Abstract
The idea that recruitment of the transcriptional machinery to a promoter suffices for gene activation is based partly on the results of “artificial recruitment” experiments performed in vivo. Artificial recruitment can be effected by a “nonclassical” activator comprising a DNA-binding domain fused to a component of the transcriptional machinery. Here we show that activation by artificial recruitment in yeast can be sensitive to any of three factors: position of the activator-binding elements, sequence of the promoter, and coding sequences downstream of the promoter. In contrast, classical activators worked efficiently at all promoters tested. In all cases the “artificial recruitment” fusions synergized well with classical activators. A classical activator evidently differs from a nonclassical activator in that the former can touch multiple sites on the transcriptional machinery, and we propose that that difference accounts for the broader spectrum of activity of the typical classical activator. A similar conclusion is reached from studies in mammalian cells in the accompanying paper [Nevado, J., Gaudreau, L., Adam, M. & Ptashne, M. (1999) Proc. Natl. Acad. Sci. USA 96, 2674–2677].
We and others have argued that recruitment is a principal mechanism of gene activation in yeast, bacteria, and other organisms (1–4). According to this idea, a transcriptional activator localizes the transcriptional machinery to a site (a specific promoter) that is dictated by the DNA-binding address of the activator. One of us has suggested that “regulated localization” of this kind underlies many other biological processes as well (e.g., signal transduction) and has argued that the strategy comprises a highly “evolvable” system for integrating physiological signals (5).
So-called “artificial recruitment” experiments played an important role in formulating these ideas. In such experiments, gene transcription is elicited in the absence of an activating region similar or identical to those found on classical activators such as Gal4, Pho4, or Gcn4. Artificial recruitment can be effected by a fusion protein bearing a DNA-binding domain (e.g., LexA or Gal4) covalently linked to a component of the transcriptional machinery; examples of the latter include TATA box-binding protein (4, 6–9), certain TAFs (4, 10, 11), TFIIB (10), Sin4 (12) Gal11 (13–16), Srb2 (15, 16), Srb6 (17), and Srb11 (18). The latter four of these are components of the RNA polymerase II (PolII) holoenzyme as is, according to one report, TFIIB (19). Artificial recruitment can also be mediated by a noncovalent interaction between a DNA-tethered peptide and the transcriptional machinery. For example, interaction of one member of the Myc-Max heterodimerization domain, tethered to DNA, with the other member fused to a component of the machinery, can trigger transcription in yeast (7). A fortuitous interaction between the Gal4 dimerization domain and Gal11P, a mutated derivative of the holoenzyme subunit Gal11 (14), was also found to trigger activation, and in that case further analysis indicated that the strength of the novel protein–protein interaction, measured in vitro, was directly correlated with the level of transcription observed in vivo (15). That interaction can, with the appropriately modified molecules, trigger transcription in bacteria as well (20). These activator recruitment experiments comprise one of two kinds of “activator bypass” experiments; in the other kind, performed in vitro, it was found that the effect of activators can be mimicked simply by increasing the concentration of the transcriptional machinery (ref. 3 for yeast and ref. 21 for bacteria).
“Classical activators,” as we use the term, refers to natural transcriptional activators such as Pho4, Gal4, and Gcn4, as well as to hybrid molecules that bear a DNA-binding domain fused to a classical activating region (e.g., LexA + Gal4, which bears the LexA DNA-binding domain fused to an activating region excised from Gal4). Thus the “nonclassical activators,” proteins that can effect artificial recruitment, may be regarded as substitution mutants that bear, in place of a natural activating region, a component of the transcriptional machinery. Classical activating regions are believed to contact multiple surfaces of that machinery (see refs. 1 and 2), whereas “nonclassical” activators bearing holoenzyme subunits such as Gal11 or the Srb proteins are believed to insert into the holoenzyme.
