Abstract
We show that the typical “nonclassical” activator, which comprises a fusion protein bearing a component of the transcriptional machinery fused to a DNA-binding domain, activates transcription in mammalian cells only weakly when tested with an array of promoters. However, as found in analogous “artificial recruitment” experiments performed in yeast, these activators work synergistically with “classical” activators. The effect of the classical activator in such experiments requires that it be tethered to DNA, a requirement that cannot be overcome by expression of that classical activator at high levels. The effect of the one nonclassical activator that does elicit significant levels of transcription when working alone (i.e., that bearing TATA box-binding protein) is strongly influenced by promoter architecture. The results, consistent with those of analogous experiments in yeast [see the accompanying paper: Gaudreau, L., Keaveney, M., Nevado, J., Zaman, Z., Bryant, G. O., Struhl, K. & Ptashne, M. (1999) Proc. Natl. Acad. Sci. USA 96, 2668–2673], suggest that classical activators, presumably by virtue of their abilities to interact with multiple targets, have a functional flexibility that nonclassical activators lack.
So-called “activator-bypass” experiments, performed in yeast and bacteria, have played an important role in formulating the idea that many transcriptional activators work by recruiting the transcriptional machinery to the promoter (1–4). These experiments show that the effect of an activator can be dispensed with—and the activator has no further effect—provided that that machinery can be brought to the promoter by some other means. One way to demonstrate this effect in vitro is by using high concentrations of bacterial RNA polymerase or of the yeast RNA polymerase II holoenzyme plus auxiliary factors (2, 3). In an alternative strategy, called “artificial recruitment,” the machinery is brought to the promoter either by an arbitrary protein–protein interaction between the machinery and a DNA-tethered peptide or by the action of a fusion protein comprising a DNA-binding domain and a component of the transcriptional machinery (5, 6). Gal4 + Srb2, an example of such a fusion protein, bears the holoenzyme component Srb2 fused to the Gal4 DNA-binding domain. It is believed to insert its Srb2 moiety into the holoenzyme; binding to a Gal4 site then recruits the holoenzyme to the nearby promoter (see ref. 2). Artificial recruitment has been demonstrated in vitro and in vivo (refs. 3–6; ref. 7 and references therein).
Proteins such as Gal4 + Srb2 are called nonclassical activators to distinguish them from classical activators that bear natural activating regions; the latter, in the eukaryotic case, are believed to contact multiple surfaces of the transcriptional machinery (1, 2). In contrast, the nonclassical activators, as indicated by the description of Gal4 + Srb2, are believed to interact in a much more restricted fashion with that machinery.
In the accompanying paper (7) we report a striking difference between classical activators such as Gal4 or Pho4 and nonclassical activators such as Gal4 + Srb2 in their abilities to activate transcription in yeast: the effects of the latter were often strongly influenced by promoter architecture (i.e., promoter sequence and/or position of the activator binding sites), as well as by downstream sequences, whereas classical activators were impervious to these factors as tested. In virtually every case tested, however, the nonclassical activator worked synergistically with a classical activator bound to DNA nearby. Moreover, the synergistic effect of the classical activator depended on its being tethered to DNA, arguing that all of the activation we observe arises from recruitment. In the experiments described here, we test a series of yeast and human nonclassical activators in mammalian cells, using an array of reporters. Our findings parallel those made in the yeast experiments and reinforce the conclusion that classical activators have a functional flexibility that nonclassical activators lack.
MATERIALS AND METHODS
Cell Culture.
HeLa cells were obtained from the American Type Cell Culture (ATCC) and maintained in DMEM (GIBCO) supplemented with 10% vol/vol fetal bovine serum (GIBCO) and 1% penicillin/streptomycin (GIBCO).
HeLa Transient Transfections.
HeLa cells were plated in six-well culture plates for 24 hr before transfect at a density 5–6 × 105 cells/well. The transfections were carried out either by calcium phosphate precipitation (8) or by the lipofectamine method, according to the manufacturer’s recommendations (Promega). For the calcium phosphate method, the cells were fed with DMEM supplemented with 10% vol/vol serum and 1% penicillin/streptomycin 2–3 hr before adding the DNA. Forty to forty-eight hours after transfection, cell extracts were prepared and chloramphenicol acetyltransferase (CAT) or luciferase assays were performed. The total amount of DNA transfected by this method is 10–11 μg. For normalization of transfection efficiencies, a β-galactosidase expression plasmid (pCMV-lacZ, Promega) was included in each transfection experiment. For the lipofectamine transfections, the total amount of DNA transfected was 1.0–2.2 μg. Each experimental number represents the average of at least three independent experiments. A complete description of each plasmid construction used in this study is available on request.
