Abstract
Eukaryotic translation initiation factor 4G (eIF4G) plays a crucial multimodulatory role in mRNA translation and decay by interacting with other translation factors and mRNA-associated proteins. In this study, we isolated eight different RNA aptamers with high affinity to mammalian eIF4G by in vitro RNA selection amplification. Of these, three aptamers (apt3, apt4, and apt5) inhibited the cap-dependent translation of two independent mRNAs in a rabbit reticulocyte lysate system. The cap-independent translation directed by an HCV internal ribosome entry site was not affected. Addition of exogenous eIF4G reversed the aptamer-mediated inhibition of translation. Even though apt3 and apt4 were selected independently, they differ only by two nucleotides. The use of truncated eIF4G variants in binding experiments indicated that apt4 (and probably apt3) bind to both the middle and C-terminal domains of eIF4G, while apt5 binds only to the middle domain of eIF4G. Corresponding to the difference in the binding sites in eIF4G, apt4, but not apt5, hindered eIF4G from binding to eIF4A and eIF3, in a purified protein solution system as well as in a crude lysate system. Therefore, the inhibition of translation by apt4 (and apt3) is due to the inhibition of formation of initiation factor complexes involving eIF4A and eIF3. On the other hand, apt5 had a much weaker affinity to eIF4G than apt4, but inhibited translation much more efficiently by an unknown mechanism. The five additional aptamers have sequences and predicted secondary structures that are largely different from each other and from apt3 through apt5. Therefore, we speculate that these seven sets of aptamers may bind to different regions in eIF4G in different fashions.
Keywords: eIF4G, cap-dependent translation, RNA aptamer, SELEX, initiation factor complex, translation initiation
INTRODUCTION
Eukaryotic mRNAs (excluding those in organelles) are modified at their 5′-ends by a structure termed the cap, which contains the sequence m7GpppN (where N is any nucleotide) (Shatkin 1976). The cap plays a key role in facilitating the binding of the ribosomal 40S subunit to the 5′-end of mRNA (Shatkin 1976) via interaction with eukaryotic translation initiation factor 4F (eIF4F) (for review, see Hershey and Merrick 2000). eIF4F is composed of three subunits termed eIF4E, eIF4A, and eIF4G. eIF4E recognizes the cap, while eIF4A is an RNA-dependent ATPase that is thought to unwind the secondary structure present in the 5′-untranslated region of mRNAs. eIF4G serves as a multifunctional adaptor protein for binding to other eIFs including eIF3, which recruits the ribosomal 40S subunit, leading to the formation of the 43S pre-initiation complex (Imataka et al. 1998; for review, see Hershey and Merrick 2000). Although eIF4F has long been known as a biochemical preparation, it has never been demonstrated as a separate entity in vivo, nor has evidence been presented that such an entity binds to mRNA prior to binding of mRNA to the ribosome. The order of assembly of the 48S initiation complex is currently unknown. Mammals possess two functional isoforms of eIF4G (eIF4GI and eIF4GII), which share 46% sequence identity and are functionally similar (Gradi et al. 1998). Unless otherwise stated, in this study eIF4G refers to eIF4GI.
With its multiple binding proteins, eIF4G participates in and modulates a variety of intracellular processes (Prevot et al. 2003). For example, besides the well-known interaction with eIF4E, eIF4A, and eIF3, yeast eIF4G interacts with eIF1 and eIF5 (He et al. 2003). While the role of eIF4G is well known in the initial formation of the eIF4F and the formation of the pre-initiation complex at the mRNA cap site, it may have an additional role in the final formation of the initiation complex at the start codon. eIF4G also interacts with the MAP kinase-activated protein kinase (Mnk1) and the poly(A)-binding protein (PABP). Mnk1 phosphorylates and activates eIF4E via binding to eIF4G (Pyronnet et al. 1999). PABP binds to eIF4G, which induces circularization of mRNA (Wells et al. 1998) to facilitate recycling of the post-termination ribosome directly to the initiation site of the mRNA (Tarun and Sachs 1996). The activity of eIF4G has also been reported in translation termination (Uchida et al. 2002) and the pioneering round of translation required for mRNA quality control (Lejeune et al. 2004).
The mammalian eIF4G protein can be divided into three domains: the N-terminal (N; amino acid positions 1–674; hereafter referred to as 1–674), middle (M; 675–1079), and C-terminal (C; 1080–1600) domains (for review, see Hershey and Merrick 2000; amino acid numbering refers to Byrd et al. 2002). The N domain binds to PABP and eIF4E, whereas the M domain binds to eIF3 (Korneeva et al. 2000) and eIF4A (Fig. 1; Imataka and Sonenberg 1997). The C domain contains additional binding sites for eIF4A and Mnk1 (Fig. 1). It is known that a truncated eIF4G (590–1130) is sufficient to catalyze cap-dependent translation in vitro and that the C domain modulates translation efficiency (Morino et al. 2000). The M domain also contains an arginine-rich sequence (683–736) and a HEAT domain, which is involved in RNA binding (Lomakin et al. 2000; Marcotrigiano et al. 2001). Indeed, the M domain of eIF4G interacts with several mRNAs, including β-globin mRNA and the internal ribosomal entry site (IRES) of the encephalomyocarditis virus (EMCV) (Lomakin et al. 2000; Marcotrigiano et al. 2001).
FIGURE 1.
The domain structures of eIF4G. eIF4G1–1600 is full-length eIF4G. eIF4G712–1600 was used as a target molecule in SELEX. eIF4G186–1600 and eIF4G712–1010 were used in the translation inhibition assay and the filter retention assay, respectively. eIF4G1080–1600 was used in the SPR experiment. Two binding sites of eIF4A are amino acids 712–1010 and 1241–1451. The binding site of eIF3 is 1010–1105. The binding sites of PABP, eIF4E, and Mnk1 are also shown. All sites of binding are shaded and indicated below eIF4G1–1600.
