Abstract
In pre-steady-state, fast-quench kinetic analysis, the tertiary-stabilized hammerhead ribozyme “RzB” cleaves its substrate RNA with maximal measured k obs values of ∼3000 min−1 in 1 mM Mn2+ and ∼780 min−1 in 1 mM Mg2+ at 37°C (pH 7.4). Apparent pKa for the catalytic general base is ∼7.8–8.5, independent of the corresponding metal hydrate pKa, suggesting potential involvement of a nucleobase as general base as suggested previously from nucleobase substitution studies. The pH-rate profile is bell-shaped for Cd2+, for which the general catalytic acid has a pKa of 7.3 ± 0.1. Simulations of the pH-rate relation suggest a pKa for the general catalytic acid to be ∼9.5 in Mn2+ and >9.5 in Mg2+. The acid pKa's follow the trend in the pKa of the hydrated metal ions but are displaced by ∼1–2 pH units in the presence of Cd2+ and Mn2+. One possible explanation for this trend is direct metal ion coordination with a nucleobase, which then acts as general acid.
Keywords: acid-base, divalent ions, hammerhead ribozyme, ribozyme kinetics
INTRODUCTION
Most natural ribozymes catalyze phosphate-group transfer or peptide-bond formation (Takagi et al. 2001; Fedor and Williamson 2005). The small (∼40–90 nucleotides [nt]), nucleolytic ribozymes—hairpin, hammerhead, hepatitis delta virus (HDV), Varkud satellite (VS), and glmS—generate 5′-hydroxyl and 2′,3′-cyclic phosphate termini because they utilize the adjacent 2′ hydroxyl as nucleophile in their cleavage mechanisms. Recent studies show that divalent metal ions can be dispensed with for several of the ribozymes. For example, in hammerhead ribozyme HH16.1, divalent metal ions can be replaced with high concentrations of monovalent ions, but reaction rates are decreased by 25-fold (Murray et al. 1998). For catalysis by the VS ribozyme (Murray et al. 1998; sequence from Guo and Collins 1995) in the presence of Li+ and Mg2+ together, a greater reaction rate was measured than when the same reaction was carried out in Mg2+ alone. For hairpin ribozymes, neither cleavage nor ligation requires a divalent metal ion (Nesbitt et al. 1997). The inference drawn from such results is that the divalent ions act via “delocalized” or nonspecific interactions to stabilize the functional fold of the molecule for these ribozymes.
The hammerhead ribozyme (HHRz) catalyzes site-specific RNA cleavage and ligation during the replication cycle of some plant viral satellite RNAs. While there is a wealth of information about the structure, sequence requirements, mechanism, and biotechnology applications of hammerhead ribozymes, many questions remain regarding the molecular basis for HHRz reactivity. The minimal structure consists of three helices that intersect at a conserved catalytic core of 11 nt. Crystallographic and solution studies reveal a Y-shaped dominant conformation (Pley et al. 1994; Tuschl et al. 1994; Scott et al. 1995; Penedo et al. 2004). Under standard in vitro assay conditions (10 mM Mg2+), a well-behaved, minimal hammerhead ribozyme can be expected to cleave its substrate at a rate of ∼1 min−1. These same ribozymes show poor intracellular activity due to their requirement for high concentrations of divalent metal ions. Recent studies highlighted the importance of peripheral sequences in the natural and engineered HHRzs for their improved in vitro activity under physiological Mg2+ conditions (<2 mM) and for their activity inside cells (De la Pena et al. 2003; Khvorova et al. 2003; Saksmerprome et al. 2004; Yen et al. 2004). Natural hammerhead ribozyme from schistosomes have shown exceptional cleavage rates (∼870 min−1) when assayed in vitro under reaction conditions that included very high Mg2+ concentrations (200 mM) and high pH (8.5) (Canny et al. 2004).
We previously described artificial tertiary stabilizing motifs (TSMs) in the context of trans-cleaving versions of natural HHRz derived from peach latent mosaic viroid (PLMVd) and from Schistosoma mansoni (Saksmerprome et al. 2004). In the present work, we have probed the catalytic mechanism of one of these artificial TSM-HHRzs, designated RzB (Fig. 1A) by examining the metal and pH dependence for self-cleavage of the ribozyme. Very high rates in the range 100–3000 min−1 are observed in submillimolar divalent ions, but only when the TSM is kept intact. This structural requirement highlights the importance of the tertiary interactions in enhancing the activity of RzB in low metal ion concentrations. The pH dependence of the reaction as a function of metal ion suggests a nucleobase-catalyzed acid-base mechanism, consistent with recent nucleobase substitution studies that supported G8 and G12 as general acid-base catalysts in the cleavage mechanism of a minimal hammerhead ribozyme (Han and Burke 2005).