Reporter Gene Assays.
CAT assays were as described previously (8). All normalized CAT activity was calculated as follows: CAT activity = acetylated chloramphenicol/total chloramphenicol; normalized CAT activity = CAT activity/β-galactosidase activity. The CAT activity was quantified by using the phosphorimager Fuji system and each number represents the average of at least three independent experiments. The luciferase assays were carried out according to the manufacturer’s recommendations (Promega). Normalized luciferase activities were calculated as relative luciferase units/β-galactosidase activity, and each number represents the average of at least three independent experiments.
RESULTS
Nonclassical Activators and Tat.
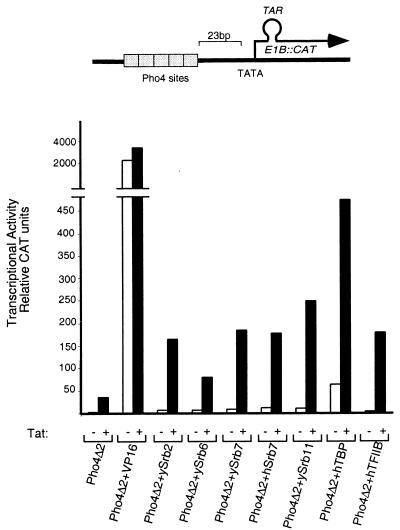
The reporter for this experiment bears five binding sites for the yeast activator Pho4 upstream of the E1B promoter modified so as to harbor a TAR element downstream of the transcription start site (Fig. 1). The figure shows expression of the reporter cotransfected with constructs expressing either a nonclassical activator alone (e.g., a fusion protein bearing a component of the transcriptional machinery fused to Pho4Δ2, the Pho4 DNA-binding domain), Tat alone, or a combination of the two. Also shown is the substantial expression elicited by Pho4Δ2 + VP16, a classical activator, fused to the Pho4 DNA-binding domain. The transcriptional machinery components include yeast Srbs (designated ySrb2, -6, -7, and -11), human (h) Srb7, human TATA box-binding protein (TBP) and hTFIIB. ySrb2 and 6 have no reported human homologues.
Figure 1.
Various DNA-tethered components of the transcriptional machinery can synergize with the HIV-1 activator Tat. All the Pho4 derivatives were expressed from the cytomegalovirus (CMV) enhancer-promoter. The reporter template contained five Pho4-binding sites upstream of the E1B TATA element and a TAR sequence downstream of the start site. DNA (4 μg) encoding Pho4-based effectors was cotransfected with (black bars) or without (white bars) 4 μg of a Tat expression plasmid (also expressed from the CMV enhancer-promoter), 2 μg of the reporter template and, as an internal control, 1 μg of a plasmid encoding lacZ expressed from the CMV promoter. The calcium phosphate precipitation method was used for transfecting HeLa cells with the indicated plasmids. Results of CAT assays are shown.
The figure shows that each nonclassical activator, with the exception of Pho4Δ2 + hTBP (which activated some 25- to 30-fold) activated only some 2- to 3-fold when working on its own. Activation by Tat alone was also barely measurable (20-fold). The figure also shows, however, that in every case the nonclassical activator worked synergistically with Tat, an effect that depended on the presence of a TAR site (not shown). A synergistic effect between DNA-tethered hTBP and Tat has been reported previously (9, 10). We repeated the experiment of Fig. 1 using as a template the HIV-1 LTR promoter bearing four Gal4 sites and as activators Gal4 + hSrb7, Gal4 + ySrb7, and Gal4 + ySrb11. The results were very similar to those shown here, that is, the fusion proteins bearing an Srb moiety activated very little if at all on their own, but they worked highly synergistically with Tat (data not shown).
Nonclassical Activators and Sp1, CTF, and E2F1.