RNA aptamers are synthetic molecules that bind to targets with high affinity. RNA aptamers are obtained from random sequences containing ∼1014 variants by systematic evolution of ligands by exponential enrichment (SELEX) (Tuerk and Gold 1990). Since the introduction of SELEX, a variety of RNA and DNA aptamers have been created, their use for characterizing target molecules has been demonstrated, and some are currently undergoing clinical trials as novel therapeutics (Thiel 2004). We have generated RNA aptamers to mammalian eIF4A (Oguro et al. 2003), eIF4E (Mochizuki et al. 2005), and eIF1A (A. Oguro and Y. Nakamura, unpubl.). These aptamers are of relatively large mass, bind strongly to each target, and efficiently inhibit cap-dependent translation initiation in the rabbit reticulocyte lysate (RRL) translation systems. Méthot et al. (1996) have developed RNA aptamers to eIF4B, which bind to the RNA recognition motif of eIF4B and prevent the interaction between the ribosome and eIF4B. It is clear that a set of RNA aptamers specific to individual eIFs and specific to each domain of an eIF would be of great value for the study of the mammalian translation initiation mechanism since much of the mechanism remains to be investigated and initiation of translation is anything but simple. To study the multifunctional role of eIF4G in translational control and cellular processes, we developed here translation-inhibitory RNA aptamers against eIF4G. Interestingly, two distinct classes of RNA aptamers were obtained. One binds to both the middle and C-terminal domains of eIF4G and interferes with eIF4G binding to eIF4A and eIF3. The other aptamer binds to the middle domain of eIF4G, does not affect the formation of the eIF4G complex with eIF4A and eIF3, but does severely inhibit cap-dependent translation initiation.
RESULTS
In vitro selection of eIF4G aptamers
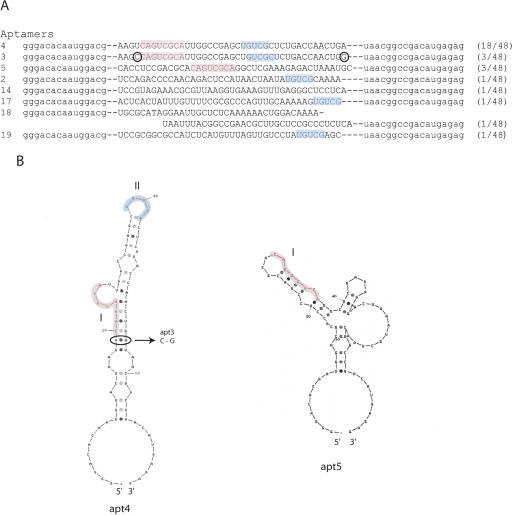
RNA aptamers were obtained against GST-tagged human eIF4G by affinity RNA selection. Full-length eIF4G is unstable as a recombinant protein in vitro. Therefore we used an M-C fragment of eIF4G truncated at position 712 (eIF4G712–1600) for selection. This fragment includes the binding sites for eIF4A and eIF3 (Fig. 1). GST-tagged eIF4G712–1600 was immobilized on glutathione-Sepharose resins. In vitro selection was initiated with a pool of 5×1014 different RNA molecules randomized over 40 nt. To eliminate RNA aptamers that bind to the resin, the 40N RNA pool was pre-incubated with empty glutathione-Sepharose resin, and unbound material was used for selection. After 12 rounds of selection, 48 individual RNA sequences were cloned and sequenced. Of these, one sequence (apt4) appeared 18 times, while two sequences (apt3 and apt5) appeared three times each. All other sequences appeared only once (Fig. 2A). The three RNAs that appeared multiple times (i.e., apt3, apt4, and apt5) were studied in detail and shown to inhibit cap-dependent in vitro translation as described below.
FIGURE 2.
In vitro selected RNA sequences and predicted secondary structures. (A) Eight different sequences of RNA selected by SELEX. Consensus motifs I and II are colored in red and blue, respectively. The circled C and G in apt3 indicate where this sequence differs from apt4. The numbers in parentheses indicate the frequency of which the sequence was selected. The primer regions are indicated in lowercase. (B) Predicted secondary structures of apt4 and apt5. The consensus motifs I and II are colored in red and blue, respectively. The circled AU base pair of apt4 corresponds to GC in apt3.
The sequence of apt3 and apt4 differs only at two positions (see Fig. 2A, marked by circles). These sequences contained two conserved motifs, with a CAGUCGCA (motif I) and UGUCG (motif II). In addition, motif I appears in apt5, and motif II appears in apt2, apt17, and apt19 (see Fig. 2A). Apt4 and Apt5 sequences adopt unique RNA secondary structures as predicted by MFOLD software (Fig. 2B; Zuker 1989, 2003). On the other hand, apt3, a variant of apt4 at two residues, is predicted to form four potential structures, one of which is equivalent to apt4.
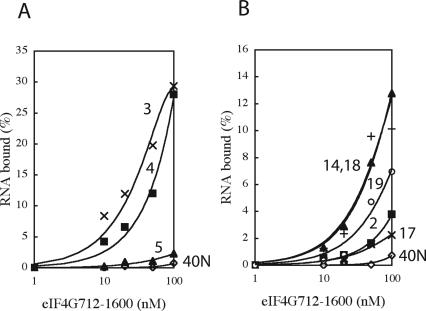
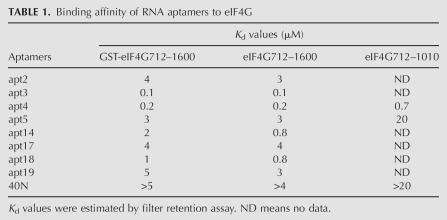
Next, these RNA species were labeled with [α-32P]CTP and examined for their ability to bind to eIF4G712–1600 (GST-tagged) by a nitrocellulose filter retention assay. All RNAs tested (aptamer numbers 2, 3, 4, 5, 14, 17, 18, and 19) bound efficiently to eIF4G712–1600 (Fig. 3). Apt3 bound most efficiently to eIF4G, and its dissociation constant (K d) was estimated at 0.1 μM (Table 1). The affinity of apt4 was similar to that of apt3. The other RNAs also bound efficiently to eIF4G712–1600, as compared to the 40N RNA pool control. The possible involvement of the GST tag sequence in aptamer binding was excluded since all these RNAs bound just as efficiently to the GST-free eIF4G712–1600 and failed to bind to GST alone (Table 1).
FIGURE 3.
Nitrocellulose filter retention of RNAs as a function of eIF4G712–1600 concentration. 32P-labeled RNAs were incubated with GST-eIF4G712–1600 for 30 min at room temperature. These mixtures were then filtered through a nitrocellulose membrane. Retained radioactivity on the filter was quantified by scintillation counting. Numbers adjacent to each curve indicate the names of aptamers. 40N is the random RNA pool.
TABLE 1.