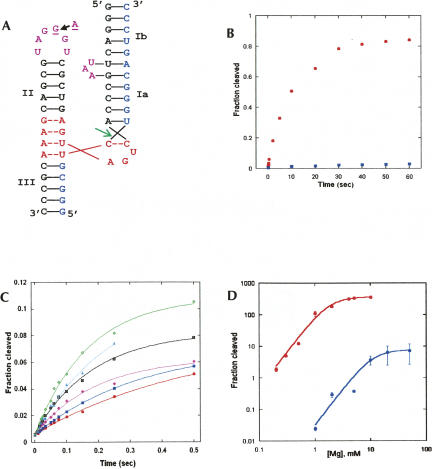
FIGURE 1.
Cleavage kinetics. (A) Predicted structures of RzB and RzB0. Red, catalytic core; blue, substrate strand; purple, ribozyme strand loop II and Bulge I; green arrow, cleavage site. The A-to-G mutation that arose during selection of RzB (11) is indicated within loop II. (B) Kinetic profile of RzB (red circles) and RzB0 (blue squares) in 1 mM Mg2+at 37°C (pH 7.4). (C) Millisecond kinetics of RzB at 37°C (pH 7.4) in 1 mM Mg2+ (red circles), 2 mM Mg2+ (blue squares), 4 mM Mg2+ (pink diamonds), 5 mM Mg2+ (black squares), 10 mM Mg2+ (green plus signs), and 20 mM Mg2+ (cyan triangles). (D) Mg2+-binding profiles of RzB (red circles) and RzB0 (blue circles). Reported error is the uncertainty of the fit of each plot as calculated in KaleidaGraph 3.5. Reactions for RzB in sub-millimolar magnesium and all data for RzB0 were manually controlled (Supplemental Fig. 1A).
RESULTS
Millisecond timescale cleavage in low Mg2+
The cis-cleaving HHRz from PLMVd exhibits rapid cleavage (k obs > 2 min−1) at submillimolar concentrations of magnesium, similar to the behavior observed for other naturally occurring hammerhead ribozymes (Khvorova et al. 2003; Canny et al. 2004; Saksmerprome et al. 2004). Upon extended incubation, the reaction approached 70% completion (Saksmerprome et al. 2004). Ribozyme RzB (Fig. 1A) is a trans-cleaving derivative of the PLMVd hammerhead ribozyme, in which loop II differs from the natural sequence by the introduction of a single A-to-G mutation, and the structural context of the tertiary contact is altered by converting the natural terminal loop I into a bulge within the ribozyme strand of stem I of RzB (Saksmerprome et al. 2004). Previous studies of this ribozyme using manual mixing methods demonstrated that RzB cleaves its target rapidly at submillimolar concentrations of magnesium at 37°C (pH 7.4). Furthermore, the reaction goes to a greater extent (>90%) than the parent molecule from which it was derived (Saksmerprome et al. 2004).
In manually controlled reactions in 1 mM Mg2+, RzB cleaves >80% of its substrate in the first 30 sec, while a control ribozyme RzB0 lacking tertiary stabilizing structural elements (UAA bulge in stem I is deleted) cleaved <2% of its substrate during this time (Fig. 1B). Therefore, to access the very earliest events in the reactions catalyzed by ribozyme RzB, initial cleavage rates were remeasured in rapid-quench reactions. Catalytic activity is vigorous on a millisecond timescale, with >10% substrate cleaved within the first 0.5 sec in 10 mM Mg2+ and ∼5% in 1 mM Mg2+ (Fig. 1C). Curve fitting with a single-exponential equation satisfactorily describes the kinetic behavior during the initial ∼0.5 sec (Fig. 1C). Increasing Mg2+ concentration yields an increase in the apparent first-order rate constant during this early phase until it saturates by 10 mM Mg2+ (k max = 376 ± 21 min−1) (Fig. 1D). The kinetic profile in the presence of Mg2+ is multiphasic when evaluated over the full time course, and cleavage during the second phase of the reaction (beyond 0. 5 sec) is not yet saturated at 10 mM Mg2+ (data not shown). The kinetic behavior of the control ribozyme, RzB0, was also biphasic at elevated Mg2+ concentrations (10–50 mM) (Supplemental Fig. 1B), and approaches saturation in the first phase at 50 mM Mg2+ (k max = 7.5 ± 0.3 min−1) (Fig. 1D).
The initial rate data for both ribozymes can be fit to a two-state model for Mg2+ binding, with Hill coefficients of 2.0 ± 0.3 for RzB and 2.2 ± 0.1 for RzB0 (Fig. 1D). The similarity in the Hill coefficients suggests a similar degree of cooperativity in Mg2+ binding by both constructs. Importantly, the concentration at which cleavage rate is half maximal, [Mg2+]1/2, decreases from 11.7 mM for RzB0 to only 1.9 mM for RzB. Interestingly, the [Mg2+]1/2 value for RzB is less than one-twentieth that observed for the natural Schistosoma hammerhead ribozyme (∼40 mM) (Canny et al. 2004).