In this paper, we describe a series of experiments that reveal severe limitations on the ability of nonclassical activators to work at certain genes in yeast. These limitations do not apply to the action of classical activators as tested here. We propose that this greater flexibility of action of classical activators is a result of their abilities to interact with multiple and perhaps alternative components of the transcriptional machinery. Our results suggest why one form of recruitment—that effected by classical activators—was exploited extensively in the construction of gene regulatory networks whereas the other—that effected by nonclassical activators—was not. This conclusion is reinforced by the results of artificial recruitment experiments in mammalian cells described in the accompanying paper (22).
EXPERIMENTAL PROCEDURES
Genetic Methods.
Yeast transformation procedures were as described previously (14, 23). Cells were grown in synthetic complete growth media to an OD of 0.6 to 1.0, harvested and assayed for β-galactosidase activity either as described in Tzamarias & K.S. (23) or as in Barberis et al. (14). Acid phosphatase activity was assayed as described previously (24). Both β-galactosidase and phosphatase assays represent the average of at least three independent experiments. Details of plasmid constructions are available on request. All the Gal4 and LexA constructs were fused at the carboxyl terminus of these DNA-binding domains, whereas the Pho4 fusions constructs were fused to the amino terminus of Pho4Δ2 (24). The yeast strains used in this study are described in Table 1.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| FT4a | a, ura3-52, his3Δ200, trp1Δ63, leu2∷PET56 | (23) |
| JPY9 | α ura3-52, his3Δ200, leu2Δ1, trp1Δ63, lys2Δ385, gal4Δ11 | (14) |
| YAG4 | α ura3-52, his3Δ200, leu2Δ1, trp1Δ63, lys2Δ385, gal4Δ11, URA3∷pJP188 | This study |
| YAG22 | α ura3-52, his3Δ200, leu2Δ1, trp1Δ63, lys2Δ385, gal4Δ11, URA3∷pJP169 | This study |
| YAG23 | α, ura3Δ5, his3-11, his3-15, Δtrp1, leu2-3, leu2-112, canR, pho4∷ura3Δ5, Δpho80∷HIS3 | (16) |
| YAG35 | α ura3-52, his3Δ200, leu2Δ1, trp1Δ63, lys2Δ385, gal4Δ11, URA3∷pLRG330 | This study |
| YAG100 | α ura3-52, his3Δ200, leu2Δ1, trp1Δ63, lys2Δ385, gal4Δ11, URA3∷pLRG277 | This study |
| YAG101 | α ura3-52, his3Δ200, leu2Δ1, trp1Δ63, lys2Δ385, gal4Δ11, URA3∷pJP185 | This study |
| ZZY41 | α, ura3-52, his3Δ200, Δtrp1, leu2-1, Δlys2, gal4, gal80, URA3∷pZZ41 | This study |
Primer Extension Analysis.
Primer extensions were carried out essentially as described by J. Ma and M.P. (25). RNA (30 μg), isolated from each respective yeast strain bearing the indicated LexA fusion, was used and hybridized to a 32P-labeled lacZ oligonucleotide. The resulting cDNAs were then resolved on a 10% acrylamide gel as described by L.G. et al. (3).
RESULTS
Various DNA-Tethered polII Holoenzyme and TFIID Components Can Activate Transcription of Target Genes.