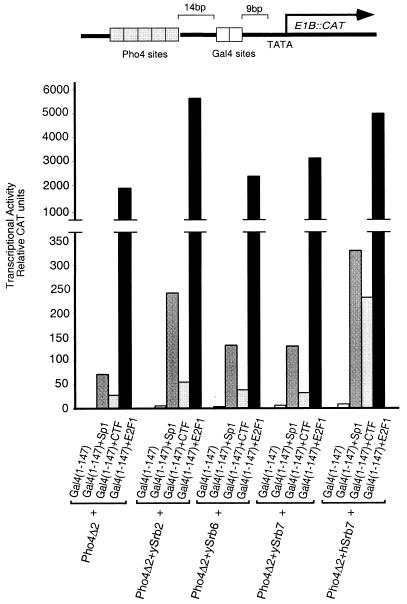
The reporter for this experiment bears two groups of upstream binding sites, one recognized by the DNA-binding domain of the yeast protein Gal4 and the other by that of Pho4. Fig. 2 shows that, as above, ySrb2, -6, and -7, and hSrb7 fused to Pho4Δ2, activated only weakly, whereas the classical activators, CTF, Sp1, and E2F1 fused to the Gal4 DNA-binding domain, activated expression to different but significant extents (15-, 30-, and 1,000-fold, respectively) when working singly. The figure also shows that all combinations of classical with nonclassical activators worked synergistically. In these experiments, the fusion protein bearing hSrb7 (the only human component tested in this experiment) worked most efficiently (i.e., synergized most efficiently) with each of the classical activators tested.
Figure 2.
Nonclassical activators can synergize with DNA-tethered E2F1, CTF, and Sp1. All the Pho4 derivatives were expressed from the CMV enhancer-promoter and all the Gal4 derivatives were expressed from the SV40 enhancer-promoter. DNA (4 μg) encoding Pho4-based effectors was cotransfected with 4 μg of DNA encoding Gal4 derivatives. The reporter template (2 μg) was used and, as an internal control, 1 μg of a plasmid encoding lacZ expressed from the CMV promoter. The calcium phosphate precipitation method was used for transfecting HeLa cells with the indicated plasmids, and results of CAT assays are shown.
These experiments have also been performed with two templates bearing Gal4 sites upstream of LexA sites and with classical activators bearing Gal4 and nonclassical activators bearing LexA DNA-binding domains (these two templates were driven from the E1B promoter). The results were very similar to those shown in Fig. 2 (data not shown), indicating that the observed synergistic effects do not depend on the specific identity of the DNA-binding domains nor on the positioning (upstream or downstream) of the classical activator binding sites relative to those for the nonclassical activators. In other results not shown, we found that DNA-tethered VP16, another acidic activator, also worked synergistically with hSrb7 and ySrb11, the two nonclassical activators tested.
Effect of a Classical Activating Region Lacking a DNA-Binding Domain.
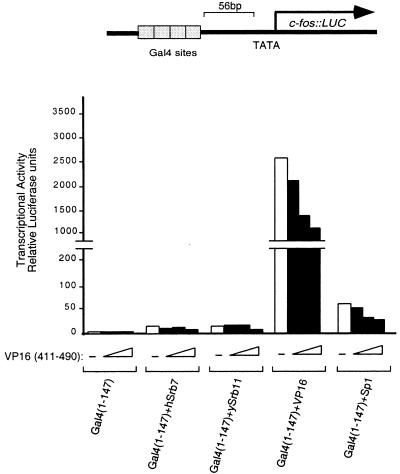
The reporter for this experiment bears four upstream Gal4-binding sites and, consistent with the results of Figs. 1 and 2, this reporter was not markedly stimulated by nonclassical activators bearing hSrb7 or ySrb11 (2- to 3-fold). The reporter was activated markedly by DNA-tethered VP16 and somewhat less so by DNA-tethered Sp1 (Fig. 3). The figure also shows the effects of three different amounts of DNA directing expression of a VP16 lacking a DNA-binding domain. In no case did this fragment activate on its own or when combined with a nonclassical activator. The only detectable effect of that fragment apparent from the figure was in fact inhibition (squelching), a result seen most clearly when the fragment was in the presence of DNA-tethered VP16 or Sp1. In experiments not shown, we found no effect in these experiments when cells were cotransfected with levels of VP16-expressing plasmids 2-fold lower than the lowest level shown here.
Figure 3.
