., in prep.). An intriguing unanswered question is why moving the BCL to the 5′-end provides no translational enhancement in the absence of subgenomic RNA2.
Requirement for ribosome scanning from the 5′-end of the RNA
Cap-independent translation of other uncapped RNAs is almost universally mediated by internal ribosome entry, independent of the 5′-end (Hellen and Sarnow 2001). In contrast, we have several lines of evidence indicating that BTE-mediated cap-independent translation relies on 5′-end-dependent scanning. First, addition of a stable stem–loop at the extreme 5′-end of the RNA abolished BTE-mediated translation (Guo et al. 2001). Secondly, addition of an upstream ORF in the BYDV 5′-UTR, out of frame with the main reporter ORF, inhibited translation initiation at the luciferase ORF start codon (Fig. 4) without hindering cap-independent translation of the uORF itself. A third line of evidence is the reduction of translation that resulted from addition of unstructured sequence (portions of the TMV Ω sequence) to the extreme 5′-end of the RNA. These results indicate that the distance of the 5′-end from the BCL can be important. Together the above results strongly support the notion that translation initiation requires ribosome scanning from the 5′-terminus of the uncapped RNA. Using an indirect approach, in which replication of a defective viral RNA is the reporter for translation of a viral replication protein, Fabian and White (2006) recently discovered a similar 5′-end-dependent scanning mechanism for the cap-independent translation of TBSV RNA. The 3′-translation enhancer of TBSV bears no resemblance to a BTE except that, like a BTE, it must base-pair to the 5′-UTR. Thus, this unconventional translational mechanism may apply to all 3′-TE-bearing viruses.
An oscillating kissing stem–loop model for ribosome delivery to the mRNA
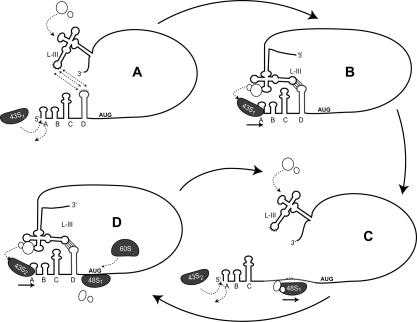
Based on the data presented here and what is known about 5′-end-dependent translation, we propose the following model in which the BTE mediates cap-independent translation by an oscillating scanning mechanism (Fig. 9): First, the BTE binds translation initiation factors and/or the 40S ribosomal subunit. Separately, the BCL–BTE long-distance kissing interaction brings the 3′-BTE in close proximity with the 5′-terminus of the RNA. Base-pairing may occur prior to, but is not required for, recruitment of the translation initiation complex. The 40S subunit may be recruited either to the BTE first or directly to the 5′-end of the mRNA. In the former case, the recruited ribosome and associated factors would jump from the BTE to the 5′-end; in the latter case, the recruited translation factors would be delivered, via the BCL–BTE kissing interaction, to the ribosome entering at the 5′-end.
FIGURE 9.
Proposed model for BTE-mediated recruitment of the 40S ribosomal subunit and associated initiation factors to the 5′-end and oscillation of the kissing stem–loop interaction caused by ribosome scanning. Entry of the first 43S complex of 40S subunit and associated factors (43S1) and long-distance kissing interaction is shown in panels A and B. Base-pairing may occur prior to, but is not required for, the recruitment of the translation machinery. After the first ribosome enters to the 5′-end (B), it would disrupt the BCL during scanning (C) and transiently prevent the kissing interaction. As the first 43S subunit continues toward the AUG, the SL-D reforms and the second 43S complex (43S2) is recruited (D). As subsequent 43S subunits enter the RNA, the kissing stem–loop complex would continue to oscillate between structures shown in panels C and D. Small circles represent translation initiation factors that interact with the BTE. An alternative possibility is that the 43S complex is recruited to the BTE. Upon base-pairing of the BTE to the BCL, the 43S complex would then jump from the BTE to the 5′-end, where scanning would ensue as in panel C (see text for details).
