Abstract
Two distinct functions have been proposed for the serine–arginine (SR)-rich family of splicing factors. First, SR proteins are essential splicing factors and are thought to function by mediating protein–protein interactions within the intron during spliceosome assembly. Second, SR proteins bind to exonic enhancer sequences and recruit spliceosome components to adjacent introns. The latter activity is required for splice-site recognition and alternative splicing. Until now it has not been possible to determine whether the requirement for SR proteins in the basic splicing reaction is a secondary consequence of their exon-dependent recruitment function. Here we show that RNA substrates containing only 1 nt of exon sequence can undergo the first step of the splicing reaction in vitro and that this activity requires SR proteins. Thus, we provide direct evidence that SR proteins have both exon-independent and exon-dependent functions in pre-mRNA splicing.
The accurate removal of introns from metazoan pre-mRNAs requires the recognition of weakly conserved splice sites by components of the spliceosome (1–3). A family of essential splicing factors characterized by the presence of a serine–arginine (SR)-rich domain are critical components of this recognition process. SR proteins bind to specific RNA sequences through one or more RNA-binding domains and interact with other splicing factors through an arginine–serine (RS)-rich protein-interaction domain (4, 5).
When initially identified, SR proteins were shown to act early in the splicing reaction (6–11) and to be required for the earliest step in spliceosome assembly (12). However, SR proteins also are thought to act later in the splicing reaction, possibly in the recruitment of the U4/6⋅5 tri-snRNP complex to the spliceosome (13). In vitro complementation studies revealed that high concentrations of SR proteins could substitute for the functions of U1 snRNP or the essential splicing factor U2AF, which bind to the 5′ and 3′ splice sites, respectively (14–17). These studies led to the proposal that SR proteins can functionally substitute for U1 snRNP or U2AF by directly binding to the 5′ splice site and recruiting U2 snRNP to the spliceosome and/or by mediating other protein–protein interactions during spliceosome assembly. These proposals were based on two observations. First, the SR protein SF2/ASF was shown to promote the binding of U1 snRNP to pre-mRNA (18, 19) and to bind to a 5′ splice site in vitro (20). Second, SR proteins were shown to interact with each other and with specific spliceosomal proteins in the yeast two-hybrid system and in vitro (18, 21, 22). The observation that SR proteins could interact simultaneously with the U1 snRNP 70-kDa protein and the 35-kDa subunit of U2AF in the yeast two-hybrid system was consistent with the proposal that SR proteins mediate interactions between factors bound to the 5′ and 3′ splice sites (21, 23). However, the observation that SR proteins bind specifically to the 5′ site is controversial, and there is no direct evidence that SR proteins mediate protein–protein interactions within the intron.
The conclusion that SF2/ASF binds specifically to 5′ splice sites in vitro (18, 20) was not supported by subsequent studies of other splicing substrates (19). Moreover, a systematic RNA–protein cross-linking study carried out with functional splicing complexes detected SR proteins only within exons, not within the intron or at the 5′ or 3′ splice site (24). Thus, it is unclear whether SR proteins bind to a specific intron sequence in a functionally significant manner. In addition, it is difficult to envision how SF2/ASF could bind to the 5′ splice site and promote the recruitment of U1 snRNP to the same sequence. The simultaneous binding of the two components to the 5′ splice site seems unlikely.
Although SR protein interactions have been investigated extensively, the functional significance of these interactions between SR-containing proteins within the intron has not been demonstrated. In fact, the recent identification of an alternative set of 5′–3′ splice-site-bridging proteins in yeast and mammals raised doubts concerning the role of SR proteins within the intron (25, 26). Because the conservation of splicing factors between budding yeast and metazoa is remarkably high, the yeast spliceosome has been viewed as a prototype of the metazoan splicing machinery. However, there are no obvious yeast orthologs of either SR proteins or U2AF35 (27), essential components of the SR protein-bridging model. Because exonic enhancers and SR proteins are unknown in budding yeast, it was suggested that SR proteins may function exclusively in exon-dependent splicing in metazoa (25).