Binding affinity of RNA aptamers to eIF4G
RNA aptamers inhibit cap-dependent translation in vitro
The effect of RNA aptamers on the in vitro translation of a 5′-capped chloramphenicol acetyltransferase (CAT) mRNA was examined using an RRL system. In this system, a bicistronic mRNA contains open reading frames for CAT and luciferase (LUC) separated by an HCV-IRES (cap-CAT/HCV/LUC) (Fig. 4A). Whereas the translation of CAT is cap-dependent, translation of LUC is directed by the HCV IRES and is therefore cap independent. HCV IRES-directed translation initiation is independent of eIF4G (Pestova et al. 1998), and rabbit eIF4G is 87% identical to the human eIF4G. Translation of both CAT and LUC was observed in RRL in the presence of [35S]methionine, [35S]cysteine, and increasing amounts of 40N (control) (Fig. 4B). Addition of apt3, apt4, and apt5 RNAs inhibited cap-dependent CAT translation in a dose-dependent manner, while cap-independent LUC translation was not inhibited appreciably (Fig. 4B). Apt5 was the strongest inhibitor of translation, reducing the CAT translation product to 26% by the addition of only 2 μM apt5. Under the same conditions, apt3 (data not shown) and apt4 inhibited CAT translation less efficiently than apt5, but when apt4 was increased to 8 μM in RRL, 89% of the translation was inhibited (Fig. 4B). The preferential block of cap-dependent translation by apt4 and apt5 is evident when calculating the relative value of CAT synthesis versus LUC synthesis (Fig. 4B). These in vitro translation experiments, as well as those shown below, were performed independently at least three times for each assay, allowing us to confirm the reproducibility of this given set of data. The other aptamers (aptamer numbers 2, 14, 17, 18, and 19) did not inhibit CAT translation (data not shown).
FIGURE 4.
Translation inhibition by the aptamers in the RRL translation system. A bicistronic mRNA, cap-CAT/HCV/LUC, was used as the reporter gene. Translation of CAT and LUC is initiated with a cap structure and HCV-IRES, respectively. The products were radioactively labeled by incorporation of a mixture of [35S] methionine and [35S] cysteine. The final concentrations of aptamers added were 0 ∼ 8 μM. Each experiment was conducted independently three to five times to confirm the reproducibility. The representative (and reproducible) results are shown. (A) Schematic diagram of the capped CAT/HCV/LUC mRNA. (B) Translation products. The products were analyzed by SDS-PAGE followed by fluorography. The relative translation of CAT (CAT%) and LUC (LUC%) was based on the amount of translated CAT and LUC, respectively, when the aptamer concentration was zero. (C) Prevention of translation inhibition by aptamers by adding exogenous eIF4G186–1600. (Lane 1) apt4 is 0 μM; (lane 2) apt4 is 6 μM; (lane 3) addition of 0.63 μM exogenous eIF4G after translation was inhibited with 6 μM apt4; (lane 4) apt5 is 0 μM; (lane 5) apt5 is 0 μM and 0.63 μM exogenous eIF4G; (lane 6) apt5 is 1 μM; (lane 7) addition of 0.63 μM exogenous eIF4G after translation was inhibited with 1 μM apt5. (D) Translation inhibition of a cap-LUC mRNA that was used as a reporter gene in the RRL translation system. The final concentrations of the aptamers added were 0 ∼ 12 μM.
To examine whether inhibition of translation by apt4 and apt5 was due to the direct inactivation of eIF4G by these aptamers, we performed two experiments. First, a recombinant human eIF4G186–1600 protein, which is able to initiate cap-dependent translation, was added to the RRL reaction mix (Morino et al. 2000). As shown in Figure 4C (lanes 3 and 7), addition of this exogenous eIF4G prevented the apt4/apt5-directed inhibition. While CAT translation was reduced to 42% upon the addition of 6 μM apt4, the additional presence of 0.63 μM eIF4G186–1600 fully restored CAT translation. Similarly, while the addition of 1 μM apt5 reduced CAT translation to 3.2%, the addition of 0.63 μM eIF4G186–1600 again fully restored translation. Under these conditions, 0.63 μM exogenous eIF4G186–1600 only slightly stimulated CAT translation in RRL in the absence of aptamers (Fig. 4C, lane 5).
To exclude the possibility that apt4/apt5 RNAs target specifically the CAT sequence leading to the inactivation of translation of this mRNA, we performed RRL translation using a capped LUC mRNA (cap-LUC) instead of the bicistronic cap-CAT/HCV/LUC mRNA. As shown in Figure 4D, translation of LUC mRNA was inhibited to 9.8% and 30% by 8 μM apt4 and 1 μM apt5, respectively, showing that these aptamers inhibit translation independently of the mRNA sequence.
Aptamer binding sites in eIF4G
To determine the binding sites of apt4 and apt5 in eIF4G, the M-domain fragment of eIF4G between positions 712 and 1010 (eIF4G712–1010) was produced as a recombinant protein and used in the filter-retention assay. Apt4 bound to eIF4G712–1010 with a K d value of 0.7 μM, which is threefold higher than for eIF4G712–1600 (Table 1). Although apt5 retained binding ability to eIF4G712–1010, the affinity was reduced by one order of magnitude as compared to its binding to eIF4G712–1600. These results were suggestive of possible involvement, directly or indirectly, of M and C domains in the aptamer binding, but were not sufficient to pinpoint the site.
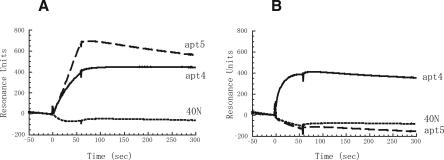
Hence, we prepared the C-domain fragment of eIF4G between positions 1080 and 1600 (eIF4G1080–1600) (see Fig. 1), and its binding to apt4 and apt5 was monitored in real time with a BIAcore 2000 instrument based on the surface plasmon resonance (SPR) technique. 3′-Poly(A)-tailed RNAs were immobilized to the streptavidin sensor chip via 5′-biotinylated oligo(dT), and the formation of eIF4G-coupled complexes on this matrix was monitored as SPR signals. No positive signal was observed in a blank flow cell (data not shown). [Note that the background Resonance Units (RUs) of RNAs and 5′-biotinylated oligo(dT) immobilized on chips were subtracted in the sensorgrams.] eIF4G712–1600 (MC fragment) and eIF4G1080–1600 (C fragment) were injected at a flow rate of 20 mL/min for 60 sec and dissociated for 200 sec by injecting a blank solution at the same flow rate. A set of sensorgrams for the eIF4G fragment association with apt4, apt5, and N40 random (control) is shown in Figure 5, in which the eIF4G fragment injection time was set as time zero. As expected, SPR signals on the apt4 and apt5 sensor chips appeared upon eIF4G712–1600 injection, while no positive signal was observed on an N40 random sensor chip. On the other hand, eIF4G1080–1600 injection gave rise to the appearance of SPR signals for the apt4 sensor chip but no SPR signal for the apt5 sensor chip. These results indicate that apt4 binds to both M and C domains of eIF4G, while apt5 binds to the M domain but not to the C domain.
FIGURE 5.
Sensorgrams of apt4 and apt5 binding to eIF4G712–1600 and eIF4G1080–1600. The sensor chip immobilized with apt4, apt5, or 40N random sequenced RNA was injected with a 0.5 μM solution of (A) eIF4G712–1600 or (B) eIF4G1080–1600 at time 0 for 60 sec. Experimental conditions and procedures are described in Materials and Methods.
