Abstract
Vaccinia virus early, intermediate, and late stage genes are sequentially transcribed by the viral RNA polymerase within the cytoplasm of infected cells. We found that the 34- and 45-kDa polypeptides encoded by vaccinia virus ORFs A8R and A23R, respectively, were necessary to reconstitute transcription of a template with an intermediate stage promoter. Coexpression of the A8R and A23R genes in Escherichia coli was required for in vitro activity. In addition, the two polypeptides copurified, indicating their association as protein subunits of a vaccinia virus intermediate transcription factor. This factor, which we named VITF-3, complemented three viral proteins—namely, the RNA polymerase, capping enzyme, and a 30-kDa protein called VITF-1 that is also a subunit of the RNA polymerase—and an unidentified cell factor called VITF-2. Expression of the A8R and A23R genes occurred between 1 and 5 h after vaccinia virus infection and was not prevented by an inhibitor of DNA replication, consistent with a role for VITF-3 in specifically regulating intermediate transcription in vivo. The vaccinia virus A8R and A23R genes are highly conserved among vertebrate poxviruses, but no other viral or cellular homologs were identified.
The majority of DNA viruses replicate in the nucleus of the cell and divert the cellular transcription system for viral gene expression. Poxviruses differ from this norm in that they replicate and transcribe their genomes within the cytoplasm of infected cells and encode most of the proteins required for the synthesis of viral macromolecules (1). Studies with vaccinia virus, the prototype of the large poxvirus family, indicated the presence of three classes of genes that are expressed in succession (2, 3). The early proteins are synthesized before DNA replication and the intermediate and late proteins are made after this event. Subsequent investigations provided evidence for a unique consensus promoter sequence for each gene class (4–6) and for cognate stage-specific transcription factors that work with a viral multisubunit RNA polymerase (7–10).
The ordered expression of vaccinia viral genes is mediated by stage-specific transcription factors. The vaccinia virus early transcription factor (VETF) (11, 12) and the 94-kDa protein that provides RNA polymerase with early promoter specificity (13) are synthesized at late times after infection and are packaged in virus particles along with the multisubunit RNA polymerase and additional enzymes. Therefore, the transcription of early genes occurs immediately after infection and does not require de novo protein or DNA synthesis. The intermediate transcription factors (VITFs) are synthesized de novo but DNA replication is not required, implying that they are products of viral early genes (14). DNA replication is needed, however, to provide a template for transcription of both intermediate and late genes. Three vaccinia virus late transcription factors are encoded by intermediate mRNAs, accounting for the DNA replication requirement for their synthesis (15).
Taken together, the data support a cascade model for poxvirus gene regulation in which critical early, intermediate, and late stage transcription factors are products of late, early, and intermediate genes, respectively. The isolation of VETF was simplified by its presence in purified vaccinia virus particles (7). Three vaccinia virus late transcription factors were discovered by a reverse genetic approach (15) and two others by fractionation of infected cell extracts (16–18). The characterization of VITFs, however, has depended entirely on fractionation procedures. Reconstitution studies established roles for the viral RNA polymerase and capping enzyme (19), a 30-kDa protein called VITF-1 that also serves as a subunit of RNA polymerase (20), and an unidentified cell factor called VITF-2 (21) for in vitro transcription of intermediate genes. Recently, the existence of another intermediate stage-specific transcription factor was reported (22). Using a reverse genetic approach, we now demonstrate that this factor is composed of subunits encoded by two vaccinia virus genes of previously unknown function. The genes are expressed early after virus infection and are conserved in other members of the family, consistent with a cascade model for poxvirus gene regulation.
MATERIALS AND METHODS
Plasmid Construction.
The A8R ORF was amplified by PCR from vaccinia virus strain Western Reserve genomic DNA using the primers 5′-AGTCTTACATATGTTCGAACCAGTACCAGATCTTAATTTGG (NdeI site underlined) and 5′-TGAGTCGTCTATGGATCCTAACTCCAGTCCTA (BamHI site underlined). The amplified fragment was digested with BamHI and NdeI and ligated to BamHI- and NdeI-digested plasmid pET11a (Novagen) to form pA8R. The same PCR fragment was ligated to BamHI- and NdeI-digested pET14b (Novagen) to create pHisA8R, in which six histidines precede the A8R ORF. A similar protocol was followed to amplify the A23R ORF with the primers 5′- CGTTAGTAACGCCATATGGATAATCTATTTACC and 5′-ACCCTAGTCGTTGGATCCATTTCTGAATC and to amplify the F11L ORF with the primers 5′-GTTTCACATATGGGGTTTTGCATTCCATCGAGAT and 5′-CAGGGGATGGATCCTTGGCAACACCCATTTATTG. The PCR products were digested with BamHI and NdeI and ligated to pET11a to create the plasmids pA23R and pF11L, or to plasmid pET14b to generate pHisA23R and pHisF11L. The BglII–BamHI fragment of pHisA23R containing the A23R ORF with a six-histidine tag at the N terminus under the control of the bacteriophage T7 promoter was cloned into the BamHI site of pA8R. In this way, we obtained pA8R-HisA23R in which each ORF is under control of a T7 promoter and transcribed in the same direction but only the A23R ORF has a six-histidine tag. We used similar strategies to construct pA8R-HisF11L, pHisA8R-A23R, and pHisA8R-F11L. Escherichia coli strains DH5α and BL21(DE3)pLysS were used for transformation and recombinant protein synthesis, respectively.