The role of SR proteins in exon-dependent splicing is better established (see refs. 5 and 28–30 for reviews). SR proteins bind to exonic splicing enhancers (31–33) and recruit the splicing machinery to the adjacent intron through protein–protein interactions (33–35). Exonic splicing enhancers initially were thought to function exclusively in pre-mRNAs that are alternatively spliced (see ref. 28 for review). However, recent studies have revealed the presence of SR protein-binding sites within the exons of constitutively spliced pre-mRNAs (24, 36), and these binding sites can function as splicing enhancers (36). It is likely that SR proteins bind to sequences found in most, if not all, exons in constitutively spliced pre-mRNAs. Thus, because all of the pre-mRNAs used for in vitro splicing studies with SR proteins contain exon sequences, the possibility that the SR proteins function entirely through the exon-dependent recruitment pathway cannot be ruled out. If this is the case, sequences within metazoan introns may not be sufficient for recognition by the spliceosome.
To address this possibility we synthesized and tested splicing substrates containing as little as a single nucleotide. Our substrate design was based on the observation that a pre-mRNA lacking the 3′ splice site AG and the downstream exon can undergo the first step of the splicing reaction: cleavage at the 5′ splice site and lariat formation (37, 38). Thus, lariat formation does not require the downstream exon, so the role of the upstream exon in 5′ splice-site recognition and the requirement for SR proteins can be examined. Remarkably, an RNA substrate containing only 1 nt of 5′ exon sequence and the downstream intron is capable of undergoing the first step of the splicing reaction in vitro, and this reaction requires SR proteins. Thus, we provide direct, functional evidence that SR proteins are required for the splicing reaction independent of their ability to bind exon sequences.
MATERIALS AND METHODS
RNA.
The template for the AdML substrate lacking the 3′ splice site AG and the downstream exon was synthesized by PCR as described (37). All other AdML-derived substrate templates (31 nt, 15 nt, 9 nt, 9 nt scrambled, 6 nt, 3 nt, 2 nt, or 1 nt of the upstream exon but lacking the downstream exon and the 3′ splice site) were generated similarly by PCR using pAdMLpar (a gift from R. Reed, Harvard Medical School) and the appropriate set of primers (primer sequences are available upon request). PCR was also used to generate the templates for the β-globin and ftz minimal substrates containing 6 and 3 nt, respectively, of the upstream exon but lacking the 3′ splice site AG and the downstream exon. 32P-labeled RNAs were synthesized either with T7 RNA polymerase or with T4 polynucleotide kinase and gel-purified before use. Recent experiments demonstrated that T7 RNA polymerase produces 5′-end heterogeneity during in vitro transcription from certain templates (39). To test for 5′ heterogeneity substrates containing 1 or 2 exon nucleotides (Fig. 1A) were synthesized by T7 RNA polymerase in the presence of [γ-32P]GTP, PAGE-purified, and subjected to complete RNase A digestion. A quantitative analysis of the digestion products demonstrated that the substrate containing 1 nt of exon sequence has less than 1% of 5′ heterogeneity, whereas the substrate containing 2 nt of exon sequence has approximately 8% of 5′ heterogeneity.
Figure 1.
As few as 3 exon nucleotides are required for the first step of the splicing reaction. (A) Substrates derived from the AdML pre-mRNA lack a downstream exon and the 3′ splice site. (B) Efficient intron lariat formation was observed for substrates containing three or more exon-1 nucleotides during a 1-hr incubation in nuclear extracts.
In Vitro Splicing.
Splicing reactions were as described previously (40), containing 20% vol/vol HeLa cell nuclear extract. Reactions (25 μl) were incubated at 30°C for 1.5 hr, PAGE-fractionated, and analyzed by phosphorimager radiography (Fuji). S100 complementation experiments were performed as described above except that 40% HeLa cell cytoplasmic extract (S100) was used. Reactions were complemented with varying amounts of baculovirus-produced SR proteins.
Recombinant Proteins.
Recombinant baculoviruses expressing 9G8, 9G8RS, SC35, and SF2/ASF and their purification were as described previously (41, 42). His-tagged SF2/ASF(FF-DD) containing a double Phe-to-Asp mutation within RNP-1 of the RNA-binding domain (43) was generated by PCR amplification of pET-19b (a gift from A. Krainer, Cold Spring Harbor Laboratory) and inserted into pFASTBAC (GIBCO/BRL) to generate recombinant baculoviruses. After infection of Sf9 cells, proteins were purified under native conditions on Ni-NTA (Qiagen, Chatsworth, CA) as described previously (42). 9G8RS and SF2/ASFRS were a gift from B. Graveley (Harvard University). Briefly, 9G8RS contains the zinc-knuckle and RS domain of 9G8 fused to MS2, and SF2/ASFRS contains a fragment of Escherichia coli DNA gyrase B fused to the RS domain of SF2/ASF. SF2/ASFRS was expressed in Sf9 cells by using recombinant baculoviruses and purified under native conditions as described (42).