Although we cannot distinguish between these possibilities, the latter model is supported by observations that the 43S ribosomal complex alone is known to bind the 5′-end of uncapped mRNAs in the absence of any other initiation factors in vitro (Pestova and Kolupaeva 2002; Jivotovskaya et al. 2006). However, additional initiation factors are needed for scanning on structured 5′-UTRs (Pestova and Kolupaeva 2002) and, with the exception of eIF4E, are essential for 5′-end-dependent translation of artificially uncapped RNAs (Gunnery et al. 1997; Tarun and Sachs 1997; De Gregorio et al. 1998). Initial binding of the ribosome only at the 5′-end explains (1) why the BTE does not act as an IRES and (2) the low but detectable level of in vitro translation of uncapped mRNAs containing the BF insertion in the BTE. Binding of the ribosome to the 5′-end, followed by delivery of translation factors from the 3′-UTR, resembles a model proposed by Gazo et al. (2004) for cap-independent translation mediated by the 3′-cap-independent translation element in STNV RNA (which does not resemble a BTE). Although evidence of long-distance base-pairing is lacking, there is direct evidence that the 40S subunit binds the STNV 5′-UTR (Browning et al. 1980) and that the 3′-element binds eIF4F (Gazo et al. 2004).
Regardless of the mechanism of recruitment to the RNA, once the ribosome has entered the 5′-end of the mRNA, with associated factors it scans in the 3′ direction toward the start codon. The scanning 43S complex must transiently disrupt the kissing interaction when it reaches the BCL, and then progress processively to the first start codon. Discontinuous scanning or shunting of the ribosome is unlikely because there is no specific sequence or structure in the 5′-UTR required for 3′-BTE-mediated translation, other than ?A3B2 tlsb?> the BTE-complementarity sequence that is normally, but not necessarily (Fig. 3D), in a kissing stem–loop. In contrast, known examples of ribosome shunting in the 5′-UTR require specific secondary structures and upstream ORFs (Ryabova and Hohn 2000). Moreover, the BTE–BCL interaction supported translation of the uORF even though the BCL was within the uORF (Fig. 4B). In this construct, the BTE mediates cap-independent translation initiation before the scanning 40S ribosomal subunit reaches the BCL. During elongation, the 80S ribosome moves through and downstream from the BCL to translate the uORF. All the above observations are inconsistent with a shunting mechanism during 5′-UTR scanning.
The disruption of the long-distance base-pairing by the scanning ribosome would temporally prevent the next ribosome from entering (or beginning to scan on) the RNA. According to our model, after the 43S complex passes the BCL, the stem–loop re-forms and the BTE rapidly re-pairs to the BCL to deliver more initiation factors to the next ribosome waiting at the 5′-end. (As mentioned above, the BCL can function within an ORF, meaning it can function even between transient disruptions by elongating 80S ribosomes.) The cycle of BCL disruption and re-formation would repeat continuously, causing a structural oscillation of the kissing loop complex that regulates scanning of each 40S ribosome from the 5′-end. This would be a novel means of regulating ribosome entry to viral RNA. It resembles the model proposed for TBSV translation (Fabian and White 2006), with the yet-to-be resolved difference of where the ribosome is recruited to first. Regardless of mechanistic details, control of translation by the 3′-UTR may allow viral proteins that interact with the 3′-UTR, such as the replicase, to regulate (switch off) translation initiation at the 5′-end (for a detailed model, see Barry and Miller 2002).
The BTE as a sensor for long-distance RNA–RNA interactions
Finally, the ability to separate long-distance base-pairing interaction from core cap-independent translation function of the BTE may have a practical application. Because dengue virus sequences entirely unrelated to BYDV in structure and function allow the BTE to mediate cap-independent translation, 3′-BTE-mediated translation can be used as a functional bioassay to determine whether any RNA sequences or structures of interest are capable of interacting across long distances. By simply placing the potentially interacting sequences of interest in opposite UTRs of a reporter gene and fusing the BTE to the 3′-end of the 3′-UTR, one can test whether the sequences of interest interact by assessing the level of cap-independent translation conferred in wheat germ extract as in Figure 8. This would provide a rapid, quantitative bioassay to complement physical methods for detecting long-distance RNA–RNA interactions.
MATERIALS AND METHODS
Plasmid constructs
The BYDV reporter plasmids were derived from p5′UTR-LUC-TE869 and pTE105-LUC (Guo et al. 2000), referred to here as LUC869 and 5′BTE-LUC, respectively. All mutations were generated by site-directed mutagenic PCR using two complementary primers with the designated mutations as described in Guo et al. (2001). For mutations within the 5′-UTR of the BYDV reporter plasmids, the mutated PCR fragment, spanning BYDV nucleotides 1–142, was digested with NotI and BssHI and cloned into NotI–BssHI-digested pLUC869 or p5′BTE-LUC. For mutations within the 3′-UTR of BYDV reporter plasmids, the mutated PCR product, spanning BYDV nucleotides 4809–5677, was digested with Acc65I and SmaI and ligated into an Acc65I–SmaI-digested pLUC898 vector.