Effect of RNA aptamers on eIF4G binding to eIF4A and eIF3
Since the eIF4G712–1600 fragment contains binding sites for eIF4A and eIF3 (see Fig. 1), we examined whether apt4 and apt5 RNAs might interfere with the interaction of eIF4G with eIF4A and/or eIF3. The eIF4A protein family contains three members: eIF4AI and eIF4AII participate in translation initiation (Hershey and Merrick 2000), while eIF4AIII is an exon-junction component (Ferraiuolo et al. 2004; Shibuya et al. 2004).
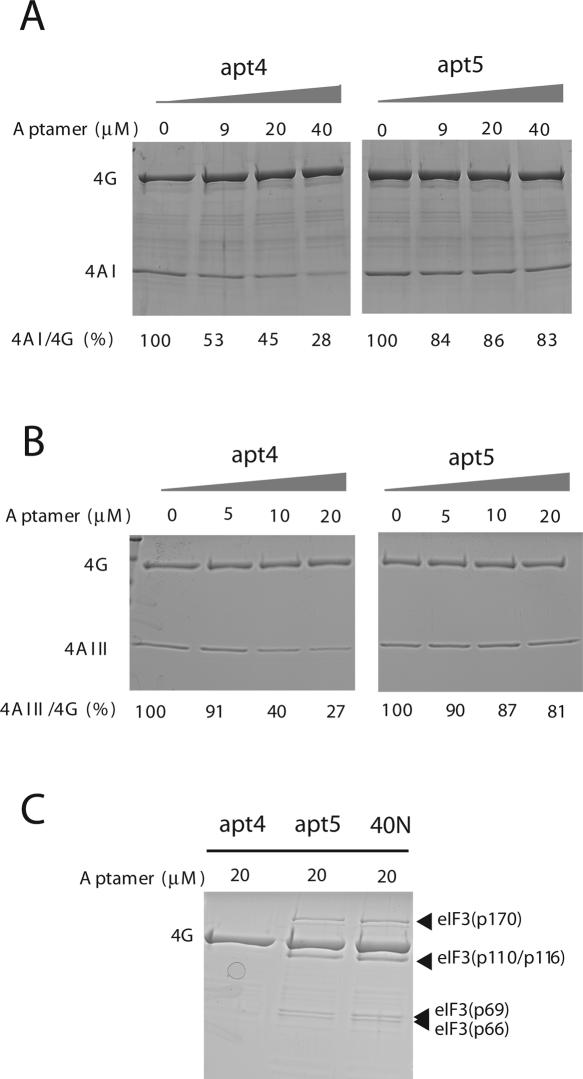
The eIF3 complex consists of 12 different subunits. The effect of aptamers on the binding activity of eIF4G to purified human eIF4AI, human eIF4AIII, or mouse eIF3 was tested. The GST-tagged human eIF4G712–1600 was immobilized on glutathione-Sepharose resin and incubated first with eIF4AI in the presence of increasing amounts of apt4 or apt5 RNAs. After washing, eIF4G-bound eIF4AI was recovered with elution buffer and quantified after SDS-polyacrylamide gel electrophoresis (PAGE) and Coomassie Brilliant Blue (CBB) staining. As shown in Figure 6A, the amount of eIF4AI bound to eIF4G-immobilized resin decreased proportionately to the amount of apt4 added, while the amount of eIF4G did not decrease appreciably by the addition of aptamer. To normalize deviation between separate pull-down experiments, the relative ratio of eIF4AI versus eIF4G (internal control) was calculated. The data clearly pointed out that the eIF4G–eIF4AI interaction was abolished by apt4, while apt5 did not interfere appreciably with the eIF4G–eIF4AI interaction.
FIGURE 6.
Effects of aptamers on interactions between (A) eIF4G-eIF4AI, (B) eIF4G-eIF4AIII, and (C) eIF4G-eIF3 in vitro. GST-eIF4G712–1600 was immobilized with glutathione-Sepharose resin. Mixtures of the aptamers and eIF4AI, eIF4AIII, or eIF3 were added to the immobilized GST-eIF4G712–1600. eIF4G, eIF4AI, eIF4AIII, and eIF3 were separated by SDS-PAGE and detected by CBB staining. The 4AI/4G and 4AIII/4G ratio obtained in the absence of aptamer was set as 100%. Each experiment was conducted independently three to five times to confirm the reproducibility. The representative (reproducible) results are shown.
The same pull-down experiment was performed with human eIF4AIII. eIF4AIII, which is 65% identical to eIF4AI, binds exclusively to the M domain of eIF4G (Li et al. 1999). eIF4AI and eIF4AIII do not bind simultaneously to the same molecule of eIF4G (Li et al. 1999). eIF4G–eIF4AIII interaction was also prevented by apt4 but not by apt5 (Fig. 6B). It is noteworthy that the inhibitory effect of apt4 to the eIF4G–eIF4AIII interaction is slightly but significantly greater than that to the eIF4G–eIF4AI interaction (Fig. 6, cf. A and B). This could be explained by the fact that eIF4G contains two binding sites for eIF4AI while only one for eIF4AIII, such that the common binding site in the M domain was hindered by apt4. These aptamer-challenging experiments in Figure 6 were performed independently at least three times, confirming the reproducibility.
Finally, purified eIF3 was incubated with the eIF4G-immobilized resin in the presence of 20 μM apt4, apt5, or 40N, and the bound proteins were recovered and analyzed as above. As shown in Figure 6C, two faint, but reproducible, bands corresponding to the major components of eIF3, p170 and p116/p110 (as well as p69 and p66), disappeared from the eIF4G complex in the presence of apt4, but remained unaffected in the presence of apt5 or 40N.
These findings suggest that apt4 occupies, at least in part, the binding sites for eIF4A and eIF3 in eIF4G, thereby inhibiting the formation of the eIF4G complex with these two components, resulting in inhibition of cap-dependent translation. In contrast, apt5 binds to the M domain in a fashion that does not inhibit interaction of the M domain of eIF4G with eIF4A or eIF3, but nevertheless severely inhibits translation by an unknown mechanism.