RESULTS
Intron Sequences Are Sufficient for the First Step of the Splicing Reaction.
Splicing substrates bearing varying lengths of the upstream exon but lacking the downstream exon and the 3′ splice site AG dinucleotide (Fig. 1A) were synthesized and tested in an in vitro splicing reaction (Fig. 1B). The progressive truncation of exon sequences from 31 to 3 nt did not reduce significantly the efficiency of cleavage at the 5′ splice site and lariat formation (Fig. 1B). Primer extension experiments (data not shown) and the gel mobilities of the lariat-containing RNAs (Figs. 1B and 3 C and D) revealed that the authentic branch point and 5′ splice site were used with each of the exon truncation substrates.
Figure 3.
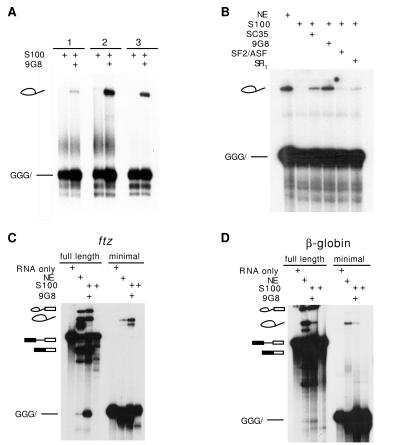
SR proteins are required for the first step of the splicing reaction with RNA substrates containing 3, 2, or 1 exon nucleotide. (A) The SR protein 9G8 (200 nM) can complement S100 extract-dependent lariat formation of AdML RNA substrates containing 3, 2, or 1 exon nucleotide. (B) The SR proteins SC35 (500 nM), ASF/SF2 (200 nM), and a preparation of total SR proteins from nuclear extracts (100 ng/25 μl reaction) can also complement S100-dependent lariat formation of an AdML minimal substrate containing 3 exon-1 nucleotides. (C and D) The SR protein 9G8 (200 nM) complements S100 extracts to reconstitute both steps of splicing for exon-containing substrates and the first step of splicing for minimal substrates derived from two other pre-mRNAs, ftz (C) and β-globin (D).
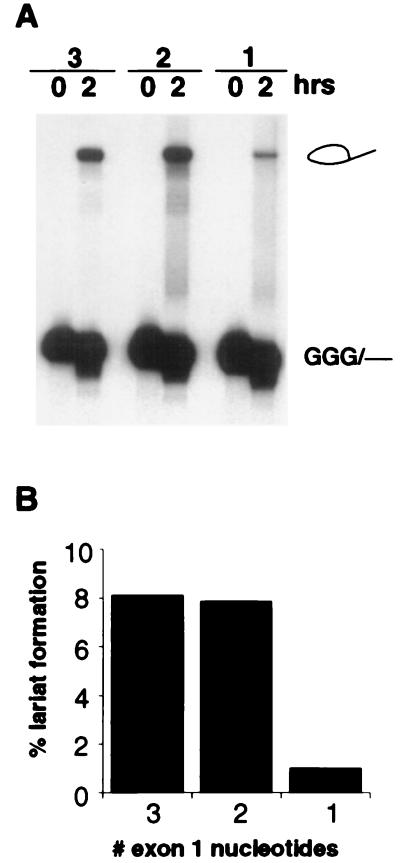
To further delineate the exon sequences required for lariat formation, substrates containing 3, 2, or 1 exon nucleotides (referred to as +3, +2, and +1 RNAs) were tested (Fig. 2). No significant difference was observed in the efficiency of lariat formation between the +3 and +2 RNAs. Remarkably, lariat formation was also observed with the +1 RNA containing only a single exon nucleotide, although the efficiency of this reaction was approximately 10-fold less than that observed with the +2 RNA (Fig. 2B). That any lariat formation could be observed with the +1 RNA shows that intronic recognition signals are sufficient for the assembly of the splicing machinery and recognition of the correct 5′ splice site and branch-point sequence.