Construction of Recombinant Vaccinia Viruses.
NcoI–BamHI DNA fragments containing the A8R, A23R, or F11L ORFs with six histidine codons were inserted into the pVOTE.1 transfer vector (23) adjacent to the bacteriophage T7 promoter and E. coli lac operator sequences to produce pVOTE.1-HisA8R, pVOTE.1-HisA23R, and pVOTE.1-HisF11L, respectively. BS-C-1 cells were infected with vT7lacOI (24), a recombinant vaccinia virus containing the bacteriophage T7 RNA polymerase gene and the E. coli lac repressor gene, and transfected with the pVOTE plasmids. The recombinant viruses vHisA8R, vHisA23R, and vHisF11L, containing the original A8R, A23R, and F11L genes plus an inducible copy of a histidine-tagged A8R, A23R, or F11L ORF, were plaque purified and amplified.
Expression of A8R, A23R, and F11L ORFs by Recombinant Vaccinia Viruses.
HeLa S3 cells were infected with 10 plaque-forming units of recombinant virus per cell in the presence of 1 mM isopropyl β-d-thiogalactopyranoside. After 20 h, the cells were harvested and cytoplasmic extracts were prepared as described (22). The histidine-tagged protein from 1 liter of infected HeLa cells was purified on a metal-affinity column as described below for recombinant proteins expressed in E. coli. The proteins eluted with 2 ml of imidazole buffer were dialyzed against buffer A [40 mM Tris⋅HCl, pH 8.0/15% (vol/vol) glycerol/0.5 mM phenylmethylsulfonyl fluoride] plus 0.2 mM EDTA, 2 mM DTT, and 0.05 M NaCl.
Preparation of Bacterial Extracts and Purification of Recombinant Protein.
A 130-ml culture of induced bacteria was centrifuged and the pellet was resuspended in buffer A containing 0.15 M NaCl, 5 mM imidazole, and 0.1% Triton X-100. Bacteria were disrupted by four freeze–thaw cycles and the suspension was then forced through a 20-gauge needle to shear the bacterial DNA. Further steps were carried out at 4°C. The insoluble residue was removed by centrifugation at 100,000 × g for 45 min, and the supernatant was mixed with 0.5 ml of Talon metal-affinity resin (CLONTECH) for 2 h. The suspension was centrifuged and the supernatant was discarded. The resin was then poured into a column and successively washed with 20 column volumes each of buffer A plus 0.15 M NaCl and 5 mM imidazole; buffer A plus 1 M NaCl and 5 mM imidazole; and buffer A plus 0.15 M NaCl, 5 mM imidazole, and 40 mM glycine. The proteins remaining bound to the resin were then eluted with 2.5 ml of buffer A plus 0.15 M NaCl and 0.1 M imidazole. The eluate was then loaded on a Poros HQ column (PerSeptive Biosystems, Framingham, MA), and the bound proteins were eluted with a 20 column volume linear 0.15–1 M NaCl gradient in buffer A plus 0.2 mM EDTA and 2 mM DTT. Fractions with transcription factor activity were pooled and dialyzed against buffer A plus 0.2 mM EDTA, 2 mM DTT, and 0.15 M NaCl. The dialyzed proteins were then loaded on a Poros heparin column (PerSeptive Biosystems) and eluted with a 20 column volume linear 0.15–1 M NaCl gradient in buffer A plus 0.2 mM EDTA and 2 mM DTT.
Gel Filtration.
The A8R-HisA23R protein complex synthesized in E. coli and purified by Talon chromatography was loaded on a 1.6 × 60 cm Sephacryl S300 column (Pharmacia) that had been equilibrated with 0.15 M NaCl/40 mM Tris⋅HCl, pH 8.0/5 mM imidazole/0.5 mM phenylmethylsulfonyl fluoride/5% glycerol.
Transcription Assays.