Rapid cleavage in transition metals
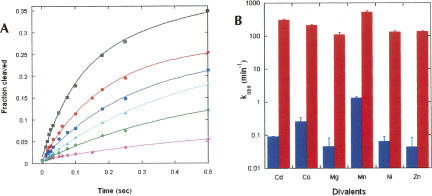
Previous studies of substrate cleavage by minimal hammerhead ribozymes demonstrated that 10 mM transition metals only moderately supported catalysis, yielding cleavage rates of <0.25 min−1 (Dahm and Uhlenbeck 1991; Dahm et al. 1993). In contrast, in the present study, substrate cleavage by hammerhead ribozyme RzB in each of several different transition metal ions (1 mM) gave cleavage rates of >100 min−1 at 37°C (pH 7.4) (Fig. 2A,B; Supplemental Fig. 2A). The greatest cleavage rates were observed for 1 mM Mn2+ (537 ± 57 min−1). Control ribozyme RzB0 behaved similarly to minimal ribozymes in previous studies (Dahm et al. 1993), yielding cleavage rates in the range of 0.04–1.34 min−1 depending on the divalent ion (Fig. 2B; Supplemental Fig. 2B). Tertiary stabilization therefore confers >400-fold activation relative to minimal hammerhead ribozymes at 1 mM transition metal ions.
FIGURE 2.
Metal ion dependence of RzB cleavage on a millisecond timescale. (A) Reactions carried out at 37°C (pH 7.4) in 1 mM Mg2+ (pink diamonds), 1 mM Ni2+ (green plus signs), 1 mM Zn2+ (aqua triangles), 1 mM Co2+ (blue squares), 1 mM Cd2+ (red circles), and 1 mM Mn2+ (black squares). Calculation of k obs was achieved by fitting each time course (through the first 0.5 sec) to a single-exponential equation, except the one for Mn2+, for which a double-exponential equation was used. (B) Bar graph of kobs values for RzB (red) and RzB0 (blue). Reported error for RzB is the uncertainty of the fit of each plot and for RzB0 (Supplemental Fig. 2B) was calculated from duplicate data sets.
Cleavage in alkaline earth metal ions
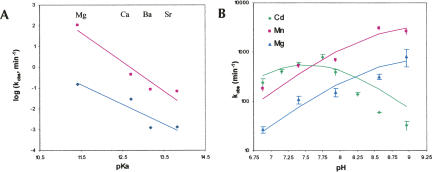
All of the transition metal ions tested support higher catalytic rates than those obtained in Mg2+ (Fig. 2A,B; Supplemental Fig. 2A). In contrast, the cleavage rates obtained in the group IIA (alkaline earth) metals were all much lower than those obtained in Mg2+, ranging from 0.072 ± 0.002 in 1 mM Ba2+ to 0.46 ± 0.02 in 1 mM Ca2+ (Fig. 3A). Two major differences between the alkaline earth metals and the transition metals are the absence of d-orbital electrons and the larger ionic radii in the alkaline earth metals. The absence of d-orbital electrons affects the pKa of the metal ion hydrate, while the larger ionic radius decreases the metal's ionic potential—defined as the ratio of the formal charge to the ionic radius of the ion. The latter effect decreases the net Coulombic interactions of the metal with neighboring ions through electrostatic interactions. No correlation was observed between activity and ionic potential for either alkaline earths or transition metals (data not shown). However, there was an inverse correlation between pKa of group IIA metal ion hydrates with RzB cleavage activity at pH 7.4 (slope of −1.4, R 2 = 0.92) (Fig. 3A). A slope of –0.95, R 2 = 0.86, was previously observed for a similar plot of pKa of the metal ion hydroxide vs. log(k obs) for a minimal hammerhead ribozyme (Fig. 3A; Dahm and Uhlenbeck 1991; Dahm et al. 1993). The negative slopes in the above plots could be explained either by the reduced concentration of the Brønsted base form of the metal hydrate ion, or by the known reduction in Lewis acidity (inversely related to pKa) of the heavier metals within a group in the periodic table. The pH dependence of the cleavage reaction (below) support the latter model.
FIGURE 3.