Figure 2.
A single exon nucleotide is required for the first step of the splicing reaction. (A) Comparison of intron lariat formation efficiency for substrates containing 3, 2, or 1 exon nucleotide. (B) Histogram quantifying the data in A.
The 10-fold decrease observed between the +2 and +1 RNAs could be a consequence of the recently discovered 5′ end heterogeneity observed for certain substrates synthesized by T7 RNA polymerase (39). However, a 5′ end analysis of the +1 RNA indicated less than 1% heterogeneity (Materials and Methods; data not shown) and therefore cannot account for the 10% activity observed for lariat formation. Most likely, the drop in activity is a result of the expected decrease in the affinity of U1 snRNP for the 5′ splice site. The 5′ end of U1 snRNA interacts with the 5′ splice site intron sequence, as well as with the 2 nt in the upstream exon (GG, Fig. 1A). Thus, the +1 and +2 RNAs differ from each other by a single base pair interaction between U1 snRNA and the substrate. The free energy of base pair interactions can be calculated by using well established parameters for nearest-neighbor base-stacking contributions (44, 45). According to this calculation the difference in the stability of the duplex formed between U1 snRNA and the +1 or +2 RNA is 1.6 kcal/mol or ≈14-fold at reaction temperature. Thus, the approximately 10-fold reduction in splicing efficiency observed with the +1 RNA can be accounted for by the weaker interaction between U1 snRNA and the 5′ splice site/exon junction.
We conclude that spliceosome assembly and the formation of a covalent bond between the 5′ splice-site and the branch-site adenosine nucleotide do not require exon sequences beyond the essential phosphodiester linkage defining the junction between exon and intron.
SR Proteins Are Required for Lariat Formation with the +1 RNA.
Having shown that an RNA containing only a single exon nucleotide is sufficient for lariat formation, it was possible to directly test the hypothesis that SR proteins have an exon-independent function in splicing. To accomplish this, RNA substrates containing only 1–3 nt of exon sequence were tested in S100 extracts deficient in SR proteins. As shown in Fig. 3A, lariat formation was not observed with any of these RNAs in the S100 extracts, suggesting that SR proteins indeed are required for splicing activity with these minimal exon substrates. This possibility was confirmed by showing that the addition of the recombinant human SR protein 9G8 resulted in high levels of lariat product with all three minimal exon substrates (Fig. 3A). Other recombinant SR proteins such as SF2/ASF and SC35 also activated the first step of the splicing reaction, showing that the activity of these substrates does not require a specific SR protein (Fig. 3B).
To address the question of whether +3 substrates from pre-mRNAs other than AdML behaved similarly, we tested derivatives of β-globin and ftz pre-mRNAs. As demonstrated in Fig. 3 C and D, each substrate was capable of undergoing lariat formation in nuclear extracts. In addition, lariat formation was not observed with these substrates in S100 extracts, but could be observed upon the addition the SR protein 9G8. We conclude that SR proteins play a critical role in the basic splicing reaction of pre-mRNA substrates containing as little as 1 exon nucleotide.
An RNA Binding Domain Is Required for Efficient SR-Dependent Lariat Formation.