The dengue virus reporter plasmids were derived from a p5′DEN-LUC-3′DEN reporter construct (Holden and Harris 2004). The DEN-BTE was cloned by ligating a PCR-generated product, spanning BYDV nucleotides 4809–5045, and digested with XbaI restriction sites introduced in the 5′-ends of the PCR primers, into XbaI-digested DEN vector. An SmaI restriction site was introduced at the 3′-end of the BYDV sequence for the linearization of the chimeric plasmid for in vitro transcription. The mutations within the 5′ and 3′ cyclization sequence of the DEN reporter construct were performed as described by You and Padmanabhan (1999). All constructs were verified by automated sequencing at the Nucleic Acids Facility of Iowa State University on an ABI377 sequencer (Applied Biosystems).
In vitro transcription and in vitro translation
All RNAs were transcribed in vitro from SmaI-linearized plasmids using the T7 Megascript kits for uncapped RNA and T7 Mmessage machine kits for capped RNA (Ambion). The DEN reporter constructs without the 3′-BTE were linearized with XbaI prior to transcription. Prior to luciferase readings, 0.2 pmol of RNA transcripts was added to the wheat germ extract (Promega) in a final reaction volume of 12.5 μL and translated for 1 h at room temperature; 1–5 μL of the translation reaction was added to 50 μL of the Luciferase Assay Reagent (Promega), and the luciferase activity was immediately measured on a Turner Designs TD-20/20 luminometer. All experiments were performed in triplicate and repeated in at least three independent experiments. For radiolabeling of in vitro translated products, 10 μCi of 35S-labeled methionine was added to the translation reaction. Total protein from the translation mix was separated on a precast 4%–12% polyacrylamide gel (Invitrogen), which was then dried, exposed to a PhosphorImager screen for 24 h, and scanned on a STORM 840 PhosphorImager (Molecular Dynamics).
In vivo translation
Oat protoplasts were prepared from cell suspension culture as described (Dinesh-Kumar et al. 1992). For the transient expression assay, 1 pmol of the RNA transcript was electroporated into ∼106 cells. To normalize electroporation variation, we included 0.1 pmol of capped, polyadenylated Renilla RNA reporter construct as an internal control. Luciferase activity was measured 4 h after electroporation using the Promega Stop-N-Glo system. First, the cells were harvested and resuspended in 50 μL of Passive Lysis Buffer. The cells were centrifuged for 10 min at 15,000g. Protoplast lysate supernatant (5–10 μL) was added to 50 μL of the Luciferase Assay Reagent II, and the firefly luciferase activity was immediately measured on a Turner Designs TD-20/20 luminometer. Next, 50 μL of Sto-N-Glo buffer was added, and the Renilla luciferase activity was determined. For each sample, the measured firefly luciferase activity was normalized against the measured Renilla luciferase activity. The relative luciferase activity of the wild-type construct was defined as 100%. All experiments were performed in triplicate and repeated in at least three independent experiments.
The functional mRNA half-life was approximated using the equation t 1/2 2 = 2 P(∝) 2 • 2 ln 22/aR 0 (Meulewaeter et al. 1998b), where P(∝) is the saturation level of LUC in relative light units (RLU), a is the slope (RLU/min) from the time point following the lag phase at which the first translation product is completed, and R 0 is the initial RNA input. Best fitting curves to the experimental data points were generated using GraphPad software.
Northern blot analysis
Total RNAs from protoplasts inoculated with 10 μg of the full-length BYDV RNA were extracted 48 h post-inoculation using Trizol reagent (Invitrogen) as per the manufacturer's instruction. RNAs were analyzed by Northern blot hybridization as described in Koev et al. (2002). A 32P-labeled probe complementary to the 3′-terminal 1.5 kb of the full-length viral RNA was used to detect the BYDV genomic and subgenomic RNA accumulation.