Overexpression of a non-DNA-tethered activating region does not enhance transcriptional activation elicited by nonclassical activators. All the Gal4 derivatives were expressed from the SV40 enhancer-promoter, and the VP16 activating region (residues 411–490) was expressed from the CMV promoter. DNA (0.5 μg) encoding each indicated Gal4 derivative (except for that encoding Gal4 + VP16, in which case 0.2 μg was used) was cotransfected with (black bars) or without (white bars) increasing amounts (0.25, 0.5, and 1 μg) of DNA encoding the VP16 activation domain. The reporter template (0.5 μg) was used and, as an internal control, 0.2 μg of a plasmid encoding lacZ expressed from the CMV enhancer-promoter. The lipofectamine method was used for transfecting HeLa cells with the indicated plasmids, and results of luciferase assays are shown.
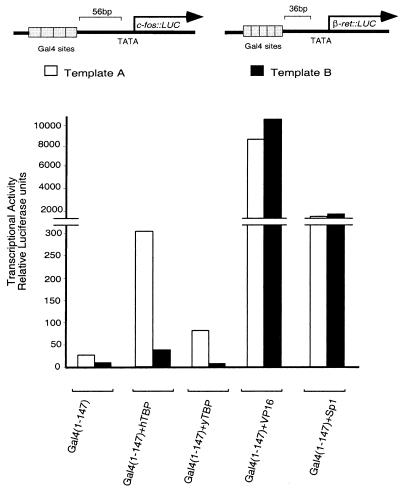
Template Effect on Activation by a Nonclassical Activator.
Two reporters are used here, both of which bear Gal4-binding sites upstream (56 bp in A and 36 in B) of TATA and downstream regions taken from two different mammalian genes (c-fos and β-retinoic acid receptor, respectively) (11). Fig. 4 shows that Gal4 + hTBP and Gal4 + yTBP worked significantly more efficiently on template A than on B. In contrast, Gal4 + Vp16 worked equally efficiently on the two templates, as did (but at a lower level) Gal4 + Sp1. In an experiment not shown, lower levels of the VP16 activator elicited lower but equal levels of expression from the two reporters, and higher levels of the Sp1 activator elicited higher but equal levels of expression from the two promoters.
Figure 4.
Transcriptional activation elicited by DNA-tethered TBP is affected by the reporter template used. All the Gal4 derivatives were expressed from the SV40 enhancer-promoter. DNA (0.5 μg) encoding the nonclassical Gal4 derivatives and Gal4 + Sp1, or 0.25 μg of DNA encoding Gal4 + VP16, was cotransfected with 0.5 μg of either template A (white bars) or template B (black bars) and, as an internal control, 0.2 μg of a plasmid encoding lacZ expressed from the CMV enhancer-promoter. The lipofectamine method was used for transfecting HeLa cells with the indicated plasmids, and results of luciferase assays are shown.
Our findings with fusion proteins bearing hTBP agree with two previous reports that such fusion proteins activate synergistically with Tat. Those studies also noted a failure of fusions bearing TBP to synergize with DNA-tethered Sp1 (9, 10), a result we have also observed (data not shown). The level of transcription elicited in our experiments by Gal4 + hTBP, compared with that elicited by Gal4 + VP16, is lower than that previously reported (9, 10, 12) but, consistent with our overall findings, the difference may be attributable to the different reporters used in the two sets of experiments.
DISCUSSION
In contrast to the results of experiments performed in yeast in ref. 7 (accompanying paper), we find that with the exception of DNA-tethered TBP (9, 10, 12), none of our nonclassical activators elicits a significant level of transcription on its own. That failure was observed in numerous experiments by using reporters bearing derivative of the different promoters (TK, SV40, E1B, HIV-1LTR, c-fos, β-ret.). Similar to the results in yeast, however, each of the nonclassical activators, with the exception of DNA-tethered ySrb6, worked synergistically with one or another classical activator. The one nonclassical activator that worked detectably on its own—that bearing yeast or human TBP—behaved similarly to several nonclassical activators studied in yeast. That is, it preferred markedly one of two templates differing in promoter sequence whereas, also as observed in yeast, classical activators showed no such preference.