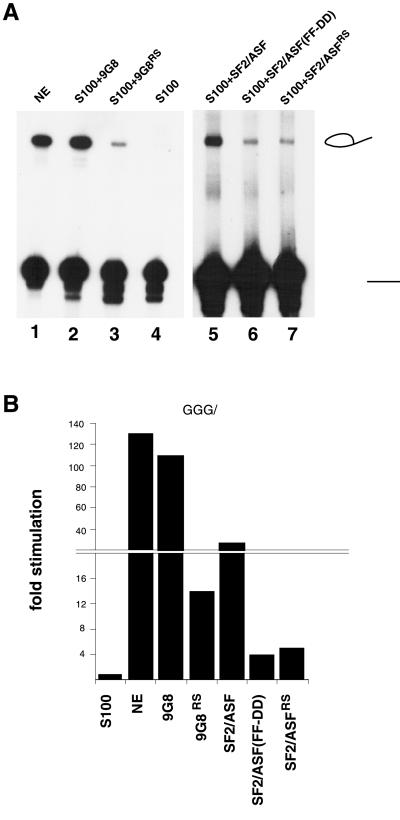
SR proteins are modular in structure, containing at least one RNA-binding domain and an RS domain. The RNA-binding domain is necessary for site-specific binding to RNA (4, 5), whereas the RS domain is required for specific protein–protein interactions (18, 21, 22). To determine whether the RNA-binding domain of SR proteins is required for lariat formation with the minimal exon substrates, we tested mutants of 9G8 and SF2/ASF lacking a functional RNA-binding domain in S100 complementation experiments. A fusion protein consisting of the bacteriophage MS2 protein fused to the SR domain of 9G8 can promote lariat formation with the +3 RNA (Fig. 4A, lane 4). However, the efficiency of lariat formation was considerably less than that observed with intact 9G8 (Fig. 4A, lane 2). Similarly, an RNA-binding mutant of SF2/ASF(FF-DD), which contains a double Phe-to-Asp mutation within the RNA-binding domain (43), as well as another fusion protein in which the SR domain of SF2/ASF is fused to the E. coli DNA gyrase B can activate lariat formation with the +3 RNA in S100 extracts (Fig. 4A, lanes 6 and 7). Again, the efficiency of lariat formation with these proteins lacking a functional RNA-binding domain is low compared with that observed with intact SF2/ASF (Fig. 4A, lane 5). Quantitation of the data revealed that the intact SR proteins activate lariat formation 30- to 100-fold over background at a concentration of 200 nM. By contrast, proteins lacking a functional SR domain activated lariat formation by only 5- to 15-fold at a concentration of 2 μM (Fig. 4B). The addition of an RNA-binding domain alone did not activate lariat formation in S100 extracts (data not shown). Thus, RNA binding significantly increases the activity of SR proteins with the minimal exon substrates. The low levels of activity observed at high concentrations of hybrid protein may be a consequence of the ability of the RS domain to promote protein–protein interactions between spliceosome components bound to the intron.
Figure 4.
A functional RNA-binding domain is required for maximal efficiency of SR-mediated lariat formation with a minimal exon substrate. S100 extracts were complemented with 9G8 (200 nM), a 9G8 hybrid protein (9G8RS) containing the RNA-binding domain of the bacteriophage MS2 in place of the natural RRM (2,000 nM) (42), SF2/ASF (200 nM), SF2/ASF (FF-DD) containing a double Phe-to-Asp mutation within the RRM (800 nM) (43), and an SF2/ASF hybrid protein (SF2/ASFRS) containing a portion of gyrase in place of the RRM (2,000 nM). (B) Histogram showing the quantitation of the data in A.
DISCUSSION
The data presented here show that SR proteins are required for the first step of the splicing reaction with RNA substrates containing only a 1-nt exon. Thus, we show that SR proteins have at least two distinct functions in pre-mRNA splicing: one that requires exon sequences and the other that does not. A number of studies are consistent with this conclusion (4, 5). However, because all of these studies were carried out with exon-containing substrates, one cannot rule out the possibility that the exon-independent function of SR proteins was masked by their exon-dependent function.
The exon-dependent function of SR proteins is to recruit the splicing machinery to the adjacent intron (29), and this mechanism is used in both alternative splicing (28) and for splice-site recognition in constitutively spliced pre-mRNAs (36). The mechanism by which SR proteins promote lariat formation in the +1 to +3 substrates described here is not understood. They could facilitate interactions between the 5′ and 3′ splice sites across the intron as originally proposed (21, 23) or they could mediate other protein–protein interactions during spliceosome assembly. Consistent with both possibilities, we have shown that the RS domain alone is sufficient for low levels of splicing of the +1 to +3 substrates in S100 extracts. Thus, it is likely that SR proteins promote protein–protein interactions within the intron during spliceosome assembly, and this function can be provided partially by the RS domain alone. However, the efficiency of lariat formation increases by approximately 50-fold if the SR protein contains a functional RNA-binding domain, indicating that RNA binding is essential for maximal splicing activity within the intron.