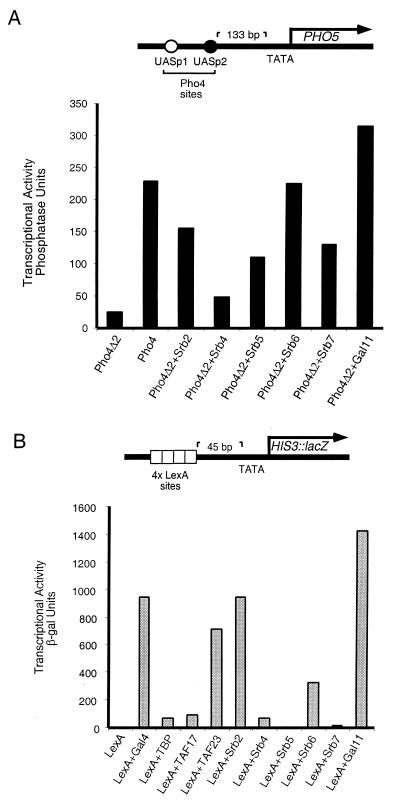
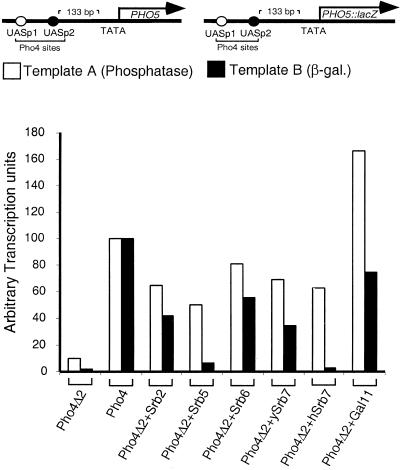
As shown in Fig. 1, several different nonclassical activators can activate a yeast reporter as efficiently, or nearly so, as classical activators. Fig. 1A shows that, on a promoter bearing Pho4-binding sites, Pho4Δ2 + Srb2, +Srb6, and +Gal11, each of which bears a holoenzyme component in place of the natural activating region of Pho4, activated about as efficiently as did Pho4. B shows that, on a template bearing LexA sites, LexA + TAF23, +Srb2, and +Gal11 worked about as efficiently as did LexA + Gal4. Note, however, that for certain of the nonclassical activators there is no consistent pattern of activity when comparing the responses of the two reporters. Thus, for example, on the reporter of B, the Srb2 fusion protein worked more efficiently than did its Srb6 counterpart, whereas the opposite order of activity was observed on the reporter of A. To take another example, whereas the Srb5 and 7 fusions worked reasonably efficiently on the template in A, both were virtually inactive on the template of B. Interpretation of these results is complicated by the fact that there are several variables distinguishing the experiments of A and B. These include the use of different DNA-binding domains and corresponding binding sites on DNA, different distances of those sites from the respective genes and from each other, and different sequences of the respective promoters and downstream regions. The following experiments test effects of each of these variables.
Figure 1.
Gene activation by various nonclassical activators in yeast. (A) Transcriptional activation elicited by Pho4 derivatives at the PHO5 gene. All the Pho4 fusions were expressed from the PHO4 promoter on a high-copy (2 μ) vector (24). Activity was measured as acid phosphatase units produced by the product of the PHO5 gene directly off its chromosomal locus under high phosphate conditions in a pho80 strain (YAG23). Wild-type Pho4 was carried on an ARS-CEN plasmid because overexpression of Pho4 causes severe growth defects (24). (B) Transcriptional activation elicited by LexA derivatives at an HIS3-based reporter template. All the LexA fusions were expressed from the ADH1 promoter on a high-copy (2 μ) vector. Transcriptional activity was measured as β-galactosidase units produced by the lacZ gene product in yeast strain FT4a. The reporter construct was carried on a 2 μ plasmid.
Position of the Activator-Binding Sites.