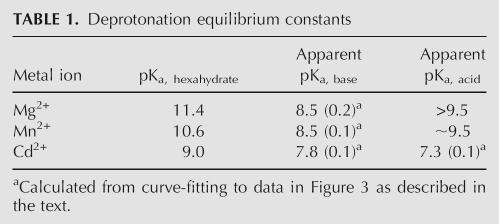
RzB cleavage rate dependence on group IIA metal ions and pH. (A) Hammerhead cleavage rates in group IIA metals as a function of their pKa. RzB measurements were made in 1 mM metal ions at 37°C (pH 7.4) (pink squares; fit to linearity, R 2 = 0.92), and minimal hammerhead (data taken from Dahm and Uhlenbeck 1991) data were taken in 10 mM metal ions at 25°C (pH 7.5) (indigo diamonds; fit to linearity, R 2 = 0.86). (B) pH-rate profile of RzB at 37°C, in 1 mM Mg2+ (blue triangles), Mn2+ (pink squares), or Cd2+ (green diamonds). All experiments were performed in the quench flow device, and all reactions included 10 μM EDTA and 50 mM Tris buffer at the respective pH. Individual kinetic traces are shown in Supplemental Figure 3 (A–C). Values for pKa were calculated using Equation 4 or Equation 5 (see Materials and Methods) to be 8.5 ± 0.2 (k max = 975 ± 179 min−1) for Mg2+ and 8.5 ± 0.1 (k max = 4380 ± 594 min−1) for Mn2+. The two pKa values for Cd2+ were calculated to be 7.8 ± 0.1 and 7.3 ± 0.1 (k max = 3870 ± 589 min−1). The interpretation given in the text assumes that the higher pKa value represents the general base for the reaction.
Dependence of rate on pH
To determine whether the rapid cleavage observed in transition metals is linked to their pKa values, we examined the pH dependence of RzB activity in three divalent ions whose pKa values differ from each other by ∼1 unit: 11.4 for Mg2+, 10.6 for Mn2+and 9.0 for Cd2+ (Fig. 3B; Supplemental Fig. 3A–C). When substrate cleavage by RzB was monitored over the pH range of 6.88–8.96 in the presence of either 1 mM Mg2+ or 1 mM Mn2+, the initial rate of cleavage increased log-linearly with pH with a slope of 0.78. Such a pH-rate correlation is expected if a single proton transfer is involved in the rate-limiting step that is being monitored. Fitting the pH-rate profile to a theoretical curve corresponding to a single proton transfer event (Bevilacqua 2003) yields pKa values close to 8.5 for both the Mg2+-dependent (8.5 ± 0.2) and the Mn2+-dependent (8.5 ± 0.1) reactions (Table 1). We assign this pKa to the general base on the assumption that higher pH would populate the deprotonated form of this active site moiety and make it more available to abstract a proton, while acknowledging that there is intrinsic ambiguity in this assignment (Bevilacqua 2003). The fact that both reactions share a common pKa value that differs markedly from the pKa of the metal ion hydrates suggest that the metal ion hydrates are not acting directly as general base in these reactions. In 1 mM Mn2+ at pH 8.56, ribozyme RzB achieves an unprecedented k obs value of ∼51 sec−1 (3060 ± 260 min−1). Very rapid cleavage is also observed in 1 mM Mg2+ (pH 8.96), where the cleavage rate exceeds 13 sec−1 (810 ± 320 min−1 at pH 8.96). Previous work on hammerhead and VS ribozymes reported rates as high as 600–800 min−1 in 200 mM Mg2+ (pH 8.5) (Canny et al. 2004; Zamel et al. 2004). The rates that we obtained here for RzB were measured in much lower concentrations of divalent metal ions, yet they are as fast as, or faster than, the catalytic rates of any previously characterized ribozyme.
TABLE 1.
Deprotonation equilibrium constants
The pH-rate profile of RzB in the presence of Cd2+ exhibits the classical bell shape indicative of two simultaneous protonation–deprotonation events taking place in the transition state (Fig. 3B). Such behavior is well known in protein enzymes such as ribonuclease A, where one active site histidine acts as general base to deprotonate the 2′ hydroxyl, and a second histidine acts as general acid to protonate the leaving group 5′ hydroxyl (Raines 1998; Pinard et al. 2001). The pH-rate profile of RzB in the presence of Cd2+ was therefore fit to a theoretical curve corresponding to two proton transfer events (Fig. 3B). The pKa of the general base (caveat vide supra) in Cd2+-activated catalysis (7.8 ± 0.1) (Table 1) is again different from that of the hydrated ion, and it is similar (within fivefold hydroxide ion concentration) to the pKa value of the general base obtained in the presence of the other two metals. The recurrence of a common pKa value for the general base, in the presence of three different cations, support a model in which the role of the general base is played by a chemical moiety other than a hydrated metal ion, such as a nucleobase functional group.
The pKa of the general acid in Cd2+-activated catalysis is 7.3 ± 0.1, which is 1.7 pKa units lower than the pKa of the metal hydrate. The pH activity profiles in the presence of Mg2+ and Mn2+ do not exhibit any bell-shaped features (Fig. 3B). Cleavage rates begin to plateau at high pH (8.56) in Mn2+, likely due to a combination of saturation of an active site residue and the beginnings of alkaline denaturation. Simulations of pH-rate plots suggest the pKa of the general acid to be >9.5 in Mg2+ and ∼9.5 in Mn2+ (data not shown). These values are difficult to confirm experimentally because nonspecific deprotonation denatures RNA at high pH (>9). The trend in the pKa value of the general acid in different metals follows the same trend as the pKa of the individual metal hydrates, but it is lower by ∼1–2 pH units in the cases of Cd2+ and Mn2+.
