Abstract
The interaction of the chaperonin GroEL14 with its cochaperonin GroES7 is dynamic, involving stable, asymmetric 1:1 complexes (GroES7⋅GroEL7–GroEL7) and transient, metastable symmetric 2:1 complexes [GroES7⋅GroEL7–GroEL7⋅GroES7]. The transient formation of a 2:1 complex permits exchange of free GroES7 for GroES7 bound in the stable 1:1 complex. Electrophoresis in the presence of ADP was used to resolve free GroEL14 from the GroES7–GroEL14 complex. Titration of GroEL14 with radiolabeled GroES7 to molar ratios of 32:1 demonstrated a 1:1 limiting stoichiometry in a stable complex. No stable 2:1 complex was detected. Preincubation of the asymmetric GroES7⋅GroEL7–GroEL7 complex with excess unlabeled GroES7 in the presence of ADP demonstrated GroES7 exchange. The rates of GroES7 exchange were proportional to the concentration of unlabeled free GroES7. This concentration dependence points to an associative mechanism in which exchange of GroES7 occurs by way of a transient 2:1 complex and excludes a dissociative mechanism in which exchange occurs by way of free GroEL14. Exchange of radiolabeled ADP from 1:1 complexes was much slower than the exchange of GroES7. In agreement with recent structural studies, this indicates that conformational changes in GroEL14 following the dissociation of GroES7 must precede ADP release. These results explain how the GroEL14 cavity can become reversibly accessible to proteins under in vivo conditions that favor 2:1 complexes.
Molecular chaperones are proteins that have evolved to assist efficient protein folding, trafficking, and assembly of proteins in vivo. The most widely studied molecular chaperones are the so-called chaperonin proteins from Escherichia coli, GroEL14 and GroES7. GroEL14 is a tetradecamer of 14 identical 57-kDa subunits arranged in two seven-member, doughnut-shaped rings that are stacked back-to-back to yield a cylindrical structure with identical ends (1). GroES7 is a single ring of seven identical 10-kDa subunits (2). GroES7 uses a large mobile loop from each of its subunits to bind to and regulate the activity of GroEL14 (3, 4). GroES7 is required for the successful refolding of polypeptides by GroEL14 under conditions where spontaneous folding does not occur (5). During this stringent folding, the partially folded target protein binds to one end of the GroEL14 cylinder. GroES7 can then bind to the same end and displace the target protein into the protected space that is formed by the GroEL14 cylindrical chamber and the overlying GroES7 dome. The release of the target protein from this complex requires the dissociation of the GroES7. The interactions with GroES7, the release of target proteins, and the required conformational changes in GroEL14 depend on ATP binding and hydrolysis and the release of the ADP and inorganic phosphate that are formed.
One difference among mechanisms proposed to explain the details of chaperonin-assisted folding relates to the stoichiometry of the GroES7–GroEL14 complexes formed during the cycle of chaperonin-assisted refolding. Electron microscopy and chemical crosslinking have demonstrated the existence of both 1:1 GroES7–GroEL14, and 2:1 (GroES7)2–GroEL14 complexes under various conditions (6–8). The 2:1 complexes were further shown to be active in the refolding of RuBisCO (9). Ultracentrifugation studies have also demonstrated that 1:1 and 2:1 complexes could be quantified under various equilibrium conditions in the presence of ADP or nonhydrolyzable ATP analogs such as adenosine 5′-[γ-[S]thio]triphosphate (ATPγS) or adenosine 5′-imido triphosphate (10). This conclusion is supported by direct binding studies using fluorescently labeled GroES7, and additional conditions were determined for the formation of 2:1 complexes (11). The results indicated that at the concentrations in the E. coli cell (12) (2.6 μM GroEL14, 8 mM ATP, and GroES7/GroEL14 = 2), 2:1 GroES7–GroEL14 complexes likely predominate. However, others have questioned the significance of the symmetric 2:1 complexes (13, 14).
Recent structural (4) and functional (15) studies have visualized the 1:1 GroES7–GroEL14 complex and demonstrated a mechanism requiring only 1:1 complexes in the functional cycle, and it was shown that ATP binding to the ring trans to the GroES7 dome was sufficient to discharge the GroES7. Although this particular model is depicted as involving an asymmetric 1:1 complex, considerations of symmetry have led to the suggestion that it would be possible to envision this mechanism as a special case of the more general situation involving a 2:1 complex (16, 17).