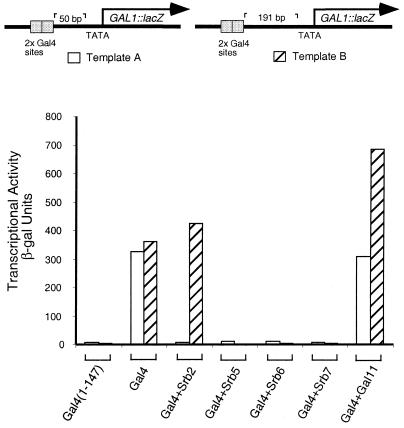
In the experiment of Fig. 2, the reporters differ in the positions of the two Gal4 binding sites: on template A they are positioned 50 bp upstream and on template B 191 bp upstream of the TATA element. All the activators used in this experiment bear a Gal4 binding domain. The figure shows that, whereas native Gal4 worked equally efficiently on the two promoters, the Srb2 fusion worked very efficiently on template B but virtually not at all on template A. The various other Srb fusions tested failed to work on either template. The Gal11 fusion activated about as well as did wild-type Gal4 on template A and about twice as efficiently as did wild-type Gal4 on template B. The inactivity observed for certain of these fusions is not accounted for by instability of the respective protein, because each has been found to activate significantly when working on a different reporter (see for example Fig. 1B, 3A, and 5). We note that in both templates the activator-binding site–TATA separations are relatively small; at larger separations, of course, all the activators would work less efficiently.
Figure 2.
Position of the activator-binding sites influences transcriptional activation elicited by nonclassical activators. All the Gal4 constructs (except Gal4 + Gal11) were expressed from the ADH1 promoter on a high-copy (2 μ) vector. Gal4 + Gal11 was expressed from a β-actin promoter and carried on an ARS-CEN plasmid (15). Transcriptional activity was measured as β-galactosidase units produced off chromosomally integrated templates in yeast strains YAG101 (template A; white bars) and YAG4 (template B; hatched bars).
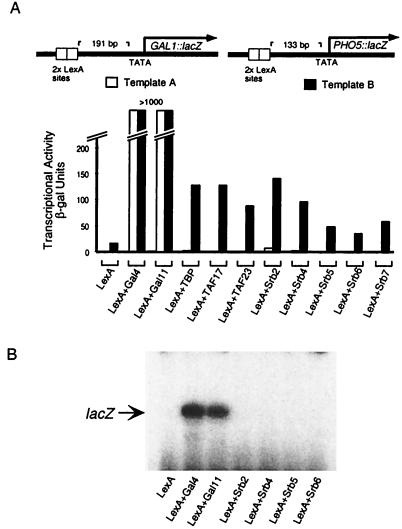
Figure 3.
The promoter sequence influences transcriptional activation elicited by nonclassical activators. (A) Transcriptional activation elicited by LexA derivatives elicited at chromosomally integrated (at the ura3–52 locus) reporter templates bearing either the GAL1 or PHO5 promoters. Both templates bear two LexA binding sites upstream of the respective TATA element, but on template A the DNA separating these sites from the ATG is taken from the GAL1 gene, and the corresponding sequence on template B is from the PHO5 gene. The distances between the LexA sites and the TATA elements are those found naturally at the GAL1 gene (191 bp; template A) and the PHO5 gene (133 bp; template B). All the LexA constructs were expressed from the ADH1 promoter on a 2 μ vector. Transcriptional activity was measured as β-galactosidase units in yeast strains YAG22 (template A; white bars) and YAG35 (template B; black bars). (B) Primer extension analysis. The cDNA-labeled lacZ denotes transcripts from the GAL1∷lacZ reporter of YAG22.
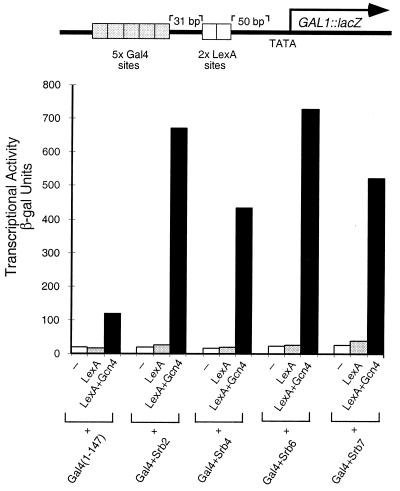
Figure 5.