Effect of aptamers on the eIF complex formation in RRL
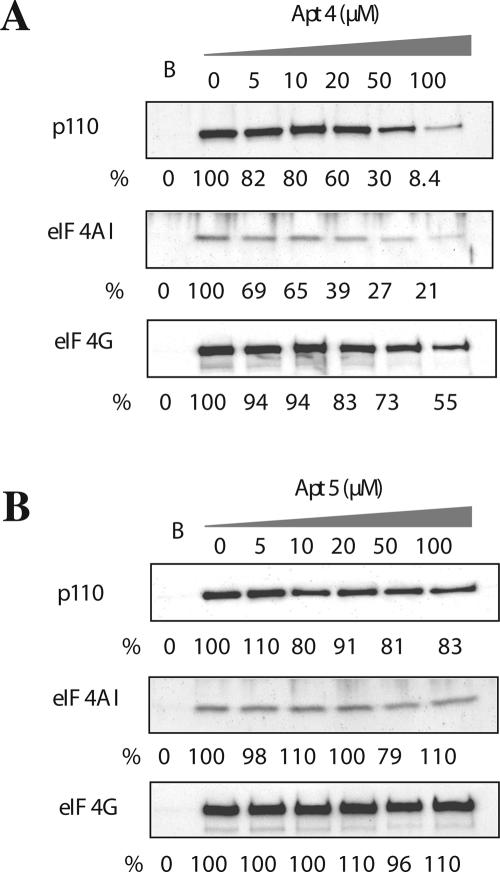
Since apt4 inhibited eIF4G's interaction with eIF4A and eIF3 in the purified recombinant protein system above, we examined the ability of the eIF4G aptamers to inhibit the binding of eIF4A and eIF3 to the eIF4E–eIF4G complex by using the m7GTP-Sepharose pull-down assay. As expected, eIF4G and its binding partners, eIF4AI and p110 (component of eIF3), were precipitated by the m7GTP-Sepharose resin in the absence of aptamers (Fig. 7). In the presence of increased amounts of apt4, the fraction of eIF4AI bound to eIF4G decreased substantially, and this decrease was followed by a gradual decrease in p110 binding (Fig. 7A). Although the amount of eIF4G complexed with m7GTP-Sepharose resin seemed to decrease in proportion to the concentration of added apt4, this decrease was much less compared to eIF4AI and p110. Probably apt4 might induce a conformational change in eIF4G, which disfavors the interaction between eIF4G and eIF4E. Alternatively, because of the close distance between the binding sites for eIF4E (proximal to the N–M-domain junction) and apt4 (M domain) on eIF4G (see Fig. 1), apt4 might weakly but physically interfere with the eIF4E binding. Under these conditions, the same range of apt5 additions affected neither the interaction of eIF4G with eIF4AI and eIF3, nor the interaction of eIF4G with eIF4E (Fig. 7B). These findings suggest that the translational inhibition by apt4 is due to the disabled interaction of eIF4G with eIF4A and eIF3, while the molecular basis of apt5 inhibition is unknown.
FIGURE 7.
Effects of the aptamers on interactions of eIF4G with eIF3 and with eIF4A in the RRL translation system. Endogenous eIF3, eIF4A, and eIF4G in RRL were pulled down with m7GTP-Sepharose resin and were detected by Western blotting following SDS-PAGE. Anti-p110 antibody was used for detection of eIF3. “B” indicates the blank test in which the Sepharose resin was used instead of the m7GTP-Sepharose resin. Apt 4 (A) and apt 5 (B) were added to final concentrations of 0 ∼ 100 μM. Each experiment was conducted independently three to five times to confirm the reproducibility. The representative (reproducible) results are shown.
DISCUSSION
In this study, eight distinct RNA aptamers were generated for human initiation factor eIF4G712–1600 by in vitro RNA selection, and their affinity to eIF4G was confirmed by a filter retention assay. Of these, three different high-affinity RNA aptamers—apt3, apt4, and apt5—inhibited in vitro translation, whereas the others did not. Apt3 and apt4 are grouped as one type since they differ at only two nucleotides and show essentially the same properties. Apt4 and apt5 inhibit cap-dependent in vitro translation in an RRL system, while they do not inhibit cap-independent (HCV IRES-directed) translation. The latter finding is consistent with previous data that HCV IRES-dependent translation does not require eIF4G (Pestova et al. 1998). Two lines of experimental data favor the conclusion that inhibition of translation by apt4 and apt5 is due to direct targeting and inactivation of eIF4G. First, addition of exogenous eIF4G prevented the inhibition. Since translation was fully restored, one might speculate that the eIF4G • aptamer complex does not remain bound to either the ribosome or other translation factors exerting a dominant-negative inhibitory effect. It, nevertheless, cannot be excluded completely that the aptamer inactivated a translation factor but eIF4G and exogenous eIF4G functioned to titrate the aptamer as a decoy. Second, apt4/apt5-mediated translational inhibition was independent of the mRNA sequence used.
The predicted secondary structures of apt4 and apt5 are very different except for a conserved motif I (CAGUCGCA) located in the stem-to-bulge region of both structures (see Fig. 2B). It is remarkable that although apt5 binds to eIF4G less efficiently than apt4; it is a more potent inhibitor of cap-dependent translation (Table 1; Figs. 3 and 4). Corresponding to the difference in the primary sequences and the secondary structures, apt4 binds to both M and C domains of eIF4G, while apt5 binds to the M domain of eIF4G. The M domain of eIF4G encodes two important binding sites for eIF4A and eIF3. Therefore, it is conceivable that apt4 inhibits cap-dependent translation by blocking the binding of eIF4A and eIF3 to eIF4G. However, the required amount of apt4 for efficient blocking of eIF4A/eIF3 binding was significantly higher than that required for efficient inhibition of in vitro translation (cf. Fig. 4 and Figs. 6 and 7). This apparent difference could be explained by assuming that if the formation of eIF4A–eIF4G–eIF3 complex is rate limiting, even its partial inhibition by apt4 would result in severe inhibition of translation. Alternatively, either site in eIF4G's M or C domain for binding to eIF4A might be functionally important and more sensitive to the inhibitory effect of the aptamer as compared with the other binding site in eIF4G.
The X-ray structure of the phylogenetically conserved M portion of human eIF4GII has been determined at 2.4 Å resolution, revealing a crescent-shaped domain consisting of 10 α-helices arranged as five HEAT repeats (Marcotrigiano et al. 2001). HEAT repeat proteins participate in a wide variety of cellular processes that are dependent on assembling large complexes (Andrade and Bork 1995). A previous genetic mutation analysis revealed that two distinct molecular surfaces support interactions with eIF4A and the EMCV IRES (Marcotrigiano et al. 2001). Since the majority of eIF4G mutations defective in eIF4A binding are normal in binding to IRES and vice versa, we speculate that apt4 binds to the intrarepeat face of the crescent structure that is responsible for recognition of eIF4A (Marcotrigiano et al. 2001). The observed simultaneous inhibition of eIF4A and eIF3 by apt4 suggests that the two binding sites are overlapping. This view is supported by the recent findings that short M-domain fragments of eIF4GI (positions 737–1010) and eIF4GII (positions 745–1003) retain binding capacity to eIF4A and eIF3 (Lomakin et al. 2000; Marcotrigiano et al. 2001).