These studies have led to the view that the chaperonin cycle only involves the asymmetric complex AGroEL7–BGroEL7⋅ADP7⋅GroES7. Single-turnover experiments (18) showed that the bound GroES7 underwent complete exchange with free GroES7 on the time scale of ATP hydrolysis. Two exchange mechanisms, associative and dissociative, can be envisaged. In the associative mechanism, binding of ATP and GroES7 to the trans ring (AGroEL7) of the asymmetric complex leads to the transient formation of an unstable, pseudosymmetrical complex GroES7⋅ATP7⋅AGroEL7–BGroEL7⋅ADP7⋅GroES7. Dissociation of the ligands from the BGroEL7 and ATP hydrolysis on the AGroEL7 ring leads to the regeneration of the asymmetric complex. In the dissociative mechanism, the departing GroES7 is thought to dissociate before the association of the incoming GroES7. The dissociative mechanism thus involves the transient formation of an unadorned GroEL14 complex. Electron microscopy and cross-linking studies of mixtures of GroEL14 and GroEL14–GroES7 in the presence of MgATP consistently show a high population of pseudosymmetrical, football-shaped particles, and the almost complete absence of unadorned GroEL7–GroEL7 particles, observations that are consistent with an associative mechanism (6–8).
These considerations make it interesting to consider the disparate views of the importance of complexes with different stoichiometries and to understand how the accessibility of the GroEL14 cavity can be modulated under conditions that apparently favor the 2:1 complexes. It has recently been demonstrated by using direct binding measurements with fluorescently labeled GroES7 that both 1:1 and 2:1 complexes could be formed with appropriate combinations of nucleotides (11). Thus, 2:1 complexes could be formed in the presence of ATP and high KCl, although only 1:1 complexes could be formed in the presence of ADP or adenosine 5′-imido triphosphate individually. Subsequent addition of the other nucleotide to preformed 1:1 complexes to give solutions with mixed nucleotides resulted in formation of 2:1 complexes, suggesting that an asymmetric distribution of nucleotides on the two rings favored a 2:1 complex. In the present study we have explored the stability of the asymmetric AGroEL7–BGroEL7⋅ADP7⋅GroES7 complex, imposing a symmetrical distribution of the nucleotide using only ADP. As before, no stable symmetric 2:1 GroES7⋅ADP7⋅AGroEL7–BGroEL7⋅ ADP7⋅GroES7 could be detected. However, the existence of such a species can be deduced from the dependence of the rate of exchange of bound, isotopically labeled GroES7 on the concentration of free GroES7, a result which points to an associative exchange mechanism.
MATERIALS AND METHODS
Protein Purification.
GroES7 and GroEL14 were purified as described previously (19, 20). Protein concentrations were determined by the method of Bradford (21).
Polyacrylamide Gel Electrophoresis.
Some nondenaturing gel electrophoresis was performed by the method of Neuhoff et al. (22) by using 6% polyacrylamide gels at a constant 200 V for a 1-mm-thick gel. Where appropriate, gels were stained for protein with 0.05% Coomassie brilliant blue R250, 25% isopropyl alcohol, and 10% acetic acid.
Resolution of GroEL14 from GroES7–GroEL14 complexes depended on the electrophoresis conditions, and the following conditions were found to give satisfactory resolution for our experiments. Electrophoresis was done under nondenaturing conditions with 4.5% polyacrylamide gels prepared in 0.5 M Tris borate (pH 8.5) supplemented with 5 mM magnesium acetate and 2 mM ADP. The running buffer contained 90 mM Tris borate, 1 mM magnesium acetate, and 0.2 mM ADP at pH 8.5. Before electrophoresis, samples were dissolved in a buffer consisting of 25 mM Tris borate (pH 8.5), 2% 2-mercaptoethanol, 10% glycerol, 2 mM ADP, 5 mM magnesium acetate, and 0.01% bromphenol blue as a tracking guide.
Preparation of 14C-Labeled GroEL14 and GroES7.
14C-labeled GroEL14 and GroES7 were prepared by reductive methylation using sodium cyanoborohydride essentially by the method of Jentoft and Dearborn (23, 24). For a typical labeling of GroES7, 20 mM oligomer was treated with 14C-labeled formaldehyde at a ratio of formaldehyde/lysine of 1.8 using a formaldehyde solution that was 59.9 μCi/ml (3 μCi/mM; 1 Ci = 37 GBq). The solution was 20 mM in cyanoborohydride and 0.1 M sodium phosphate, pH 7.6. The sample was incubated at room temperature for 2 hours. The 150-μl sample was freed of excess label and small molecules by two successive treatments with Sephadex G50 gel filtration spin columns of 1 ml each, equilibrated with 50 mM Tris⋅HCl (pH 7.8) containing 0.5 mM DTT. GroEL14 was similarly labeled. The protein concentrations of GroES7 and GroEL14 were determined as indicated above. Each radiolabeled sample was tested and found to be active in refolding of denatured rhodanese by using the assay noted above.