DISCUSSION
Tertiary stabilization markedly affects hammerhead ribozyme kinetic behavior and metal ion sensitivity. The Mg2+ binding isotherms of RzB and RzB0 (Fig. 1D) suggest a requirement for approximately two Mg2+ ions in order to access the transition state for both ribozymes (Hill coefficients of 2 and 2.2). The large difference in observed rate constants (k max = 375 min−1 vs. 7 min−1 at pH 7.4) (Fig. 1D) suggests that these two molecules may experience different rate-limiting steps. For example, tertiary stabilization may place RzB into a predominant conformation which is further along the reaction pathway. The rate-determining step could then be either the chemical step itself or an active-site rearrangement of low activation energy, such as a rigid-body rotation of helix I, accompanied by alignment of the 2′ oxygen for in-line attack on the scissile phosphate, as previously suggested by Dunham and Scott (Dunham et al. 2003). In the case of RzB0, the slow rate may be due to an on- or off-pathway conformational change of high activation barrier, which saturates at 7.5 ± 0.3 min−1 and requires much higher Mg2+ ([Mg2+]1/2 ∼12 mM).
Ribozyme-catalyzed RNA cleavage requires deprotonation of the attacking 2′ hydroxyl by a general base and protonation of the leaving group 5′ oxyanion by a general acid. The hammerhead ribozyme appears to employ both proton transfer strategies. The pH-dependent activities in Mg2+, Cd2+, and Mn2+ indicate a common general base of pKa ∼7.8–8.5 (Table 1), which is distinct from the pKa values of all three of the metal ion hydrates. This observation would seem to rule out a model in which the decreased reactivity observed for larger divalent alkaline earth metal ions (Fig. 3A) is caused by a decrease in the concentration of the Brønsted base form; decreased Lewis acidity is the more likely explanation (see Results). It is possible that the general acids for the reactions promoted by Mg2+, Cd2+, and Mn2+ are provided by the corresponding metal ion hydrates and that their pKa's are lowered by the local environment. However, there is mounting evidence that small ribozymes use nucleotide bases for their catalytic chemistry (Nakano and Bevilacqua 2001; Pinard et al. 2001; Collins 2002; Jones and Strobel 2003; Nakano et al. 2003; Bevilacqua et al. 2004, 2005; Han and Burke 2005). For example, in previous studies of a minimal HHRz (Han and Burke 2005), simultaneous substitution of G8 and G12 with 2,6-diaminopurine (diAP) (pKa 5.1 vs. pKa 9.2 for G) yielded a bell-shaped pH-rate profile, implicating these two nucleobases in both proton transfer steps. Although these two Guanosine bases are 10–15 Å away from the cleavage site in ground-state crystal structures (Scott et al. 1995; Wedekind and McKay 1998; Blount and Uhlenbeck 2005), they can form an intramolecular crosslink with bases spanning the cleavage site (Heckman et al. 2005), providing evidence for a large-scale conformational change wherein G8 and G12 stack on the cleavage site bases. In RzB catalysis, the role of a general base can be fulfilled by either of the two guanosines. If the other guanosine serves as general acid, our observation that the pKa of the general acid varies with the metal ion used in the reaction may suggest perturbation of that guanosine's pKa by a direct (inner sphere) coordination of the metal ion to the nucleobase or by the close proximity of the metal ion's positive charge. The metal ion's proximity would be particularly stabilizing for the anionic form of the guanosine formed upon contributing a proton to the leaving group (general acid in the reaction) or positioned to abstract a proton from the attacking nucleophile (general base in the reaction), independent of the metal ion's pKa. Confirmation of this model awaits high-resolution structural data or alternative support.
It appears that different classes of metals promote catalysis by following different catalytic pathways. Mg2+ and the transition metals promote cleavage at higher rates by following a catalytic route involving a transition state with a low activation energy barrier. The rapid catalysis observed for the tertiary stabilized RzB in the presence of Mg2+ and the transition metals (k obs ≫ 100 min−1 in 1 mM cations) support a special structural or catalytic role for divalent metal ions, such as participation in the chemistry step indirectly through electrostatic interaction or coordinate bond formation with the general acid, G8 or G12. Proximity of divalent ions to these nucleobases has been seen for both the natural and minimal hammerhead molecules (A9/G10.1 metal binding site) (Scott et al. 1995; Wang et al. 1999; Hunsicker and DeRose 2000; Maderia et al. 2000; Hampel and Burke 2003; Vogt et al. 2003). The slower catalysis observed in Ca2+, Sr2+, and Ba2+ (Fig. 3A) and in Na+, spermidine, and cobalt hexammine (M. Roychowdhury-Saha and D.H. Burke, in prep.), may reflect a high-energy barrier conformational change that accompanies acquisition of the transition state. On the other hand, because these ions cannot form strong inner sphere coordination with guanosine, the low catalytic rates that they promote may simply result from loss of a catalytic mechanism by failure of the general acid to protonate the 5′ oxyanion leaving group.