Apt5 did not affect the affinity pull down of eIF4A–eIF4G–eIF3 complex in either the pure component system or in the RRL system. The interaction of eIF4G and eIF4E was not affected by apt5 since the amount of eIF4G pulled down with m7GTP-Sepharose resin was unaffected (see Fig. 7B). Although the interaction of apt5 with eIF4G is weaker than that of apt4, apt5 inhibited the translation more efficiently than apt4. These findings suggest the existence of an alternative, efficient step to inhibit translation, apart from blocking the interaction of eIF4G with eIF4A and eIF3. One possibility is that apt5 disables another factor for interacting, directly or indirectly, with eIF4G. Although the nature of this putative protein is not immediately obvious, it is a protein involved in cap-dependent translation not in the HCV IRES-driven translation. Alternatively, the apt5 binding with eIF4G is loaded into the initiation complex and causes a failure to form the functional complex. Further study is required to uncover the precise mechanism of translation inhibition by apt5.
Other aptamers—apt2, apt14, apt17, apt18, and apt19—failed to inhibit translation in the RRL even though their K d values to eIF4G are less than or roughly equal to those to apt5. Although motif II is conserved in apt2, apt17, and apt19, these five aptamer sequences and their predicted secondary structures are largely different from each other and from the structures of apt4 or apt5. Therefore, these independent isolates may bind to different regions in eIF4G in different fashions. All the aptamers isolated here might be useful for future studies of translation initiation and other intracellular processes.
MATERIALS AND METHODS
Protein expression and purification
Plasmid pGEX-6P (Amersham Bioscience) encoding human eIF4G712–1600 and the pET15b plasmid (Novagen) encoding human eIF4G712–1010 were reconstructed from constructs described previously (Morino et al. 2000). Plasmid pET15b encoding human eIF4AIII was reconstructed from a construct described previously (Li et al. 1999). GST-tagged eIF4G712–1600, GST-tagged eIF4G1080–1600, and his-tagged eIF4G712–1010 were expressed in Escherichia coli strain NovaBlue (Novagen) and BL21(DE3), respectively, and purified with glutathione-Sepharose 4B (GE Healthcare Bio-Sciences) and Ni-NTA agarose (QIAGEN), respectively, according to the manufacturers’ instructions. These samples were then applied to a Resource Q (GE Healthcare Bio-Sciences) column equilibrated with buffer A (20 mM Tris-HCl at pH 7.6, 80 mM potassium acetate, 2.5 mM magnesium acetate, 5% glycerol) and eluted by a 0%–100% gradient of buffer B (20 mM Tris-HCl at pH 7.6, 1 M potassium acetate, 5% glycerol) with ÄKTA (GE Healthcare Bio-Sciences). His-tagged human eIF4AIII was expressed using E. coli strain RosettaBlue (DE3) pLysS (Novagen) and purified using a Ni-NTA agarose column according to the manufacturer's instruction. His-tagged human eIF4AI was prepared as described previously (Oguro et al. 2003). Mouse eIF3 was purified from Krebs-2 ascites cells as described previously (Trachsel et al. 1979). GST-free eIF4G712–1600 protein was prepared from GST-tagged eIF4G712–1600 by digestion with PreScission protease (GE Healthcare Bio-Sciences). The activities of most of these recombinant proteins were confirmed by their binding ability to their relevant partners, such as efficient binding of GST-tagged eIF4G712–1600 and his-tagged eIF4G712–1010 to eIF4AI and eIF4AIII. A cDNA encoding eIF4G186–1600 was appended with a Flag sequence at the C terminus, and cloned in pGEX-6P to generate pGEX-6P-eIF4G186–1600-Flag. GST-eIF4G186–1600-Flag was bacterially expressed and purified with glutathione-Sepharose 4B (GE Healthcare Bio-Sciences) followed by anti-Flag chromatography (Sigma).
Selection-amplification of eIF4G aptamers
Preparation of the random 40N RNA pool and affinity RNA selection-amplification was carried out essentially as described previously (Oguro et al. 2003) except that the target protein was GST tagged and immobilized to glutathione-Sepharose resin instead of Ni-NTA agarose resin. The sequence of the first template was taatacgactcactatagggacacaatggacg-(40N)-taacggccgacatgagag, where 40N is 40 random nucleotide sequences. The initial RNA pool theoretically contained 1014 variants. The buffer used in selection was 80 mM potassium acetate, 2.5 mM magnesium acetate, 1 mM DTT, and 20 mM Tris (pH 7.6). eIF4G712–1600 was immobilized with glutathione-Sepharose resin and was eluted with the elution buffer containing the reduced glutathione (Sigma). After 12 rounds, cDNAs for selected RNAs were cloned into a plasmid pGEM-T Easy vector (Promega) and sequenced.
Filter retention assay
Filter-binding assays were employed using a nitrocellulose membrane (0.45 μm pore size; Millipore) essentially as described previously (Méthot et al. 1996). After 12 rounds, selected RNAs were labeled in an in vitro transcription reaction using [α-32P]CTP (800 Ci/mmol; Amersham Biosciences). The labeled RNA was incubated with GST-eIF4G712–1600, eIF4G712–1600, or eIF4G712–1010 in 50 μL of binding buffer containing 5% glycerol and 10 μg/mL tRNA for 30 min at room temperature. The samples were filtered and washed with the binding buffer. The retained radioactivity on the filter was quantitated by scintillation counting. RNA trapped in the absence of eIF4G under these conditions was negligible. The K d values were obtained from the equation RNAinitial/RNAtrapped = K d/[eIF4G]initial + 1, where RNAinitial is the initial amount of 32P-RNA, RNAtrapped the amount of 32P-RNA trapped on the filter, and [eIF4G]initial the initial concentration of eIF4G. It was assumed that all the aptamer–eIF4G complexes formed were trapped on the filter since the concentration of RNAs was much lower than the concentration of eIF4G.