Column fractions were dialyzed against buffer A supplemented with 0.05 M NaCl, 0.2 mM EDTA, and 2 mM DTT at 4°C. Transcription reactions were performed by complementation of a DEAE-cellulose 0.15 M NaCl elution fraction from cells infected with vaccinia virus in the presence of cytosine arabinonucleoside (AraC) (22) except that recombinant proteins were usually used in place of VITF-X. VITF-X was partially purified through the DEAE I, DEAE II, phosphocellulose, SP Sepharose, and single-stranded DNA agarose columns (22).
mRNA Analysis.
HeLa S3 cells (4 × 106) were infected with 10 plaque-forming units per cell of vaccinia virus in the absence or presence of 40 μg/ml AraC or 100 μg/ml cycloheximide for up to 6 h. Total RNAs were prepared by using the RNaqueous procedure (Ambion, Austin, TX) as instructed by the manufacturer. Samples (6 μg) of each RNA were resolved on a denaturing agarose gel, electrotransferred in a Transblot SD semidry cell (Bio-Rad) at 3 mA/cm2 for 30 min to a positively charged Brightstar-Plus nylon membrane (Ambion) moistened with 0.045 M Tris base/0.045 M boric acid/0.001 M disodium EDTA.
Northern blot analysis was carried out with the Northern Max-Plus kit (Ambion). PCR fragments of the A8R and A23R ORFs were prepared as described above, gel purified (Geneclean II kit, Bio 101), labeled with [α-32P]dCTP by random priming (Prime a Gene Labeling System, Promega), and purified on a G-50 microcolumn (Pharmacia). The primers 5′-CAGATCATTCGCCGATAGTGGTAAC and 5′-GGTAGTTTAGTTCGTCGAGTGAACCT were used to amplify the vaccinia virus C11R growth factor ORF for use as a probe for a known early mRNA.
RESULTS
Identification of VITF Genes.
We recently described the partial purification of an intermediate stage transcription factor from vaccinia virus-infected cells (22). The native factor, provisionally named VITF-X, had an apparent mass of ≈100 kDa as determined by gel filtration. SDS/PAGE of the most highly purified VITF-X fractions revealed bands of 200, 65, 50, 45, and 35 kDa. However, the small amounts of these polypeptides precluded further analysis. Because VITF-X was isolated from cells that were infected with vaccinia virus in the presence of an inhibitor of DNA replication but could not be detected in extracts of uninfected cells, we suspected that it was a virus-encoded early gene product. As the vaccinia virus genome contains ≈200 ORFs (25), we focused our search on those that (i) encode proteins of 30–65 kDa, (ii) have early promoter consensus sequences, (iii) are conserved in all sequenced vertebrate poxvirus genomes, (iv) have not been demonstrated to be nonessential, (v) have no described function, and (vi) lack putative transmembrane domains. The three top candidates in this list were the A8R, A23R, and F11L ORFs, which were predicted to encode proteins of 34, 45, and 40 kDa, respectively. We learned subsequently, however, that the F11L ORF is nonessential (26).
The basic strategy was to determine if recombinant proteins could substitute for VITF-X in transcription assays. Candidate ORFs were initially expressed in infected cells using a vaccinia virus expression vector in case the polypeptides needed to be posttranscriptionally modified or additional viral or cellular factors not predicted in our survey might be required. An isopropyl thiogalactopyranoside-inducible vaccinia virus/bacteriophage T7 RNA polymerase expression system provided a high yield of recombinant protein (23). In addition, six histidine codons were introduced at the N terminus of each candidate ORF to allow convenient purification of the induced proteins on a metal-affinity resin (27). A set of vaccinia viruses, each expressing one of the histidine-tagged candidate ORFs, was derived by recombination of transfer plasmids with vT7lacOI (23). The resulting viruses contained the genes encoding the bacteriophage T7 RNA polymerase and the E. coli lac repressor from vT7lacOI and the A8R, A23R, or F11L ORF under T7 promoter and lac operator control.
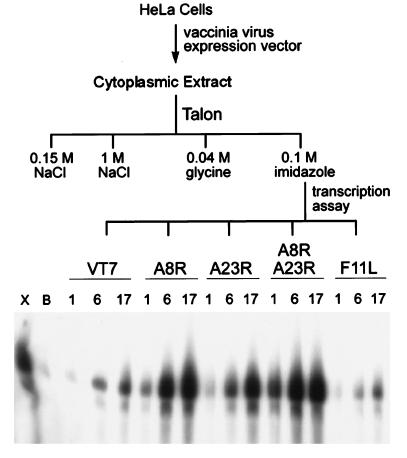
HeLa cells were infected with the parental virus (vT7lacOI) or with one or two recombinant vaccinia viruses expressing the candidate ORFs in the presence of inducer. The cells were lysed and the histidine-tagged proteins in the soluble fraction were partially purified by metal-affinity chromatography. These proteins were then tested for their ability to substitute for VITF-X in a transcription assay using a template with an intermediate stage promoter (22). While a low level of activity was obtained from cells infected with vT7lacOI or the virus expressing the histidine-tagged F11L protein, a high level of activity was obtained from cells that expressed either the histidine-tagged A8R or the A23R ORF (Fig. 1). Even higher activity was obtained from cells that expressed both A8R and A23R proteins (Fig. 1).