Preparation of [α-32P]ADP and Measurement of ADP Exchange.
[α-32P]ADP was prepared by treating 60 μl of 10.5 mM [α-32P]ATP (48 μCi/μM) in a total volume of 160 μl of a solution containing hexokinase (375 units/ml), glucose (0.2 M), MgCl2 (10 mM), and Tris⋅HCl (50 mM, pH 7.8). TLC on polyethyleneimine-cellulose using 1 M LiCl confirmed that the conversion to ADP was complete at 30 min. The sample was diluted 1:5 with 10 mM triethylammonium bicarbonate, pH 7.5, and loaded onto a DEAE-cellulose column (0.8 × 4.5 cm) previously equilibrated with the same buffer. ADP was eluted by using a gradient of 10–450 mM triethylammonium bicarbonate, pH 7.5. ADP-containing fractions were collected, their identity confirmed by chromatography, pooled, and vacuum dried.
GroEL14⋅[α-32P]ADP⋅GroES7 complexes were formed by coincubating GroEL14 (1 μM oligomer), 0.9 μM GroES7, 50 μM [α-32P]ADP (20 μCi/μM), 50 mM KCl, 5 mM Mg acetate, and 10 mM Mops⋅KOH, pH 7.2 in a final volume of 120 μl. The sample was incubated for 15 min at room temperature. The following additions were made to separate aliquots of the complex: (i) No addition; (ii) unlabeled ADP (2 mM final); (iii) GroES7 (5-fold excess over GroEL14); and (iv) unlabeled ADP (2 mM) + GroES7 (5-fold excess over GroEL14). Samples were incubated at room temperature for 2 hr, and aliquots were subjected to electrophoresis as described below. For the zero-time sample, electrophoresis was started within 5 min of mixing the samples.
Detection and Quantitation of Radiolabeled GroEL14, GroES7, and ADP.
Gel electrophoresis was performed as indicated in the individual experiments in Results and Discussion. Resulting gels were dried under vacuum onto Whatman 3MM paper. Radiolabel was detected and quantified by using a storage phosphor screen and a PhosphorImager from Molecular Dynamics. Linearity of the response of the system was evaluated for each protein by determining standard curves using increasing quantities of radiolabeled samples containing known protein concentrations. These standard curves were used to relate the PhosphorImager responses to protein concentrations. Analogous procedures were applied to quantitation of [α-32P]ADP.
Quantitation of GroES7 Exchange Using Gel Permeation Chromatography.
Labeled GroEL14–GroES7 complexes were formed by incubating 5 μM GroEL14 with 5 μM 14C[GroES7] in 50 mM Tris⋅HCl (pH 7.8) containing 10 mM MgCl2, 10 mM KCl, and 1 mM ADP. After 1 hour, unlabeled GroES7 was added to the solutions to give the desired molar ratios of GroES7/GroEL14, which diluted the complexes to 3.2 μM. Labeled GroES7 remaining in the complexes at a particular time was quantified by diluting an aliquot of the incubation mixture to 0.256 μM and injecting 100 μl onto a 7.8 × 300 mm Bio-Sep SEC 4000 HPLC gel permeation column (Phenomenex) that was developed at 0.5 ml/min with 50 mM Tris⋅HCl (pH 7.8) containing 10 mM MgCl2, 0.5 mM KCl, 0.5 mM DTT, and 50 μM EDTA. Fractions of 500 μl were subjected to scintillation counting. This procedure resulted in baseline separation between the peaks containing GroEL14 and free GroES7. The initial incubation typically led to incorporation of 0.8–0.85 GroES7/GroEL14.