Surface Plasmon Resonance assay
The SPR assays were performed according to the same coupling method as described previously (Wood 1993; Van Ryk and Venkatesan 1999) using a BIAcore 2000 instrument (BIAcore AB). The aptamer templates were amplified and tagged at the 3′-end with dA16 by PCR using 5′-template primer (5′-TAATACGACTCACTATAGGGACACAATGGACG-3′) and dT16 tagging 3′-template primer (5′-TTTTTTTTTTTTTTTTCTCTCATGTCGGCCGTTA-3′). 5′-Biotinylated dT16 oligomer was bound to the surface of the streptavidin sensor chip (BIAcore AB) of flow cells 1 and 2; 20 μg/mL 3′-A16-tagged RNA was immobilized to ∼800 RUs in flow cell 2 by complementary hybridization to the 5′-biotinylated dT16 oligomer in binding buffer at a flow rate of 20 μL/min at 25°C for 1 min by the INJECT program (BIAcore AB). A 0.5 μM solution of eIF4G712–1600 or eIF4G1080–1600 in binding buffer was passed through flow cells 1 and 2 of the sensor chip for 1 min at the KINJECT program (BIAcore AB), and bound proteins were gradually dissociated for 200 sec. The data are obtained by subtracting the signals for 5′-biotinylated dT16 fixed on the sensor chip (flow cell 1) from the signal for RNA aptamer (flow cell 2), thereby showing the net interaction between RNA and protein (Fig. 5). To regenerate the sensor chip, bound materials were completely removed by injecting 20 μL of 2 M urea at a flow rate of 20 μL/min. Close-fitting curves to the sensorgrams were calculated by global fitting curves (1:1 Langmuir binding) generated using BIAevaluation 3.0 software (BIAcore AB).
In vitro translation
Plasmid pcDNA3 CAT/HCV/LUC (Svitkin et al. 2001) was linearized with ApaI (Takara). A capped CAT/HCV/LUC mRNA was synthesized using RiboMAX in vitro transcription systems (Promega). In vitro translation was performed in the RRL (Promega) containing 100 mM potassium acetate and 0.5 mM magnesium acetate according to the manufacturer's instruction. RRL was pre-incubated at 30°C for 10 min with or without the aptamers. The cap-CAT/HCV/LUC mRNA and a mixture of [35S]methionine and [35S]cysteine (GE Healthcare Bio-Sciences) were then added. The mixtures were incubated at 30°C for 30 min, separated by SDS-PAGE. CAT and LUC translation products were quantified using a BAS-2000 PhosphorImager (FUJIFILM) and NIH image software. The percent residual CAT synthesis was established against the amount of CAT synthesized in the absence of the aptamer.
The same translation inhibition experiment was carried out with a 5′-capped LUC mRNA (cap-LUC), instead of the capped CAT/HCV/LUC mRNA. The cap-LUC mRNA was synthesized in the RiboMAX in vitro transcription systems (Promega) using LUC DNA (Promega).
Exogenous eIF4G reversed translational inhibition
In vitro translation was essentially performed according to the manufacturer's instruction provided by Promega. Mixtures of aptamers, RRL, RNase inhibitor, and RRL buffer were incubated at 30°C for 10 min. A recombinant eIF4G186–1600, cap CAT/HCV/LUC mRNA, a mixture of [35S]methionine and [35S]cysteine, and an amino acid mixture were then added. The final concentrations of apt4, apt5, and eIF4G186–1600 were 6 μM, 1 μM, and 0.63 μM, respectively. RRL buffer was added to adjust the final concentrations of potassium acetate and magnesium acetate to 100 mM and 0.5 mM, respectively. The mixtures were incubated at 30°C for 60 min and were analyzed by 12% SDS-PAGE followed by fluorography.
GST-tagged eIF4G pull-down assay
Thirty pmoles of molecules of GST-tagged human eIF4G712–1600 were added to glutathione-Sepharose resin (10 μL) for immobilization. Mixtures of the aptamers (0 ∼ 100 μM) and human eIF4AI (20 pmol), human eIF4AIII (40 pmol), or mouse eIF3 (6 pmol) were added to the immobilized eIF4G712–1600 and incubated for 30 min at room temperature. The final concentrations of eIF4AI, eIF4AIII, and eIF3 were 0.7, 1, and 0.2 μM, respectively. After washing with the binding buffer, the complexes were eluted with elution buffer containing reduced glutathione and were analyzed by SDS-PAGE followed by staining with CBB. For pull-down experiments of eIF3, the final concentration of apt4, apt5, and 40N was 20 μM. The relative efficiency of eIF4A binding to eIF4G in the presence of aptamers (compared with the absence of aptamers) was corrected by eIF4G immobilized.
m7GTP-sepharose pull-down assay
m7GTP-Sepharose pull-down was carried out essentially as described previously (Mochizuki et al. 2005). Briefly, mixtures composed of RRL (19 μL; Promega), aptamers (25 μL), RNase inhibitor (1 μL; Takara), amino acid mixture (1 μL; Promega), and RRL buffer (4 μL) were incubated at 30°C for 10 min and then incubated with m7GTP-Sepharose resin (10 μL; GE Healthcare Bio-Sciences) on ice for 2 h. The RRL buffer was added to adjust the final concentrations of potassium acetate and magnesium acetate to 100 mM and 0.5 mM, respectively. After washing with the binding buffer, proteins bound to m7GTP-Sepharose resin were pulled down and heated with SDS-sample buffer at 65°C for 15 min. The bound eIF3, eIF4AI, and eIF4G were recovered, separated by 7% SDS-PAGE, and transferred to the polyvinylidene difluoride (PVDF) membrane. The membrane was soaked with blocking buffer (5% non-fat dry milk, 10 mM Tris-HCl at pH 7.5, 100 mM NaCl, 0.1% Tween 20) for 30 min at room temperature, and incubated with anti-eIF4G rabbit polyclonal antibody (Wakiyama et al. 2000), anti-eIF4AI polyclonal antibody that had been raised in a guinea pig immunized with a recombinant eIF4AI described above (this study), and anti-p110 polyclonal antibody that had been raised in a rabbit immunized with GST-p110 (1–101) (this study), in buffer containing 20 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 0.05% Tween 20 for 1 h at room temperature. After repeated washing, the membrane was incubated with anti-mouse IgG sheep secondary antibody HRP conjugate (GE Healthcare Bio-Sciences), anti-rabbit IgG donkey secondary antibody HRP conjugate (GE Healthcare Bio-Sciences), or anti-guinea pig IgG rabbit secondary antibody HRP conjugate (Rockland) for 1 h at room temperature. The signal was detected by ECL reagent (GE Healthcare Bio-Sciences) according to the manufacturer's instruction and analyzed by LAS-1000 (FUJIFILM) using the NIH image software for quantification.
ACKNOWLEDGMENTS
We thank Colin Crist for critical reading of the manuscript and valuable comments. This work was supported in part by grants from the Ministry of Education, Sports, Culture, Science and Technology of Japan (MEXT) and the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NIBIO).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2169406.