Two possible explanations for the failure of the typical nonclassical activator to work on its own in mammalian cells, both of which we disfavor, are: (i) the nonclassical activator is simply not functional in mammalian cells; this notion is contradicted by the numerous examples of synergy between classical and nonclassical activators; (ii) a classical activator is required to perform some function beyond recruitment—inducing an isomerization, for example—that cannot be performed by a nonclassical activator. This idea is contradicted by our failure to find any helping effect of a classical activating region, expressed at several different concentrations but unattached to a DNA-binding domain, in the presence of a DNA-tethered nonclassical activating region. That is, the free activating region had no effect at low concentration and squelched transcription at higher concentration.
We propose two explanations for our results, both of which are based on the surmise that classical activators can contact multiple targets in the transcriptional machinery, whereas nonclassical activators have a much more restricted interaction with that machinery (see discussion in ref. 7). First, it seems possible that, at certain promoters at least, different components of the machinery must be recruited independently for activation. The typical nonclassical activator would be able to recruit only one of these, whereas the typical classical activator, by virtue of the multiple interactions just alluded to, would be able to recruit all the required components. According to this explanation, at certain promoters recruitment of TBP would suffice for activation, the remaining components presumably binding cooperatively with it. It is possible that at the promoters tested here a nucleosome-modifying activity must be recruited independently of the polymerase holoenzyme. We attempted to test that idea by adding the histone deacetylase inhibitor TSA (Trichostatin A) to the transfected cells, but this treatment had no effect on the activities of the nonclassical activators (not shown). Second, it seems plausible that the nonclassical activator–transcriptional machinery complex is highly constrained, and only at certain positionings of the activator binding sites (none of which might have been observed here) would a stable interaction be effected. In contrast, classical activators might be able to adjust their interactions so that a functional complex is properly recruited regardless of the kinds of variables described here and in ref. 7.
The various classical activating regions we have tested here vary somewhat in their abilities to work synergistically with nonclassical activators. Thus both Tat and E2F1 work well with all nonclassical activators tested, whereas CTF works well only with that bearing hSrb7, and Sp1 works best with those bearing ySrb2 and hSrb7. It may be significant in this regard that multiple putative targets have been described for Tat (13–24) and for other activators that, like E2F1, are acidic (25–29).
We suggest that a highly “evolvable” system of gene regulation requires that activators work on nearby genes with as few restrictions as possible with regard to precise positioning or promoter sequence (see ref. 30). Classical activating regions that can contact many surfaces on the transcriptional machinery would work at any of a wide array of promoters. This notion, taken with the results presented here and in ref. 7, both of which show a restricted effectiveness of nonclassical activators compared with their classical counterparts, would explain why the latter, but not the former, is used so widely in nature.
Acknowledgments
We thank David Bentley and Zafar Zaman for the plasmids provided. We thank members of the Ptashne laboratory for useful discussions and comments. We thank Idriss M. Bennani-Baiti for discussion of the manuscript. J.N. was supported by a postdoctoral fellowship from the Spanish Ministry of Education (F.P.I.); L.G. was supported in part by a fellowship from the Medical Research Council of Canada. This work was supported by grants from the National Institutes of Health to M.P.