RESULTS AND DISCUSSION
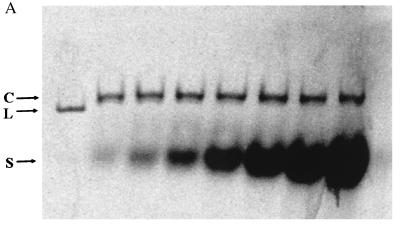
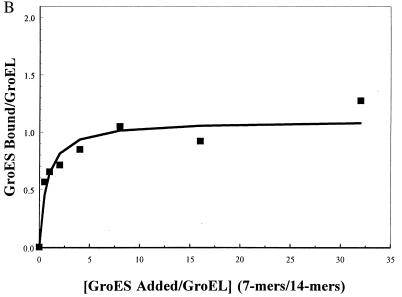
The binding of GroES7 to GroEL14 produces a complex that has altered electrophoretic properties (25). The resolution of GroEL14–GroES7 complexes from GroEL14 itself depends on the conditions used, and the method described in Materials and Methods using a Tris borate buffer was found to give clearer and more reproducible separations than earlier procedures. Fig. 1A shows the results of electrophoresis of GroES7–GroEL14 complexes formed and run in the presence of ADP. Lanes 2–8 show radiolabeled bands from GroES7 as increasing amounts of [14C]GroES7 are added to a fixed concentration of GroEL14. Lane 1 is a control that shows the position of [14C]GroEL14 (labeled L); all other lanes contain radioactivity only in GroES7. The GroES7, when added to GroEL14, only migrates in positions corresponding to free GroES7 (labeled S) and the complex marked C. No GroES7 comigrates with uncomplexed GroEL14. Lanes 2–8 represent increasing ratios of radiolabeled GroES7/GroEL14 from 0.5:1 to 32:1. Although the Coomassie stain of this gel shows some protein at the position of GroEL14 at the lower concentrations of GroES7 (data not shown), no radiolabel is detected in a position corresponding to uncomplexed GroEL14. Thus, any binding of radiolabled GroES7 in the presence of ADP leads to a shift in GroEL14 to the position designated C. Fig. 1B shows the results of quantitation of the radiolabel in the complex, which demonstrate that even at very high levels of GroES7, only 1:1 complexes are stable during electrophoresis in the presence of ADP. This conclusion is supported by the observation of insignificant amounts of radiolabel between the complex and free GroES7 in each lane of Fig. 1, suggesting that no radiolabel was dissociating from the complexes during electrophoresis. Furthermore, although a comparable decrease in electrophoretic mobility was to be expected on formation of a 2:1 complex, no stable 2:1 complexes were detected after electrophoresis. Previous fluorescence anisotropy studies indicated that there was some amount of 2:1 complex in solution at steady state under comparable conditions. Together, these results indicate that the 2:1 complexes are present, but they are in dynamic equilibrium with 1:1 complexes, and under these dilute conditions the 1:1 complexes are favored. For example, at [ADP] = 2 mM used here, and at a ratio of 5:1 GroES7/GroEL14, the previous anisotropy results suggested that there were 1.3 GroES7 per GroEL14 (11). At a ratio of 32:1 GroES7/GroEL14, 2:1 complexes are expected to predominate. However, least squares fitting of the electrophoretic binding data shown here gives a maximum binding of 1.1 ± 0.08 GroES7/GroEL14. Therefore, the results in Fig. 1 A and B demonstrate that the only stable complexes observed in electrophoresis are 1:1. These complexes do not significantly dissociate during electrophoresis. This latter conclusion was confirmed by noting that increasing the duration of electrophoresis did not change the results described here, i.e., not much dissociation occurs, even if more time is allowed (the electrophoresis in Fig. 1A was for 5 hours). Thus, it is clear that in the presence of the levels of ADP used here, 2:1 complexes are not stable.
Figure 1.
Titration of GroEL with increasing concentrations of GroES. (A) Nondenaturing electrophoresis of GroEL–GroES complexes containing radiolabeled GroES. All bands in lanes 2–8 contain 2.88 pg of unlabeled GroEL and radiolabeled GroES at increasing molar ratios of GroES/GroEL (0.5, 1, 2, 4, 8, 16, and 32, respectively). Lane 1 shows 2.88 pg of radiolabeled GroEL and is included to mark the position of uncomplexed GroEL. L, position of GroEL; C, position of GroES/GroEL complex; S, position of uncomplexed GroES. Each incubation sample contained the following concentrations: 0.24uM GroEL14, 2 mM ADP, 5 mM magnesium acetate, 50 mM KCl, and 10 mM Mops⋅KOH (pH 7.2). (B) The ratio of bound GroES/complex as a function of added GroES7/GroEL14. Line is a least-squares fit to a binding isotherm. The maximum binding corresponds to 1.1 ± 0.08 GroES/GroEL.