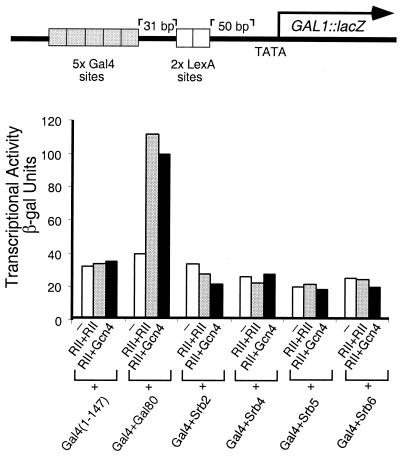
Synergistic activation of nonclassical activators with a DNA-tethered activating region. All the Gal4 and LexA constructs were expressed from the ADH1 promoter on a high-copy (2 μ) vector. Transcriptional activity was measured as β-galactosidase units produced off a chromosomally integrated template in yeast strain ZZY41.
Promoter Sequence.
In the experiment of Fig. 3, the two templates differ in the sequences between the activator-binding sites (LexA sites) and the ATG translational starts (see figure legend), and all the activators bear a LexA DNA-binding domain. The figure shows that, as in the previous experiments, the Gal11 activator (LexA + Gal11) worked very efficiently and about as well as the Gal4 activator (LexA + Gal4). In contrast, the various other nonclassical activators worked less well on template B than did the Gal4 activator, and worked virtually not at all on template A. In an experiment not shown here we found that changing the spacing of the LexA sites from 191 to 133 bp upstream of TATA had no significant effect on the results, and so we attribute the differences in activities of some of the nonclassical activators to the different sequences separating the activator binding sites from the TATA elements. Fig. 3B shows that transcription as assayed by β-galactosidase levels corresponded to that assayed by primer extension analysis.
Coding Sequences Downstream of the ATG.
In the experiment of Fig. 4 the two templates are identical except that one bears the wild-type PHO5 gene (template A) and the other bears lacZ fused downstream of the ATG in place of that gene (template B; a gift from W. Hörz). All of the other sequences, including the activator-binding sites (which are recognized by Pho4) are as found in the PHO5 promoter. In this case we measured phosphatase activity for template A and β-galactosidase activity for B and normalized the results by equating the levels elicited by native Pho4. The figure shows that none of the Srb fusions worked as well as did wild-type Pho4 at either promoter. More strikingly, each of these fusions worked better on template A than on B, and in two cases (those involving Srb5 and hSrb7), the difference is substantial. The Gal11 fusions worked well on both templates and some 2-fold better on template A.
Figure 4.
Coding sequences downstream of the ATG influence transcriptional activation elicited by nonclassical activators. All the Pho4 fusions were expressed from the PHO4 promoter on a high-copy (2 μ) vector and wild-type Pho4 was carried on an ARS-CEN plasmid (24).
In this experiment, template B was carried on a replicating plasmid whereas template A was the endogenous PHO5 chromosomal locus (see figure legend); in an experiment not shown, identical templates, but both carried on replicating plasmids, were compared with results similar to those shown in the figure.
Synergistic Activation.
In the experiment of Fig. 5 the reporter bears two separate upstream elements: one comprises five Gal4-binding sites and the other, further downstream, two LexA sites. The experiments first measured, separately, transcription elicited by an activator bearing a classical activating region bound at the LexA sites (i.e., LexA + Gcn4), and various “nonclassical” activators bound to the Gal4 sites, and then the effects of combinations of the two kinds of activators were measured. The figure shows that, whereas none of four Gal4 + Srb fusions activated transcription when working alone on the reporter, each dramatically increased activation when working together with the activator bearing the Gcn4-activating region. Thus each Srb fusion, though virtually inactive on its own, increased activation by LexA + Gcn4 some 4- to 8-fold. In experiments not shown, we have observed similar synergistic effects when these Srb fusion proteins work together with LexA + B42, an activator bearing an acidic activating region (26).
DNA Binding and Synergy.