SUPPLEMENTAL MATERIAL
Kinetic traces of RzB and RzB0 cleavage as a function of Mg2+ concentration, kinetic traces in various divalent metal ions for RzB at 10–100 msec and for RzB0 up to 10 min, and kinetic traces at various pH values for RzB in 1 mM Mg2+, Mn2+, or Cd2+ are available upon request as Supplemental Material by sending an e-mail message containing the keyword “RNA_06_RzB_Supplementary” to the corresponding author.
MATERIALS AND METHODS
Materials
DNA and RNA oligonucleotides were purchased from Integrated DNA Technologies; radiolabeled nucleotides, from ICN; and dNTPs, from Amersham Pharmacia. All other chemicals were purchased from Sigma-Aldrich at the highest purity grade available.
Methods
Kinetic measurements were performed under single-turnover conditions (40:1 excess ribozyme over substrate) by assembling reactive complexes from separately prepared ribozyme and substrate strands. Ribozyme strands were synthesized by in vitro transcription using T7 RNA polymerase from Ampliscribe kit following the supplier's protocol. Substrate strand was synthesized chemically by Integrated DNA Technologies, 5′ radiolabeled using [γ-32P] ATP and polynucleotide kinase, and gel-purified.
Unless noted otherwise, cleavage reactions were performed on a rapid quench flow device (Kintek RQF-3). Reactions were initiated by rapidly (<0.2 msec) mixing equal volumes (∼15 μL) of preassembled hammerhead complexes from chamber A (2 μM ribozyme strand + 50 nM substrate strand in 50 mM Tris·HCl and 10 μM EDTA at pH 7.4) with metal ion solution from chamber C (metal ions at 2× final concentration in 50 mM Tris·HCl and 10 μM EDTA at pH 7.4). Each solution was equilibrated separately for ∼1 min at 37°C in the sample loading loops prior to mixing. Reactions were stopped with 90 μL of quench solution (90% formamide, 50 mM EDTA, 0.005% each of xylene cyanol and bromophenol blue) supplied from chamber B. Samples corresponding to each reaction time were collected independently, electrophoresed on 15% denaturing polyacrylamide gel to separate the cleaved product band from the full-length substrate band, and exposed to a PhosphorImager screen. Band intensities were quantified by using ImageQuant software (Molecular Dynamics). The fraction cleaved (Ft) at time t was fit using KaleidaGraph 3.5 either to a single-exponential equation:
or to a bi-exponential equation,
where F 0 is the zero-point correction, F ∞ is the estimated plateau value at infinite time, α is the fraction of the cleaved population with a rate constant of k obs,1, and (1−α) is the fraction cleaved with a rate constant of k obs,2. Uncertainties were calculated either from curve fit or from analysis of duplicate measurements as indicated in the respective figure legends. Manually controlled cleavage reactions for times >10 sec were carried out under the same ribozyme–substrate complex concentrations as in quench flow experiments following the protocol as described previously (Saksmerprome et al. 2004). For kinetic traces shown in Figure 1, data were gathered to 1–2 sec. To determine initial rates, data were fit to single-exponential kinetic model (Equation 1) through 0.5 sec (0.25 sec in the case of 20 mM Mg2+), after which a second, slower phase became apparent. Data were fit to
using KaleidaGraph 3.5, where [Mg1/2] = K D (1/n). In the case of RzB, the Mg2+ binding equation was fit to k obs values observed over the range of 0.2–10 mM Mg2+.
The studies of pH dependence were done at 37°C in the presence of 1 mM divalent metal ions in 50 mM Tris.HCl buffer (pH 6.88–8.96), 10 μM EDTA. For reactions to be carried out at pH values higher than 8.5, fresh Mn2+ stock solutions (2 mM) in the pH buffer were prepared for each time point to minimize the formation of insoluble MnO2. These solutions were incubated at 37°C in the loading loop of the quench-flow apparatus for 1 min before the start of the reaction. Similarly, for pH values higher than 8.25, fresh Cd2+ stock solutions were prepared to minimize the insoluble Cd(OH)2 formation. For Mg2+ and Mn2+ reactions, the data were fit to
For Cd2+, the data were fit to
ACKNOWLEDGMENTS
Venkat Gopalan and Hsin-Yue Tsai at Ohio State University helped train M.R.S. on the use of the quench flow apparatus. Sugata Roy Chowdhury provided technical assistance. The quench flow apparatus was purchased with funds from NIH grant AI062513 and from NASA Exobiology Award NAG5-12360 to D.H.B.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.128906.