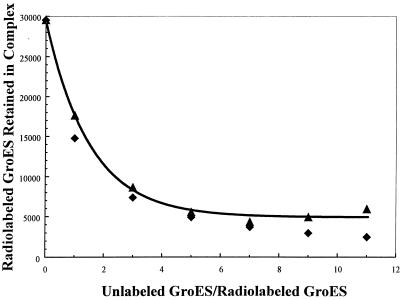
Fig. 2 shows the results of preincubation of radiolabeled 1:1 complexes with increasing ratios of unlabeled GroES7. The samples for this experiment were incubated at room temperature for 60 minutes before electrophoresis. The bands corresponding to ‘C’ in Fig. 1A were quantified, and the results clearly indicate that the 1:1 complexes can exchange their radiolabeled GroES7 when incubated in this fashion. It should be noted that GroES7 migrates considerably faster than either the complex or the free GroEL14. Therefore, the unlabeled, unbound GroES7 rapidly separates from the GroEL14 species so that the complex largely electrophoreses in the absence of unlabeled GroES7. These results show that ≈50% of the radiolabeled GroES7 can be displaced by this procedure at a molar ratio of 1:1 unlabeled to radiolabeled GroES7, with the original GroES7 being present at a molar ratio of 5:1 over GroEL14. Therefore, based on Fig. 1, the stable complexes that were initially present contained approximately 0.8:1 GroES7/GroEL14. The counts remaining in the complex after incubation with increasing amounts of added unlabeled GroES7 (Fig. 2) closely correspond to that expected for isotope dilution, indicating that equilibriation is complete after 60 min. Overall, the results confirm that the 1:1 complexes are stable and demonstrate that exchange occurs with excess GroES7.
Figure 2.
Exchange of bound radiolabeled GroES7 as a function of added unlabeled GroES. Plot shows radiolabeled GroES7 retained in GroES7–GroEL14 complex vs. the ratio of added unlabeled GroES to radiolabeled GroES7. The initial sample contained 0.34 μM GroEL14 and 1.7 μM radiolabeled GroES7 in the same buffer as in Fig. 1. The abscissa gives the ratio of added unlabeled GroES7 over the initial radiolabeled GroES7. Units of the ordinate are counts from the PhosphorImager output. Electrophoresis was performed as in Fig. 1, and the radiolabel was quantified as described in Materials and Methods. Actual data are represented by ▴; theoretical expectations for isotopic equilibration are represented by ⧫. The line is an exponential fit to the data.
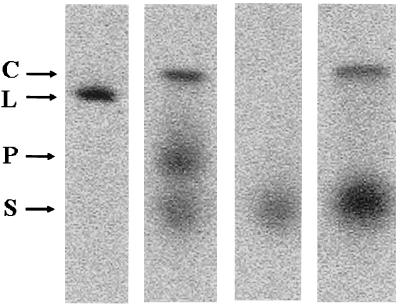
Fig. 3 demonstrates the exchange of GroES7 from stable 1:1 complexes in a different way. These results demonstrate what might be termed “exchange in passing,” because the experiment takes advantage of the fact that GroES7 migrates more rapidly on native gel electrophoresis than either the complex or GroEL14, thus allowing GroES7 that is added after initiating electrophoresis to pass the complex. While passing, the unlabeled GroES7 has an opportunity to exchange with the radiolabeled GroES7 in complex. Lanes 2, 3, and 4 contain only radiolabeled GroES7, whereas lane 1 shows the position of radiolabeled GroEL14 for comparison. Lane 3 shows the position of labeled GroES7 electrophoresed in the absence of GroEL14. Lane 2 shows the behavior of the GroEL14–GroES7 complex prepared with radiolabeled GroES7 in which electrophoresis was stopped after 1 hour, at which time the tracking dye was approximately 1.7 cm into the gel. At that time, unlabeled GroES7 was added to the sample well and electrophoresis was continued for an additional 4 hours. The added GroES7 migrates rapidly, and it overtakes and passes the stable complex. These results demonstrate that there is displacement of radiolabeled GroES7 as the unlabeled GroES7 passes the complex. The displaced GroEL14 migrates at the position labeled P, whereas the excess GroES7 that was present with the original complex is at the position labeled S. Lane 4 shows that if the complex between GroEL14 and radiolabeled GroES7 is preincubated with excess unlabeled GroES7 before electrophoresis, the only uncomplexed GroES7 migrates at the position S, demonstrating that the exchange was complete before electrophoresis.
Figure 3.