At present the binding site(s) of SR proteins within the intron of the minimal exon substrates are not known. We have not been able to detect significant binding of recombinant SR proteins to the 5′ splice site of a +2 RNA substrate in S100 extracts under conditions in which efficient lariat formation is observed (data not shown). This observation is in agreement with a previous study showing that SR proteins bind to exon but not intron sequences in functional splicing complexes (24), and suggests that the activity of SR proteins observed with the minimal exon substrates may require only nonspecific binding to RNA. This possibility is consistent with the result that a number of different SR proteins can promote lariat formation in our assays using at least three different minimal substrates, and with in vitro selection studies showing that the sequence selectivity of individual SR proteins is rather weak (46, 47).
Efficient pre-mRNA splicing of the 5′ proximal intron depends on the presence of a 5′ cap (48, 49). Because SR proteins were shown to complement cap-binding protein complex (CBC)-depleted nuclear extracts (48), it is possible that SR proteins interact with the CBC to recruit the spliceosome to the adjacent splice site. However, all of the substrates used in our splicing assays were synthesized without a 5′ cap, and additional experiments indicated that the presence or absence of a 5′ cap had less than a 2-fold effect on the splicing efficiency of the substrates tested (unpublished data). Thus, it is unlikely that an SR protein/CBC interaction is an important component of the splicing reactions described here.
The requirement of SR proteins for lariat formation with RNA substrates containing only 1 exon nucleotide also raises an interesting evolutionary question. SR proteins are not found in budding yeast, and their introns have highly conserved splice-site and branch-point recognition sequences. By contrast, metazoan introns have highly degenerate splice-site and branch-point recognition sequences, and intron recognition requires SR proteins. This difference affords the opportunity for regulating splice-site selection in metazoan pre-mRNAs, and it appears to necessitate additional mechanisms for ensuring correct splice-site recognition in constitutively spliced RNAs. These mechanisms appear to involve both the exon-dependent and exon-independent activities of SR proteins.
Acknowledgments
We are grateful to Brent Graveley for sharing critical unpublished reagents and recombinant proteins. We thank Adrian Krainer for plasmid pET-19b containing the SF2/ASF(FF-DD) mutant, Robin Reed for plasmid pAdMLpar and comments on the manuscript, Kristen Lynch for recombinant proteins, and Kevin Jarrell for comments on the manuscript. This investigation was supported by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research (K.J.H.) and National Institutes of Health Grant GM42231 (T.M.).
ABBREVIATIONS
- SR
serine–arginine
- RS
arginine–serine
References
- 1.Krämer A. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 2.Will C L, Lührmann R. Curr Opin Cell Biol. 1997;9:320–328. doi: 10.1016/s0955-0674(97)80003-8. [DOI] [PubMed] [Google Scholar]
- 3.Reed R, Palandjian L. In: Spliceosome Assembly. Krainer A R, editor. Oxford: IRL; 1997. pp. 103–129. [Google Scholar]
- 4.Manley J L, Tacke R. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 5.Fu X-D. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 6.Krainer A R, Maniatis T. Cell. 1985;42:725–736. doi: 10.1016/0092-8674(85)90269-7. [DOI] [PubMed] [Google Scholar]
- 7.Krainer A R, Conway G C, Kozak D. Genes Dev. 1990;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- 8.Ge H, Manley J L. Cell. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- 9.Fu X D, Maniatis T. Nature (London) 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- 10.Zahler A M, Lane W S, Stolk J A, Roth M B. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 11.Fu X-D. Nature (London) 1993;365:82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- 12.Staknis D, Reed R. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roscigno R F, Garcia-Blanco M A. RNA. 1995;1:692–706. [PMC free article] [PubMed] [Google Scholar]
- 14.Tarn W Y, Steitz J A. Genes Dev. 1994;8:2704–2717. doi: 10.1101/gad.8.22.2704. [DOI] [PubMed] [Google Scholar]