The experiment of Fig. 6, which uses the template used in the experiment of Fig. 5, shows that a classical activator that is not bound to DNA fails to work synergistically with DNA-bound nonclassical activators. Thus neither a peptide bearing two activating regions from Gal4 fused in tandem (RII + RII), nor a peptide bearing RII fused to the activating region of Gcn4 (RII + Gcn4) has any effect on activation by DNA-tethered Srb2, 4, 5, or 6. This failure cannot be attributed to an inherent inactivity of the activating peptides because, as shown in the figure, each activates transcription in the presence of DNA-tethered Gal80. The latter is the natural inhibitor of Gal4, which ordinarily works by binding to Gal4, recognizing sites in RII and blocking the activating function (27, 28). In this case, Gal80 has been artificially tethered to DNA, and the bifunctional activating peptide is evidently recruited to the DNA by interaction of one RII sequence with Gal80 in such a fashion as to leave the other activating region (RII in one case, and a fragment of GCN4 in the other) free to interact with the transcriptional machinery.
Figure 6.
Overexpression of non-DNA-tethered activating regions does not enhance transcriptional activation elicited by nonclassical activators. The Gal4 constructs were expressed from the ADH1 promoter on a high-copy (2 μ) vector; Gal4 + Gal80 was carried on an ARS-CEN vector. RII + RII is comprised of two tandemly fused copies of the Gal4-activating region and RII + Gcn4 comprises the same RII moiety fused to Gcn4 lacking its DNA-binding domain. Both of these activating regions were expressed from the β-actin promoter and carried on a 2 μ plasmid. Transcriptional activity was measured as β-galactosidase units in yeast strain ZZY41.
DISCUSSION
The results presented here show that the activities of some, but not all (see below), “nonclassical” transcriptional activators are strongly influenced by at least three factors: the position of the upstream DNA-binding sites for these activators, the sequence of the promoter region of the gene being transcribed, and the coding sequences downstream of the ATG. These “nonclassical” activators comprise a DNA-binding domain fused to certain proteins ordinarily found as part of the polII holoenzyme (e.g., an Srb protein) or as part of TFIID (i.e., TATA box-binding protein and a TAF protein). In striking contrast, activators bearing a “classical” activating region (such as those found in the yeast activators Gal4, Pho4, and Gcn4), fused to a DNA-binding domain, are, to the extent tested, largely insensitive to the variables described above. That is, the classical activators work efficiently under all the conditions tested. The accompanying paper (22) describes similar experiments performed in mammalian cells.
Two possible explanations for these observed differences between classical and nonclassical activators, both of which we disfavor, are as follows. First, it could be imagined that under certain conditions the nonclassical activators simply fail to recruit any part of the transcriptional machinery. This idea is contradicted by our finding that every nonclassical activator tested worked synergistically with a classical activator binding to sites nearby, and that each of these nonclassical activators activated on its own at least one of the reporters tested. A second explanation for our results might be that at certain (but not at all) promoters some activity of classical activating regions is required for transcription beyond recruitment. It is difficult to exclude this second possibility, but it is inconsistent with our finding that activating regions lacking DNA-binding domains and expressed at high levels in cells were unable to work synergistically with our nonclassical activators. Similar experiments studying the activities of genes turned on by classical activators have also shown that overproduced activating regions, unattached to DNA-binding domains, fail to increase transcription. These results are all consistent with the idea that the sole role of classical activating regions is to recruit the transcriptional machinery to the DNA (see refs. 1 and 3), a scenario further consistent with the observation that activation does not spontaneously occur when an activation domain is covalently linked to various components of the transcription machinery (4).
Why then are the effects of certain nonclassical activators subject to restrictions that do not apply to the effects of classical activators? We entertain two related scenarios.