ACKNOWLEDGMENTS
This research was funded by NIH grant no. GM067104 to W.A.M. and by Hatch Act and State of Iowa Funds. A.M.R. was funded by fellowships from the Fulbright Foundation and the Pioneer Hi-Bred Company (Johnston, Iowa). We thank Krzysztof Treder and Chris Fraser for critical reading of the manuscript and insightful comments, and Kai Tanaka for assistance with artwork.
Footnotes
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.115606.
REFERENCES
- Alvarez, D.E., De Lella Ezcurra, A.L., Fucito, S., Gamarnik, A.V. Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology. 2005a;339:200–212. doi: 10.1016/j.virol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Alvarez, D.E., Lodeiro, M.F., Luduena, S.J., Pietrasanta, L.I., Gamarnik, A.V. Long-range RNA–RNA interactions circularize the dengue virus genome. J. Virol. 2005b;79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry, J.K., Miller, W.A. A programmed −1 ribosomal frameshift that requires base-pairing across four kilobases suggests a novel mechanism for controlling ribosome and replicase traffic on a viral RNA. Proc. Natl. Acad. Sci. 2002;99:11133–11138. doi: 10.1073/pnas.162223099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, D.J., O'Donnell, B.J., Flanegan, J.B. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 2001;20:1439–1448. doi: 10.1093/emboj/20.6.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batten, J.S., Desvoyes, B., Yamamura, Y., Scholthof, K.B. A translational enhancer element on the 3′-proximal end of the Panicum mosaic virus genome. FEBS Lett. 2006;580:2591–2597. doi: 10.1016/j.febslet.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Browning, K.S., Leung, D.W., Clark, J.M., Jr. Protection of Satellite tobacco necrosis virus ribonucleic acid by wheat germ 40S and 80S ribosomes. Biochemistry. 1980;19:2276–2283. doi: 10.1021/bi00551a044. [DOI] [PubMed] [Google Scholar]
- Chiu, W., Kinney, R., Dreher, T. Control of translation by the 5′- and the 3′-terminal regions of the dengue virus genome. J. Virol. 2005;13:8303–8315. doi: 10.1128/JVI.79.13.8303-8315.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio, E., Preiss, T., Hentze, M.W. Translational activation of uncapped mRNAs by the central part of human eIF4G is 5′ end-dependent. RNA. 1998;4:828–836. doi: 10.1017/s1355838298980372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh-Kumar, S.P., Brault, V., Miller, W.A. Precise mapping and in vitro translation of a trifunctional subgenomic RNA of Barley yellow dwarf virus. Virology. 1992;187:711–722. doi: 10.1016/0042-6822(92)90474-4. [DOI] [PubMed] [Google Scholar]
- Dreher, T.W., Miller, W.A. Translational control in positive strand RNA plant viruses. Virology. 2006;344:185–197. doi: 10.1016/j.virol.2005.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgil, D., Harris, E. End-to-end communication in the modulation of translation by mammalian RNA viruses. Virus Res. 2006;119:43–51. doi: 10.1016/j.virusres.2005.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian, M.R., White, K.A. 5′ 3′ RNA RNA interaction facilitates cap- and poly(A) tail-independent translation of tomato bushy stunt virus mRNA:A potential common mechanism for Tombusviridae. J. Biol. Chem. 2004;279:28862–28872. doi: 10.1074/jbc.M401272200. [DOI] [PubMed] [Google Scholar]
- Fabian, M.R., White, K.A. Analysis of a 3′-translation enhancer in a tombusvirus: A dynamic model for RNA RNA interactions of mRNA termini. RNA. 2006. (in press) [DOI] [PMC free article] [PubMed]
- Gallie, D.R., Sleat, D.E., Watts, J.W., Turner, P.C., Wilson, T.M. The 5′-leader sequence of tobacco mosaic virus RNA enhances the expression of foreign gene transcripts in vitro and in vivo. Nucleic Acids Res. 1987;15:3257–3273. doi: 10.1093/nar/15.8.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazo, B.M., Murphy, P., Gatchel, J.R., Browning, K.S. A novel interaction of Cap-binding protein complexes eukaryotic initiation factor (eIF) 4F and eIF(iso)4F with a region in the 3′-untranslated region of satellite tobacco necrosis virus. J. Biol. Chem. 2004;279:13584–13592. doi: 10.1074/jbc.M311361200. [DOI] [PubMed] [Google Scholar]