Abbreviations: HHRz, hammerhead ribozyme; nt, nucleotides, TSM- tertiary stabilizing motif.
REFERENCES
- Bevilacqua, P.C. Mechanistic considerations for general acid-base catalysis by RNA: Revisiting the mechanism of the hairpin ribozyme. Biochemistry. 2003;42:2259–2265. doi: 10.1021/bi027273m. [DOI] [PubMed] [Google Scholar]
- Bevilacqua, P.C., Brown, T.S., Nakano, S., Yajima, R. Catalytic roles for proton transfer and protonation in ribozymes. Biopolymers. 2004;73:90–109. doi: 10.1002/bip.10519. [DOI] [PubMed] [Google Scholar]
- Bevilacqua, P.C., Brown, T.S., Chadalavada, D., Lecomte, J., Moody, E., Nakano, S. Linkage between proton binding and folding in RNA: Implications for RNA catalysis. Biochem. Soc. Trans. 2005;33:466–470. doi: 10.1042/BST0330466. [DOI] [PubMed] [Google Scholar]
- Blount, K.E., Uhlenbeck, O.C. The structure–function dilemma of the hammer head ribozyme. Annu. Rev. Biophys. Biomol. Struct. 2005;34:415–440. doi: 10.1146/annurev.biophys.34.122004.184428. [DOI] [PubMed] [Google Scholar]
- Canny, M.D., Jucker, F.M., Kellogg, E., Khvorova, A., Jayasena, S.D., Pardi, A. Fast cleavage kinetics of a natural hammerhead ribozyme. J. Am. Chem. Soc. 2004;126:10848–10849. doi: 10.1021/ja046848v. [DOI] [PubMed] [Google Scholar]
- Collins, R.A. The Neurospora Varkud satellite ribozyme. Biochem. Soc. Trans. 2002;30:1122–1126. doi: 10.1042/bst0301122. [DOI] [PubMed] [Google Scholar]
- Dahm, S.C., Uhlenbeck, O.C. Role of divalent metal-ions in the hammerhead RNA cleavage reaction. Biochemistry. 1991;30:9464–9469. doi: 10.1021/bi00103a011. [DOI] [PubMed] [Google Scholar]
- Dahm, S.C., Derrick, W.B., Uhlenbeck, O.C. Evidence for the role of solvated metal hydroxide in the hammerhead cleavage mechanism. Biochemistry. 1993;32:13040–13045. doi: 10.1021/bi00211a013. [DOI] [PubMed] [Google Scholar]
- De la Pena, M., Gago, S., Flores, R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003;22:5561–5570. doi: 10.1093/emboj/cdg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham, C.M., Murray, J.B., Scott, W.G. A helical twist-induced conformational switch activates cleavage in the hammerhead ribozyme. J. Mol. Biol. 2003;332:327–336. doi: 10.1016/s0022-2836(03)00843-x. [DOI] [PubMed] [Google Scholar]
- Fedor, M.J., Williamson, J.R. The catalytic diversity of RNAs. Nat. Rev. Mol. Cell Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- Guo, H.C.T., Collins, R.A. Efficient trans-cleavage of a stem-loop RNA substrate by a ribozyme derived from neurospora VS RNA. EMBO J. 1995;14:368–376. doi: 10.1002/j.1460-2075.1995.tb07011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel, K.J., Burke, J.M. Solvent protection of the hammerhead ribozyme in the ground state: Evidence for a cation-assisted conformational change leading to catalysis. Biochemistry. 2003;42:4421–4429. doi: 10.1021/bi020659c. [DOI] [PubMed] [Google Scholar]
- Han, J., Burke, J.M. Model for general acid-base catalysis by the hammerhead ribozyme: pH-activity relationships of G8 and G12 variants at the putative active site. Biochemistry. 2005;44:7864–7870. doi: 10.1021/bi047941z. [DOI] [PubMed] [Google Scholar]
- Heckman, J.E., Lambert, D., Burke, J.M. Photocrosslinking detects a compact, active structure of the hammerhead ribozyme. Biochemistry. 2005;44:4148–4156. doi: 10.1021/bi047858b. [DOI] [PubMed] [Google Scholar]
- Hunsicker, L.M., DeRose, V.J. Activities and relative affinities of divalent metals in unmodified and phosphorothioate-substituted hammerhead ribozymes. J. Inorg. Biochem. 2000;80:271–281. doi: 10.1016/s0162-0134(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Jones, F.D., Strobel, S.A. Ionization of a critical adenosine residue in the Neurospora Varkud Satellite ribozyme active site. Biochemistry. 2003;42:4265–4276. doi: 10.1021/bi020707t. [DOI] [PubMed] [Google Scholar]
- Khvorova, A., Lescoute, A., Westhof, E., Jayasena, S.D. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- Maderia, M., Hunsicker, L.M., DeRose, V.J. Metal–phosphate interactions in the hammerhead ribozyme observed by P-31 NMR and phosphorothioate substitutions. Biochemistry. 2000;39:12113–12120. doi: 10.1021/bi001249w. [DOI] [PubMed] [Google Scholar]