(i) One explanation would assume, quite plausibly it would seem, that nonclassical activators interact less flexibly with the transcriptional machinery than do classical activators. The latter are believed, as noted in the introduction, to contact multiple sites in the transcriptional machinery, and perhaps these activators are able to use alternative sites of interaction that allow recruitment whatever the precise spacing between the activator-binding sites and the transcription start site. In contrast, the typical nonclassical activator would be expected to insert into the holoenzyme in only one way, and hence very few activator positionings relative to the start site would permit an unconstrained interaction.
(ii) Another, or additional, explanation for our results would assume that the requirements for recruitment of a functional transcriptional machinery differ at different promoters. Depending on the precise promoter sequence, the presence of negatively acting factors including nucleosomes, etc., activation could require stabilization of one or another interaction between the transcriptional machinery and DNA and/or recruitment of subcomplexes of the transcriptional machinery, including those that would remodel chromatin (see 29, 31) or ensure transcription through pause sites (30) that would otherwise impede the process. Perhaps classical activating regions, by virtue of their abilities to interact with multiple targets, make whatever stabilizing interactions or recruit whatever complexes are necessary to ensure transcription at any promoter, whereas nonclassical activators cannot do so. We have attempted to probe these possibilities in experiments designed to measure synergy between a relatively inactive DNA-tethered Srb protein and with DNA-tethered TATA box-binding protein, with DNA-tethered Swi2/Snf2, and with DNA-tethered Gcn5, but in none of those experiments was synergy detected (data not shown).
Gal11, when fused to a DNA-binding domain, behaved more like a classical activator than a nonclassical activator in our experiments; i.e., it activated transcription to high levels virtually independent of the parameters we varied (with the possible exception of that of Fig. 4). We do not know why Gal11 behaves differently in this regard from other holoenzyme components such as Srb2, 4, 6, and 7. One possibility is that the location of Gal11 within the holoenzyme permits the flexibility necessary for the fusion proteins to function efficiently at all promoters. Another would be that more than one complex including RNA polymerase II can direct transcription, each of which contains Gal11 (14, 32, 33); accordingly, depending on the promoter configuration, a DNA-tethered Gal11 might be able to recruit one or another such complex that would work where the single complex bearing the Srb proteins would not.
Acknowledgments
We thank Wolfram Hörz, Andrés Aguilera, Alcide Barberis, Joe Pearlberg, Sebastián Chávez, and especially Yibing Wu for reagents. We thank Maryse Adam for superb technical assistance. We thank Aseem Ansari, Mario Mencia, and Zarmik Moqtaderi for useful discussions. We also thank Idriss M. Bennani-Baiti for discussion of the manuscript. L.G. was supported in part by a fellowship from the Medical Research Council of Canada, M.K. was supported by a National Institutes of Health (N.I.H.) Postdoctoral Fellowship (GM17930), J.N. was supported by a postdoctoral fellowship from the Spanish Ministry of Education (F.P.I.), and Z.Z. was supported by the European Molecular Biology Organization (EMBO) and by a Clinical Scholars Training Fellowship from the Winston Foundation. This work was supported by grants from N.I.H. to K.S. and to M.P.