ABBREVIATIONS
- CAT
chloramphenicol acetyltransferase
- CMV
cytomegalovirus
- h
human
- TPB
TATA box-binding protein
References
- 1.Struhl K. Cell. 1996;84:179–182. doi: 10.1016/s0092-8674(00)80970-8. [DOI] [PubMed] [Google Scholar]
- 2.Ptashne M, Gann A. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 3.Gaudreau L, Adam M, Ptashne M. Mol Cell. 1998;1:913–916. doi: 10.1016/s1097-2765(00)80090-8. [DOI] [PubMed] [Google Scholar]
- 4.Keaveney M, Struhl K. Mol Cell. 1998;1:917–924. doi: 10.1016/s1097-2765(00)80091-x. [DOI] [PubMed] [Google Scholar]
- 5.Farrell S, Simkovich N, Wu Y, Barberis A, Ptashne M. Genes Dev. 1996;10:2359–2367. doi: 10.1101/gad.10.18.2359. [DOI] [PubMed] [Google Scholar]
- 6.Gaudreau L, Schmid A, Blaschke D, Ptashne M, Hörz W. Cell. 1997;89:55–62. doi: 10.1016/s0092-8674(00)80182-8. [DOI] [PubMed] [Google Scholar]
- 7.Gaudreau L, Keaveney M, Nevado J, Zaman Z, Bryant G O, Struhl K, Ptashne M. Proc Natl Acad Sci USA. 1999;96:2668–2673. doi: 10.1073/pnas.96.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehming N, Thanos D, Brickman J M, Ma J, Maniatis T, Ptashne M. Nature (London) 1994;371:175–179. doi: 10.1038/371175a0. [DOI] [PubMed] [Google Scholar]
- 9.Majello B, Napolitano G, De Luca P, Lania L. J Biol Chem. 1998;273:16509–16516. doi: 10.1074/jbc.273.26.16509. [DOI] [PubMed] [Google Scholar]
- 10.He S, Weintraub S J. Mol Cell Biol. 1998;18:2876–2883. doi: 10.1128/mcb.18.5.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryant G O, Martel L S, Burley S K, Berk A J. Genes Dev. 1996;10:2491–2504. doi: 10.1101/gad.10.19.2491. [DOI] [PubMed] [Google Scholar]
- 12.Xiao H, Lis J T, Xiao H, Jeang K T. Mol Cell Biol. 1997;17:6898–6905. doi: 10.1128/mcb.17.12.6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrmann C, Rice A. Virology. 1993;197:601–608. doi: 10.1006/viro.1993.1634. [DOI] [PubMed] [Google Scholar]
- 14.Hagemeier C, Cook A, Kouzarides T. Nucleic Acids Res. 1993;21:4998–5004. doi: 10.1093/nar/21.22.4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeang K T, Chun R, Lin N H, Gatignol A, Glabe C G, Fan H. J Virol. 1993;67:6224–6233. doi: 10.1128/jvi.67.10.6224-6233.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kashanchi F, Piras G, Radonovich M F, Duvall J F, Fattaey A, Chiang C M, Roeder R G, Brady J N. Nature (London) 1994;367:295–299. doi: 10.1038/367295a0. [DOI] [PubMed] [Google Scholar]
- 17.Chiang C M, Roeder R G. Science. 1995;267:531–536. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Q A, Sharp P A. EMBO J. 1995;14:321–328. doi: 10.1002/j.1460-2075.1995.tb07006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parada C A, Roeder R. Nature (London) 1996;384:375–378. doi: 10.1038/384375a0. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Martinez L F, Mavankal G, Neveu J M, Lane W S, Ivanov D, Gaynor R B. EMBO J. 1997;15:2836–2850. doi: 10.1093/emboj/16.10.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Martinez L F, Ivanov D, Gaynor R B. J Biol Chem. 1997;272:6951–6958. doi: 10.1074/jbc.272.11.6951. [DOI] [PubMed] [Google Scholar]
- 22.Cujec T P, Cho H, Maldonado E, Meyer J, Reinberg D, Peterlin B M. Mol Cell Biol. 1997;17:1817–1823. doi: 10.1128/mcb.17.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cujec T P, Okamoto H, Fujinaga K, Meyer J, Chamberlin H, Morgan D O, Peterlin B M. Genes Dev. 1997;11:2645–2657. doi: 10.1101/gad.11.20.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chun R F, Semmes O J, Neuvent C, Jeang K T. J Virol. 1998;72:2615–2529. doi: 10.1128/jvi.72.4.2615-2629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Truant R, Xiao H, Ingles C J, Greenblatt J. J Biol Chem. 1993;268:2284–2287. [PubMed] [Google Scholar]
- 26.Goodrich J A, Hoey T, Thut C J, Admon A, Tjian R. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- 27.Xiao H, Pearson A, Coulombe B, Truant R, Zhang S, Regier J, Triezemberg T, Reinberg D, Flores O, Ingles C, Greenblatt J. Mol Cell Biol. 1994;14:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blau J, Xiao X, McKracken S, O’Hare P, Greenblatt J, Bentley D. Mol Cell Biol. 1997;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearson A, Greenblatt J. Oncogene. 1997;15:2643–2658. doi: 10.1038/sj.onc.1201451. [DOI] [PubMed] [Google Scholar]
- 30.Ptashne M, Gann A. Curr Biol. 1998;8:R812–R822. doi: 10.1016/s0960-9822(07)00508-8. [DOI] [PubMed] [Google Scholar]