- Murray, J.B., Seyhan, A.A., Walter, N.G., Burke, J.M., Scott, W.G. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- Nakano, S., Bevilacqua, P.C. Proton inventory of the genomic HDV ribozyme in Mg(2+)-containing solutions. J. Am. Chem. Soc. 2001;123:11333–11334. doi: 10.1021/ja0166850. [DOI] [PubMed] [Google Scholar]
- Nakano, S., Cerrone, A.L., Bevilacqua, P.C. Mechanistic characterization of the HDV genomic ribozyme: Classifying the catalytic and structural metal ion sites within a multichannel reaction mechanism. Biochemistry. 2003;42:2982–2994. doi: 10.1021/bi026815x. [DOI] [PubMed] [Google Scholar]
- Nesbitt, S., Hegg, L.A., Fedor, M.J. An unusual pH-independent and metal-ion-independent mechanism for hairpin ribozyme catalysis. Chem. Biol. 1997;4:619–630. doi: 10.1016/s1074-5521(97)90247-7. [DOI] [PubMed] [Google Scholar]
- Penedo, J.C., Wilson, T.J., Jayasena, S.D., Khvorova, A., Lilley, D.M. Folding of the natural hammerhead ribozyme is enhanced by interaction of auxiliary elements. RNA. 2004;10:880–888. doi: 10.1261/rna.5268404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinard, R., Hampel, K.J., Heckman, J.E., Lambert, D., Chan, P.A., Major, F., Burke, J.M. Functional involvement of G8 in the hairpin ribozyme cleavage mechanism. EMBO J. 2001;20:6434–6442. doi: 10.1093/emboj/20.22.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pley, H.W., Flaherty, K.M., McKay, D.B. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- Raines, R.T. Ribonuclease A. Chem. Rev. 1998;98:1045–1066. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- Saksmerprome, V., Roychowdhury-Saha, M., Jayasena, S., Khvorova, A., Burke, D.H. Artificial tertiary motifs stabilize trans-cleaving hammerhead ribozymes under conditions of submillimolar divalent ions and high temperatures. RNA. 2004;10:1916–1924. doi: 10.1261/rna.7159504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, W.G., Finch, J.T., Klug, A. The crystal structure of an all-RNA hammerhead ribozyme: A proposed mechanism for RNA catalytic cleavage. Cell. 1995;81:991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
- Takagi, Y., Warashina, M., Stec, W.J., Yoshinari, K., Taira, K. Recent advances in the elucidation of the mechanisms of action of ribozymes. Nucleic Acids Res. 2001;29:1815–1834. doi: 10.1093/nar/29.9.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuschl, T., Gohlke, C., Jovin, T.M., Westhof, E., Eckstein, F. A three-dimensional model for the hammerhead ribozyme based on fluorescence measurements. Science. 1994;266:785–789. doi: 10.1126/science.7973630. [DOI] [PubMed] [Google Scholar]
- Vogt, M., Hunsicker, L., Cosper, N., Scott, R., DeRose, V.J. Spectroscopic probes for metals and catalysis in the hammerhead ribozyme. Biophys. J. 2003;84:180A–181A. [Google Scholar]
- Wang, S.L., Karbstein, K., Peracchi, A., Beigelman, L., Herschlag, D. Identification of the hammerhead ribozyme metal ion binding site responsible for rescue of the deleterious effect of a cleavage site phosphorothioate. Biochemistry. 1999;38:14363–14378. doi: 10.1021/bi9913202. [DOI] [PubMed] [Google Scholar]
- Wedekind, J.E., McKay, D.B. Crystallographic structures of the hammerhead ribozyme: Relationship to ribozyme folding and catalysis. Annu. Rev. Biophys. Biomol. Struct. 1998;27:475–502. doi: 10.1146/annurev.biophys.27.1.475. [DOI] [PubMed] [Google Scholar]
- Yen, L., Svendsen, J., Lee, J.S., Gray, J.T., Magnier, M., Baba, T., D'Amato, R.J., Mulligan, R.C. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature. 2004;431:471–476. doi: 10.1038/nature02844. [DOI] [PubMed] [Google Scholar]
- Zamel, R., Poon, A., Jaikaran, D., Andersen, A., Olive, J., De Abreu, D., Collins, R.A. Exceptionally fast self-cleavage by a Neurospora Varkud satellite ribozyme. Proc. Natl. Acad. Sci. 2004;101:1467–1472. doi: 10.1073/pnas.0305753101. [DOI] [PMC free article] [PubMed] [Google Scholar]