Displacement en passant of labeled GroES7 by unlabeled GroES7 added after initiation of electrophoresis. Lane 1, labeled GroEL14 (L) to mark the position of uncomplexed GroEL14. Lanes 2–4 contain radiolabel only in GroES7: lane2, GroEL14–labeled GroES7 complex prepared as in Fig. 1 by using equimolar labeled GroES7 and incubated for 15 min at room temperature. Complex (3.6 pM) was electrophoresed for 60 min. Electrophoresis was briefly interrupted, and 100× unlabeled GroES7 in 2 mM ADP was added and the run was continued for 4 more hours. Lane 3, radiolabeled GroES7 alone to mark position of GroES7. Lane 4, GroEL14–GroES7 prepared as for lane 2 but preincubated with 100× unlabeled GroES7 before electrophoresis.
To support the electrophoretic results, the kinetics of GroES7 exchange were investigated by using gel permeation chromatography. An associative mechanism involving a 2:1 complex would be expected to show increasing rates of exchange with increasing concentrations of unlabeled GroES7, whereas the normalized exchange rates for a mechanism that proceeds by dissociation of the 1:1 complex would be independent of the concentration of added GroES7. The results shown in Table 1 are clearly in accord with the associative process. The rates strongly depend on the added GroES7 concentration, and the exchange rates increase by a factor of 40 at a 30-fold excess of unlabeled GroES7. The half-time for exchange with no added GroES7 is almost 75 hr, whereas the half-time falls to 1.9 hr in the presence of a 30-fold molar excess of unlabeled GroES7.§ As expected for an associative process in which the apparent rate constant includes the unlabeled GroES7 concentration as a factor, the observed rates increase linearly with increasing unlabeled GroES7.
Table 1.
Quantitation of GroES7 exchange by gel permeation chromatography
| Ratio [GroES7/ ([14C]GroES7/GroEL14)] | Apparent normalized rate constants for GroES7 exchange ×104 min−1 |
|---|---|
| Complex alone (no additions) | 1.6 |
| 10 | 25 |
| 20 | 40 |
| 30 | 60 |
GroEL14–GroES7 complexes were prepared using [14C]GroES7 as described in Materials and Methods. After 1 hour at 25°C, the samples were brought to the indicated ratio of GroES7/GroEL14 by the addition of unlabeled GroES7. The GroES7 remaining with the complexes was quantified using gel permeation chromatography as described in Materials and Methods. The percent remaining labeled GroES7 in complexes was quantified using gel permeation chromatography as a function of time between 0 and 235 min. The data were fit to first-order kinetics, and the derived pseudo-first-order rate constants are presented.
Under comparable conditions, the exchange of ADP is 7–10 times slower than the exchange of GroES7 from the AGroEL7–BGroEL7⋅ADP7⋅GroES7 complex (data not shown). In the structure of this complex (4), the ADP is locked into its binding site by the conformational change that accompanies GroES7 binding. This suggests an obligatory order for association and dissociation of these ligands—first on, last off—in which nucleotide binds before GroES7, whereas GroES release precedes nucleotide dissociation. The present results indicate that the events following the dissociation of GroES7 that cause the release of ADP must be slow relative to the rate of rebinding of GroES7. Otherwise the rates of ADP and GroES7 exchange would be the same, which is not what is observed.
The present results demonstrate that stable 1:1 GroES7–GroEL14 complexes form in the presence of ADP, and they do not significantly dissociate during electrophoresis or gel permeation chromatography. Furthermore, 2:1 complexes detected by fluorescence anisotropy measurements (11) are not detected as stable species under the conditions used here. However, the GroES7 in these stable 1:1 complexes can be exchanged with GroES7 in solution by an associative mechanism involving the transient formation of unstable 2:1 complexes.
A previous study (27) detected little exchange of GroES7 from 1:1 complexes at levels of GroES7 at which the present work clearly demonstrates exchange. This difference can be rationalized, because it has been shown that, in addition to excess GroES7, the formation of 2:1 complexes depends on the level of ADP present in solution (11). At the low ADP levels used in a previous studies (27), no 2:1 complexes could be detected by fluorecence anisotropy, whereas at the ADP levels used here, a significant population of 2:1 complexes (≈20%) could be observed (11). Thus, the present results are consistent with the model suggested previously indicating that GroES7–GroEL14 complexes are not stable if they are truly symmetric (i.e., ADP and GroES7 on both rings).