REFERENCES
- Andrade, M.A., Bork, P. Heat repeats in the Huntington's disease protein. Nat. Genet. 1995;11:115–116. doi: 10.1038/ng1095-115. [DOI] [PubMed] [Google Scholar]
- Byrd, M.P., Zamora, M., Lloid, R.E. Generation of multiple isoforms of eukaryotic translation initiation factor 4GI by use of alternate translation initiation codons. Mol. Cell. Biol. 2002;22:4499–4511. doi: 10.1128/MCB.22.13.4499-4511.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraiuolo, M.A., Lee, C.S., Ler, L.W., Hsu, J.L., Costa-Mattioli, M., Luo, M.J., Reed, R., Sonenberg, N. A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc. Natl. Acad. Sci. 2004;101:4118–4123. doi: 10.1073/pnas.0400933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradi, A., Imataka, H., Svitkin, Y.V., Rom, E., Raught, B., Morino, S., Sonenberg, N. A novel functional human eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, H., van der Haar, T., Singh, C.R., Ii, M., Li, B., Hinnebusch, A.G., McCarthy, E.G., Asano, K. The yeast eukaryotic initiation factor 4G (eIF4G) HEAT domain interacts with eIF1 and eIF5 and is involved in stringent AUG selection. Mol. Cell. Biol. 2003;23:5431–5445. doi: 10.1128/MCB.23.15.5431-5445.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey, J.W.B, Merrick, W.C. Pathway and mechanism of initiation of protein synthesis. In: Sonenberg N., et al., editors. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 33–88. [Google Scholar]
- Imataka, H., Sonenberg, N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka, H., Gradi, A., Sonenberg, N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneeva, N.L., Lamphear, B.J., Hennigan, F.L.C, Rhoads, R.E. Mutually cooperative binding of eukaryotic translation initiation factor (eIF) 3 and eIF4A to human eIF4G-1. J. Biol. Chem. 2000;275:41369–41376. doi: 10.1074/jbc.M007525200. [DOI] [PubMed] [Google Scholar]
- Lejeune, F., Ranganathan, A.C., Maquat, L.E. eIF4G is required for the pioneer round of translation in mammalian cells. Nat. Struct. Mol. Biol. 2004;11:992–1000. doi: 10.1038/nsmb824. [DOI] [PubMed] [Google Scholar]
- Li, Q., Imataka, H., Morino, S., Rogers, G.W., Jr., Richter-Cook, N.J., Merrick, W.C., Sonenberg, N. Eukaryotic translation initiation factor 4AIII (eIF4AIII) is functionally distinct from eIF4AI and eIF4AII. Mol. Cell. Biol. 1999;19:7336–7346. doi: 10.1128/mcb.19.11.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin, I.B., Hellen, C.U.T, Pestova, T.V. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 2000;20:6019–6029. doi: 10.1128/mcb.20.16.6019-6029.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano, J., Lomakin, I.B., Sonenberg, N., Pestova, T.V., Hellen, C.U.T, Burley, S.K. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol. Cell. 2001;7:193–203. doi: 10.1016/s1097-2765(01)00167-8. [DOI] [PubMed] [Google Scholar]
- Méthot, N., Pickett, G., Keene, J.D., Sonenberg, N. In vitro RNA selection identifies RNA ligands that specifically bind to eukaryotic translation initiation factor 4B: The role of the RNA recognition motif. RNA. 1996;2:38–50. [PMC free article] [PubMed] [Google Scholar]
- Mochizuki, K., Oguro, A., Ohtsu, T., Sonenberg, N., Nakamura, Y. High affinity RNA for mammalian initiation factor 4E interferes with mRNA-cap binding and inhibits translation. RNA. 2005;11:77–89. doi: 10.1261/rna.7108205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morino, S., Imataka, H., Svitkin, Y.V., Pestova, T.V., Sonenberg, N. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol. Cell. Biol. 2000;20:468–477. doi: 10.1128/mcb.20.2.468-477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguro, A., Ohtsu, T., Svitkin, Y.V., Sonenberg, N., Nakamura, Y. RNA aptamers to initiation factor 4A helicase hinder cap-dependent translation by blocking ATP hydrolysis. RNA. 2003;9:394–407. doi: 10.1261/rna.2161303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Shatsky, I.V., Fletcher, S.P., Jackson, R.J., Hellen, C.U.T. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes & Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevot, D., Darlix, J-L, Ohlmann, T. Conducting the initiation of protein synthesis: The role of eIF4G. Biol. Cell. 2003;95:141–156. doi: 10.1016/s0248-4900(03)00031-5. [DOI] [PubMed] [Google Scholar]
- Pyronnet, S., Imataka, H., Gingras, A.-C., Fukunaga, R., Hunter, T., Sonenberg, N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits Mnk1 to phosphorylate eIF4E. EMBO J. 1999;18:270–279. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatkin, A.J. Capping of eukaryotic mRNAs. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- Shibuya, T., Tange, T., Sonenberg, N., Moore, M.J. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat. Struct. Mol. Biol. 2004;11:346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- Svitkin, Y.V., Pause, A., Haghighat, A., Pyronnet, S., Witherell, G., Belsham, G.J., Sonenberg, N. The requirement for eukaryotic initiation factor 4A (eIF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA. 2001;7:382–394. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun, S.Z., Jr., Sachs, A.B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Thiel, K. Oligo oligarchy—The surprisingly small world of aptamers. Nat. Biotechnol. 2004;22:649–651. doi: 10.1038/nbt0604-649. [DOI] [PubMed] [Google Scholar]
- Trachsel, H., Erni, B., Schreier, M.H., Braun, L., Staehelin, T. Purification of seven protein synthesis initiation factors from Krebs II ascites cells. Biochim. Biophys. Acta. 1979;561:484–490. doi: 10.1016/0005-2787(79)90156-4. [DOI] [PubMed] [Google Scholar]
- Tuerk, C., Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DAN polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Uchida, N., Hoshino, S., Imataka, H., Sonenberg, N., Katada, T. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in cap/poly(A)-dependent translation. J. Biol. Chem. 2002;277:50286–50292. doi: 10.1074/jbc.M203029200. [DOI] [PubMed] [Google Scholar]
- Van Ryk, D.I., Venkatesan, S. Real-time kinetics of HIV-1 Rev-Rev response element interactions. Definition of minimal binding sites on RNA and protein and stoichiometric analysis. J. Biol. Chem. 1999;274:17452–17463. doi: 10.1074/jbc.274.25.17452. [DOI] [PubMed] [Google Scholar]
- Wakiyama, M., Imataka, H., Sonenberg, N. eIF4G interaction with poly(A)-binding protein stimulates translation and is critical for Xenopus oocyte maturation. Curr. Biol. 2000;10:1147–1150. doi: 10.1016/s0960-9822(00)00701-6. [DOI] [PubMed] [Google Scholar]
- Wells, S.E., Hillner, P.E., Vale, R.D., Sachs, A.B. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- Wood, S.J. DNA-DNA hybridization in real time using BIAcore. Microchem. J. 1993;47:330–337. [Google Scholar]
- Zuker, M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]