- 15.Tarn W Y, Steitz J A. Proc Natl Acad Sci USA. 1995;92:2504–2508. doi: 10.1073/pnas.92.7.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crispino J D, Blencowe B J, Sharp P A. Science. 1994;265:1866–1869. doi: 10.1126/science.8091213. [DOI] [PubMed] [Google Scholar]
- 17.MacMillan A M, McCaw P S, Crispino J D, Sharp P A. Proc Natl Acad Sci USA. 1997;94:133–136. doi: 10.1073/pnas.94.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohtz J D, Jamison S F, Will C L, Zuo P, Lührmann R, Garcia-Blanco M A, Manley J L. Nature (London) 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 19.Jamison S F, Pasman Z, Wang J, Will C, Lührmann R, Manley J L, Garcia-Blanco M A. Nucleic Acids Res. 1995;23:3260–3267. doi: 10.1093/nar/23.16.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zuo P, Manley J L. Proc Natl Acad Sci USA. 1994;91:3363–3367. doi: 10.1073/pnas.91.8.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu J Y, Maniatis T. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 22.Amrein H, Hedley M L, Maniatis T. Cell. 1994;76:735–746. doi: 10.1016/0092-8674(94)90512-6. [DOI] [PubMed] [Google Scholar]
- 23.Fu X-D, Maniatis T. Proc Natl Acad Sci USA. 1992;89:1725–1729. doi: 10.1073/pnas.89.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiara M D, Gozani O, Bennett M, Champion-Arnaud P, Palandjian L, Reed R. Mol Cell Biol. 1996;16:3317–3326. doi: 10.1128/mcb.16.7.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abovich N, Rosbash M. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 26.Berglund J A, Abovich N, Rosbash M. Genes Dev. 1998;12:858–867. doi: 10.1101/gad.12.6.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rudner D Z, Kanaar R, Breger K S, Rio D C. Proc Natl Acad Sci USA. 1996;93:10333–10337. doi: 10.1073/pnas.93.19.10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams M D, Rudner D Z, Rio D C. Curr Opin Cell Biol. 1996;8:331–339. doi: 10.1016/s0955-0674(96)80006-8. [DOI] [PubMed] [Google Scholar]
- 29.Hertel K J, Lynch K W, Maniatis T. Curr Opin Cell Biol. 1997;9:350–357. doi: 10.1016/s0955-0674(97)80007-5. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Manley J L. Curr Opin Genet Dev. 1997;7:205–211. doi: 10.1016/s0959-437x(97)80130-x. [DOI] [PubMed] [Google Scholar]
- 31.Sun Q, Mayeda A, Hampson R K, Krainer A R, Rottman F M. Genes Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- 32.Lavigueur A, La Branche H, Kornblihtt A R, Chabot B. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 33.Tian M, Maniatis T. Cell. 1993;74:105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z, Hoffmann H M, Grabowski P J. RNA. 1995;1:21–35. [PMC free article] [PubMed] [Google Scholar]
- 35.Zuo P, Maniatis T. Genes Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]
- 36.Schaal T D, Maniatis T. Mol Cell Biol. 1999;19:261–273. doi: 10.1128/mcb.19.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson K, Moore M J. Science. 1997;276:1712–1716. doi: 10.1126/science.276.5319.1712. [DOI] [PubMed] [Google Scholar]
- 38.Smith C W, Porro E B, Patton J G, Nadal-Ginard B. Nature (London) 1989;342:243–247. doi: 10.1038/342243a0. [DOI] [PubMed] [Google Scholar]
- 39.Pleiss J A, Derrick M L, Uhlenbeck O C. RNA. 1998;4:1313–1317. doi: 10.1017/s135583829800106x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hertel K J, Maniatis T. Mol Cell. 1998;1:449–455. doi: 10.1016/s1097-2765(00)80045-3. [DOI] [PubMed] [Google Scholar]
- 41.Lynch K W, Maniatis T. Genes Dev. 1996;10:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- 42.Graveley B R, Maniatis T. Mol Cell. 1998;1:765–771. doi: 10.1016/s1097-2765(00)80076-3. [DOI] [PubMed] [Google Scholar]
- 43.Cáceres J F, Krainer A R. EMBO J. 1992;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freier S M, Kierzek R, Jaeger J A, Sugimoto N, Caruthers M H, Neilson T, Turner D H. Proc Natl Acad Sci USA. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serra M J, Turner D H. Methods Enzymol. 1995;259:242–261. doi: 10.1016/0076-6879(95)59047-1. [DOI] [PubMed] [Google Scholar]
- 46.Liu H X, Zhang M, Krainer A R. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaal T D, Maniatis T. Mol Cell Biol. 1999;19:1705–1719. doi: 10.1128/mcb.19.3.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis J D, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj I W. Genes Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- 49.Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj I W. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]