Although apparently symmetric 2:1 complexes can form and appear to persist in the presence of ATP, this steady-state snapshot of the population of molecules obscures highly dynamic behavior. Pre-steady state studies of the catalytic cycle (15, 18, 28–30) indicate that the cycle proceeds unidirectionally, with the ligands GroES7 and ADP dissociating from alternate GroEL7 rings in the manner of a two stroke engine.; i.e., AGroEL7–BGroEL7⋅ADP7⋅GroES7 → GroES7⋅ATP7⋅AGroEL7–BGroEL7⋅ ADP7⋅GroES7 → GroES7⋅ATP7⋅AGroEL7–BGroEL7 → GroES7⋅ADP7⋅AGroEL7–BGroEL7 → GroES7⋅ ADP7⋅A GroEL7–BGroEL7⋅ATP7⋅GroES7 → AGroEL7–BGroEL7⋅ATP7⋅ GroES7 → AGroEL7–BGroEL7⋅ADP7⋅GroES7. Although the binding of a second GroES7 is not obligatory to operation of the cycle, at the cellular concentrations of GroEL14, GroES7, and ATP, there is every reason to expect its participation in the cycle in vivo.
The truly symmetric 2:1 complex GroES7⋅ADP7⋅AGroEL7–BGroEL7⋅ADP7⋅GroES7 invoked in the present study cannot be considered an intermediate in the chaperonin cycle for two reasons. If it were involved, a single round of ATP hydrolysis would cause the loss of only half of the radiolabled ligand (ADP or GroES7) present in the starting asymmetric complex. In reality, 100% of the radiolabeled ligand is lost (18). Second, the asymmetry of the system is maintained by the fact that ATP hydrolysis by one GroEL7 ring is inhibited so long as there is ADP present in the other GroEL7 ring (30). Alternatively, asymmetry could be maintained by the cooperative hydrolysis of ATP on one ring of GroEL14 provided that both products, ADP and Pi, remained trapped within that ring until ADP had dissociated from the other ring. However, it has not proven possible to trap 32Pi from [γ-32P]ATP in the stable asymmetric complex, although [α-32P]ADP from [α-32P]ATP can readily be trapped, as such a model would predict. Furthermore, unpublished results (cited in ref. 30) indicate that the release of Pi from the complex is coincident with ATP cleavage.
The recent x-ray structure (4) of the 1:1 complex with ADP and GroES7 bound on the same ring of GroEL14 suggests a common origin for the instability of the truly symmetric 2:1 GroES7⋅ADP7⋅AGroEL7–BGroEL7⋅ADP7⋅GroES7 complex observed here as well as for the pseudosymmetric 2:1 GroES7⋅ATP7⋅AGroEL7–BGroEL7⋅ADP7⋅GroES7 complex that is thought to form transiently during the normal catalytic cycle. Three things are coupled in the occupied GroEL14 ring: (i) binding of GroES7, (ii) binding of seven nucleotides, and (iii) changes in the orientations of the equatorial domains at the ring–ring interface. Because this interface is maintained by a tight coupling of the equatorial domains between the two rings, changes in one ring induce complementary changes in the opposite ring, with the result of reducing the symmetry that is present in unliganded GroEL14. This symmetry-breaking at the interface can explain the observed anticooperativity in binding of ligands to the two rings of GroEL14. Binding of ATP alone to the trans ring is sufficient to induce the dissociation of the ligands in the cis ring (15). However, binding of ADP to the trans ring does not induce the dissociation of the ligands from the cis ring. But, as shown here, binding of both ADP and GroES7 to the trans ring induces the departure of GroES7 from the cis ring. Thus, regardless of whether the 2:1 complexes are symmetric or merely pseudosymmetric, they are intrinsically unstable and tend to revert to asymmetric resting states.
Footnotes
The rate of GroES7 exchange observed by using chromatography to separate the GroES7–GroEL14 complex from free GroES7 is slower than that observed by using PAGE. We attribute this discrepancy to excluded volume effects in the polyacrylamide gel matrix (26).