- Gunnery, S., Maivali, U., Mathews, M.B. Translation of an uncapped mRNA involves scanning. J. Biol. Chem. 1997;272:21642–21646. doi: 10.1074/jbc.272.34.21642. [DOI] [PubMed] [Google Scholar]
- Guo, L., Allen, E., Miller, W.A. Structure and function of a cap-independent translation element that functions in either the 3′ or the 5′ untranslated region. RNA. 2000;6:1808–1820. doi: 10.1017/s1355838200001539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, L., Allen, E., Miller, W.A. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol. Cell. 2001;7:1103–1109. doi: 10.1016/s1097-2765(01)00252-0. [DOI] [PubMed] [Google Scholar]
- Hellen, C.U., Sarnow, P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes & Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- Holden, K.L., Harris, E. Enhancement of dengue virus translation: Role of the 3′ untranslated region and the terminal 3′ stem–loop domain. Virology. 2004;329:119–133. doi: 10.1016/j.virol.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Jivotovskaya, A.V., Valasek, L., Hinnebusch, A.G., Nielsen, K.H. Eukaryotic translation initiation factor 3 (eIF3) and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast. Mol. Cell. Biol. 2006;26:1355–1372. doi: 10.1128/MCB.26.4.1355-1372.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapp, L.D., Lorsch, J.R. The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- Khromykh, A.A., Meka, H., Guyatt, K.J., Westaway, E.G. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 2001;75:6719–6728. doi: 10.1128/JVI.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneller, E.L., Rakotondrafara, A.M., Miller, W.A. Cap-independent translation of plant viral RNAs. Virus Res. 2006;119:63–75. doi: 10.1016/j.virusres.2005.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koev, G., Liu, S., Beckett, R., Miller, W.A. The 3′-terminal structure required for replication of barley yellow dwarf virus RNA contains an embedded 3′ end. Virology. 2002;292:114–126. doi: 10.1006/viro.2001.1268. [DOI] [PubMed] [Google Scholar]
- Kozak, M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak, M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- Kozak, M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulewaeter, F., Danthinne, X., Van Montagu, M., Cornelissen, M. 5′- and 3′-sequences of satellite tobacco necrosis virus RNA promoting translation in tobacco. Plant J. (Erratum. 1998a;1415:169–176. 153–154). doi: 10.1046/j.1365-313x.1998.00104.x. [DOI] [PubMed] [Google Scholar]
- Meulewaeter, F., Van Montagu, M., Cornelissen, M. Features of the autonomous function of the translational enhancer domain of satellite tobacco necrosis virus. RNA. 1998b;4:1347–1356. doi: 10.1017/s135583829898092x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulewaeter, F., van Lipzig, R., Gultyaev, A.P., Pleij, C.W., Van Damme, D., Cornelissen, M., van Eldik, G. Conservation of RNA structures enables TNV and BYDV 5′ and 3′ elements to cooperate synergistically in cap-independent translation. Nucleic Acids Res. 2004;32:1721–1730. doi: 10.1093/nar/gkh338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, W.A., White, K.A. Long-distance RNA–RNA interactions in plant virus gene expression and replication. Annu. Rev. Phytopathol. 2006 doi: 10.1146/annurev.phyto.44.070505.143353. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, W.A., Liu, S., Beckett, R. Barley yellow dwarf virus: Luteoviridae or Tombusviridae? Mol. Plant Pathol. 2002;3:177–183. doi: 10.1046/j.1364-3703.2002.00112.x. [DOI] [PubMed] [Google Scholar]
- Mizumoto, H., Tatsuta, M., Kaido, M., Mise, K., Okuno, T. Cap-independent translational enhancement by the 3′ untranslated region of red clover necrotic mosaic virus RNA1. J. Virol. 2003;77:12113–12121. doi: 10.1128/JVI.77.22.12113-12121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller, T., Franch, T., Hojrup, P., Keene, D.R., Bachinger, H.P., Brennan, R.G., Valentin-Hansen, P. Hfq: A bacterial Sm-like protein that mediates RNA–RNA interaction. Mol. Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- Mujeeb, A., Clever, J.L., Billeci, T.M., James, T.L., Parslow, T.G. Structure of the dimer initiation complex of HIV-1 genomic RNA. Nat. Struct. Biol. 1998;5:432–436. doi: 10.1038/nsb0698-432. [DOI] [PubMed] [Google Scholar]