ABBREVIATION
- PolII
RNA polymerase II
References
- 1.Ptashne M, Gann A. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 2.Struhl K. Cell. 1996;84:179–182. doi: 10.1016/s0092-8674(00)80970-8. [DOI] [PubMed] [Google Scholar]
- 3.Gaudreau L, Adam M, Ptashne M. Mol Cell. 1998;1:913–916. doi: 10.1016/s1097-2765(00)80090-8. [DOI] [PubMed] [Google Scholar]
- 4.Keaveney M, Struhl K. Mol Cell. 1998;1:917–924. doi: 10.1016/s1097-2765(00)80091-x. [DOI] [PubMed] [Google Scholar]
- 5.Ptashne M, Gann A. Curr Biol. 1998;8:R812–R822. doi: 10.1016/s0960-9822(07)00508-8. [DOI] [PubMed] [Google Scholar]
- 6.Chatterjee S, Struhl K. Nature (London) 1995;374:820–822. doi: 10.1038/374820a0. [DOI] [PubMed] [Google Scholar]
- 7.Klages N, Strubin M. Nature (London) 1995;374:822–823. doi: 10.1038/374822a0. [DOI] [PubMed] [Google Scholar]
- 8.Xiao H, Friesen J D, Lis J T. Mol Cell Biol. 1995;15:5757–5761. doi: 10.1128/mcb.15.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee M, Struhl K. Mol Cell Biol. 1997;17:1336–1345. doi: 10.1128/mcb.17.3.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez-Couto E, Klages N, Strubin M. Proc Natl Acad Sci USA. 1997;94:8036–8041. doi: 10.1073/pnas.94.15.8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apone L M, Virbasius C A, Reese J C, Green M R. Genes Dev. 1996;10:2368–2380. doi: 10.1101/gad.10.18.2368. [DOI] [PubMed] [Google Scholar]
- 12.Jiang Y M, Stillman D J. Mol Cell Biol. 1992;12:4503–4514. doi: 10.1128/mcb.12.10.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Himmelfarb H J, Pearlberg J, Last D H, Ptashne M. Cell. 1990;63:1299–1309. doi: 10.1016/0092-8674(90)90425-e. [DOI] [PubMed] [Google Scholar]
- 14.Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 15.Farrell S, Simkovich N, Wu Y, Barberis A, Ptashne M. Genes Dev. 1996;10:2359–2367. doi: 10.1101/gad.10.18.2359. [DOI] [PubMed] [Google Scholar]
- 16.Gaudreau L, Schmid A, Blaschke D, Ptashne M, Hörz W. Cell. 1997;89:55–62. doi: 10.1016/s0092-8674(00)80182-8. [DOI] [PubMed] [Google Scholar]
- 17.Hengartner C J, Myer V E, Liao S M, Wilson C J, Koh S S, Young R A. Mol Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 18.Kuchin S, Yeghiayan P, Carlson M. Proc Natl Acad Sci USA. 1995;92:4006–4010. doi: 10.1073/pnas.92.9.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koleske A, Young R A. Nature (London) 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 20.Dove S L, Hochschild A. Genes Dev. 1998;12:745–754. doi: 10.1101/gad.12.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer B J, Ptashne M. J Mol Biol. 1980;139:195–205. doi: 10.1016/0022-2836(80)90304-6. [DOI] [PubMed] [Google Scholar]
- 22.Nevado J, Gaudreau L, Adam M, Ptashne M. Proc Natl Acad Sci USA. 1999;96:2674–2677. doi: 10.1073/pnas.96.6.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tzamarias D, Struhl K. Nature (London) 1994;369:758–761. doi: 10.1038/369758a0. [DOI] [PubMed] [Google Scholar]
- 24.Svaren J, Schmitz J, Hörz W. EMBO J. 1994;13:4856–4862. doi: 10.1002/j.1460-2075.1994.tb06812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma J, Ptashne M. Cell. 1987;48:847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- 26.Ma J, Ptashne M. Cell. 1987;51:113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- 27.Ma J, Ptashne M. Cell. 1987;50:137–142. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- 28.Wu Y, Reese R J, Ptashne M. EMBO J. 1996;15:3951–3963. [PMC free article] [PubMed] [Google Scholar]
- 29.Struhl K. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 30.Chavez S, Aguilera A. Genes Dev. 1997;11:3459–3470. doi: 10.1101/gad.11.24.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stargell L A, Struhl K. Mol Cell Biol. 1996;16:4456–4464. doi: 10.1128/mcb.16.8.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim Y J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 33.Chang M, Jaening J A. Nucleic Acids Res. 1997;25:4861–4865. doi: 10.1093/nar/25.24.4861. [DOI] [PMC free article] [PubMed] [Google Scholar]