References
- 1.Braig K, Otwinowski Z, Hedge R, Boisvert D C, Joachimiak A, Horwich A L, Sigler P B. Nature (London) 1994;371:578–586. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- 2.Hunt J F, Weaver A J, Landry S J, Gierasch L M, Deisenhofer J. Nature (London) 1996;379:37–45. doi: 10.1038/379037a0. [DOI] [PubMed] [Google Scholar]
- 3.Landry S J, Zeilstra-Ryalls J, Fayet O, Georgopoulos C, Gierasch L M. Nature (London) 1993;364:255–258. doi: 10.1038/364255a0. [DOI] [PubMed] [Google Scholar]
- 4.Xu Z, Horwich A L, Sigler P M. Nature (London) 1997;388:741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt M, Buchner J, Todd M J, Lorimer G H, Viitanen P V. J Biol Chem. 1994;269:10304–10311. [PubMed] [Google Scholar]
- 6.Azem A, Kessel M, Goloubinoff P. Science. 1994;265:653–656. doi: 10.1126/science.7913553. [DOI] [PubMed] [Google Scholar]
- 7.Llorca O, Carrascosa J L, Valpuesta J M. J Biol Chem. 1996;271:68–76. doi: 10.1074/jbc.271.1.68. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt M, Rutkat K, Rachel R, Pfeifer G, Jaenicke R, Viitanen P V, Lorimer G H, Buchner J. Science. 1994;265:656–659. doi: 10.1126/science.7913554. [DOI] [PubMed] [Google Scholar]
- 9.Azem A, Diamant S, Kessel M, Weiss C, Goloubinoff P. Proc Natl Acad Sci USA. 1995;92:12021–12025. doi: 10.1073/pnas.92.26.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Behlke J, Ristau O, Schonfeld H-J. Biochemistry. 1997;36:5149–5156. doi: 10.1021/bi962755h. [DOI] [PubMed] [Google Scholar]
- 11.Gorovits B M, Ybarra J, Seale J W, Horowitz P M. J Biol Chem. 1997;272:26999–27004. doi: 10.1074/jbc.272.43.26999. [DOI] [PubMed] [Google Scholar]
- 12.Lorimer G H. FASEB J. 1996;10:5–9. doi: 10.1096/fasebj.10.1.8566548. [DOI] [PubMed] [Google Scholar]
- 13.Engel A, Hayer-Hartl M K, Goldie K N, Pfeifer G, Hegerl R, Muller S, da Silva A C R, Baumeister W, Hartl F U. Science. 1995;269:832–836. doi: 10.1126/science.7638600. [DOI] [PubMed] [Google Scholar]
- 14.Hayer-Hartl M K, Martin J, Hartl F U. Science. 1995;269:836–841. doi: 10.1126/science.7638601. [DOI] [PubMed] [Google Scholar]
- 15.Rye H S, Burston S G, Fenton W A, Beechem J M, Xu Z, Sigler P B, Horwich A L. Nature (London) 1997;388:792–798. doi: 10.1038/42047. [DOI] [PubMed] [Google Scholar]
- 16.Xu, Z. & Sigler, P. B. (1999) J. Struct. Biol., in press.
- 17.Lorimer G. Nature (London) 1997;388:720–723. doi: 10.1038/41892. [DOI] [PubMed] [Google Scholar]
- 18.Todd M J, Viitanen P V, Lorimer G H. Science. 1994;265:659–666. doi: 10.1126/science.7913555. [DOI] [PubMed] [Google Scholar]
- 19.Staniforth R A, Cortes A, Burston S G, Atkinson T, Holbrook J J, Clarke A R. FEBS Lett. 1994;344:129–135. doi: 10.1016/0014-5793(94)00348-3. [DOI] [PubMed] [Google Scholar]
- 20.Clark A C, Hugo E, Frieden C. Biochemistry. 1996;35:5893–5901. doi: 10.1021/bi953051v. [DOI] [PubMed] [Google Scholar]
- 21.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 22.Neuhoff V, Cheong-Kim K S, Altland K. Electrophoresis. 1986;7:56–57. [Google Scholar]
- 23.Jentoft N, Dearborn D G. J Biol Chem. 1976;254:4359–4365. [PubMed] [Google Scholar]
- 24.Jentoft N, Dearborn D G. Anal Biochem. 1980;106:186–190. doi: 10.1016/0003-2697(80)90135-9. [DOI] [PubMed] [Google Scholar]
- 25.Langer T, Pfeifer G, Martin J, Baumeister W, Hartl F U. EMBO J. 1992;11:4757–4765. doi: 10.1002/j.1460-2075.1992.tb05581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minton A P. Methods Enzymol. 1998;298:127–149. doi: 10.1016/s0076-6879(98)95038-8. [DOI] [PubMed] [Google Scholar]
- 27.Hayer-Hartl M K, Weber F, Hartl F U. EMBO J. 1996;15:6111–6121. [PMC free article] [PubMed] [Google Scholar]
- 28.Burston S G, Ranson N A, Clarke A R. J Mol Biol. 1995;249:138–152. doi: 10.1006/jmbi.1995.0285. [DOI] [PubMed] [Google Scholar]
- 29.Ranson N A, Burston S G, Clarke A R. J Mol Biol. 1997;266:656–664. doi: 10.1006/jmbi.1996.0815. [DOI] [PubMed] [Google Scholar]
- 30.Kad N M, Ranson N A, Cliff M G, Clarke A R. J Mol Biol. 1998;278:267–278. doi: 10.1006/jmbi.1998.1704. [DOI] [PubMed] [Google Scholar]