- Oberer, M., Marintchev, A., Wagner, G. Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes & Dev. 2005;19:2212–2223. doi: 10.1101/gad.1335305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, C.P., Barry, J.K., Dinesh-Kumar, S.P., Brault, V., Miller, W.A. A sequence required for −1 ribosomal frameshifting located four kilobases downstream of the frameshift site. J. Mol. Biol. 2001;310:987–999. doi: 10.1006/jmbi.2001.4801. [DOI] [PubMed] [Google Scholar]
- Pestova, T.V., Kolupaeva, V.G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes & Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, D., Na, H., White, K.A. Structural properties of a multifunctional T-shaped RNA domain that mediate efficient tomato bushy stunt virus RNA replication. J. Virol. 2004;78:10490–10500. doi: 10.1128/JVI.78.19.10490-10500.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryabova, L.A., Hohn, T. Ribosome shunting in the cauliflower mosaic virus 35S RNA leader is a special case of reinitiation of translation functioning in plant and animal systems. Genes & Dev. 2000;14:817–829. [PMC free article] [PubMed] [Google Scholar]
- Sarnow, P., Cevallos, R.C., Jan, E. Takeover of host ribosomes by divergent IRES elements. Biochem. Soc. Trans. 2005;33:1479–1482. doi: 10.1042/BST0331479. [DOI] [PubMed] [Google Scholar]
- Scheets, K., Redinbaugh, M.G. Infectious cDNA transcripts of Maize necrotic streak virus: Infectivity and translational characteristics. Virology. 2006;350:171–183. doi: 10.1016/j.virol.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Shen, R., Miller, W.A. The 3′ untranslated region of tobacco necrosis virus RNA contains a barley yellow dwarf virus-like cap-independent translation element. J. Virol. 2004;78:4655–4664. doi: 10.1128/JVI.78.9.4655-4664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siridechadilok, B., Fraser, C.S., Hall, R.J., Doudna, J.A., Nogales, E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- Tarun, S.Z., Jr., Sachs, A.B. Binding of eukaryotic translation initiation factor 4E (eIF4E) to eIF4G represses translation of uncapped mRNA. Mol. Cell. Biol. 1997;17:6876–6886. doi: 10.1128/mcb.17.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmer, R.T., Benkowski, L.A., Schodin, D., Lax, S.R., Metz, A.M., Ravel, J.M., Browning, K.S. The 5′ and 3′ untranslated regions of satellite tobacco necrosis virus RNA affect translational efficiency and dependence on a 5′ cap structure. J. Biol. Chem. 1993;268:9504–9510. [PubMed] [Google Scholar]
- Vlot, A.C., Bol, J.F. The 5′ untranslated region of alfalfa mosaic virus RNA 1 is involved in negative-strand RNA synthesis. J. Virol. 2003;77:11284–11289. doi: 10.1128/JVI.77.20.11284-11289.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar, T., Gross, J.D., Wagner, G., McCarthy, J.E. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat. Struct. Mol. Biol. 2004;11:503–511. doi: 10.1038/nsmb779. [DOI] [PubMed] [Google Scholar]
- Wang, S., Browning, K.S., Miller, W.A. A viral sequence in the 3′-untranslated region mimics a 5′ cap in facilitating translation of uncapped mRNA. EMBO J. 1997;16:4107–4116. doi: 10.1093/emboj/16.13.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., Guo, L., Allen, E., Miller, W.A. A potential mechanism for selective control of cap-independent translation by a viral RNA sequence in cis and in trans. RNA. 1999;5:728–738. doi: 10.1017/s1355838299981979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz, C.J., Wilusz, J. Eukaryotic Lsm proteins: Lessons from bacteria. Nat. Struct. Mol. Biol. 2005;12:1031–1036. doi: 10.1038/nsmb1037. [DOI] [PubMed] [Google Scholar]
- Wu, B., White, K.A. A primary determinant of cap-independent translation is located in the 3′-proximal region of the tomato bushy stunt virus genome. J. Virol. 1999;73:8982–8988. doi: 10.1128/jvi.73.11.8982-8988.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, S., Padmanabhan, R. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 1999;274:33714–33722. doi: 10.1074/jbc.274.47.33714. [DOI] [PubMed] [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]