Figure 1.
Expression of candidate intermediate transcription factors by recombinant vaccinia virus vectors. HeLa cells were infected with the parental vaccinia virus (vT7lacOI) or vaccinia virus expression vectors encoding histidine-tagged copies of the A8R, A23R, or F11L ORF. Cytoplasmic extracts were applied to a Talon metal-affinity resin that was washed successively with 0.15 M NaCl, 1 M NaCl, 0.04 M glycine, and 0.1 M imidazole as indicated. Partially purified VITF-X (X), buffer (B), or 1-, 6-, or 17-μl samples of the dialyzed 0.1 M imidazole fractions from cells infected with vT7lacOI (vT7) or recombinant vaccinia viruses vHisA8R (A8R), vHisA23R (A23R), vHisA8R plus vHisA23R, or vHisF11L (F11L) were added to transcription reactions deficient in VITF-X. [α-32P]UMP-labeled RNAs transcribed from an intermediate promoter G-less cassette template were analyzed by denaturing PAGE and detected by autoradiography.
These experiments suggested that the A8R and A23R products or factors associated with them were involved in intermediate transcription. Two main hypotheses were considered: (i) A8R and A23R are interchangeable, each individually able to stimulate transcription; (ii) the intermediate transcription factor is an A8R–A23R complex. In the latter case, complexes might form between the histidine-tagged A8R protein and the native A23R protein in one case, and between the histidine-tagged A23R protein and the native A8R protein in the other, allowing some heterodimer binding to the metal-affinity resin. Not excluded, however, was the possibility that additional viral or cellular proteins were in these putative complexes.
Activity of Recombinant A8R and A23R Proteins Synthesized in E. coli.
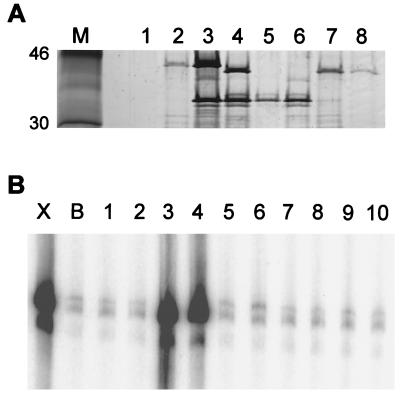
To determine whether either hypothesis was correct, we expressed the A8R, A23R, and F11L ORFs individually or in pairs and with or without N-terminal histidine tags in E. coli. Soluble proteins were bound and eluted from a metal-affinity column and analyzed by SDS/PAGE and for transcription complementation activity. The electrophoretic mobilities of the A23R and A8R polypeptides were consistent with predicted masses of ≈45 and 34 kDa, respectively (Fig. 2A). The HisA23R polypeptide migrated slightly slower than the authentic A23R polypeptide, whereas the HisA8R and authentic A8R polypeptides had similar mobilities. Importantly, the A8R protein was bound to the metal-affinity resin when it was coexpressed with the HisA23R protein, and the A23R protein was bound when it was expressed with the HisA8R protein (Fig. 2A). In contrast, neither the A8R nor the A23R protein without a histidine tag bound efficiently to the resin when expressed alone or with the HisF11L protein (Fig. 2A and data not shown). These results suggested that the A8R and A23R polypeptides exist as a complex.
Figure 2.
Metal-affinity binding and activity of candidate intermediate transcription factors expressed in bacteria. (A) Histidine-tagged proteins were partially purified by metal-affinity chromatography. A sample (20 μl) of each 0.1 M imidazole fraction was analyzed on a 4–20% polyacrylamide gel, and proteins were detected by silver staining. Marker proteins are shown in lane M. The expression plasmids contained no insert (lane 1); HisA23R (lane 2); A8R and HisA23R (lane 3); HisA8R and A23R (lane 4); HisA8R (lane 5); HisA8R and F11L (lane 6); A8R and HisF11L (lane 7); or HisF11L (lane 8). The positions of 46- and 30-kDa marker proteins are shown (Left). (B) the partially purified proteins were tested for transcription factor activity as described in the legend to Fig. 1. Lanes 1–8, 17 μl of dialyzed imidazole fraction corresponding to lanes of the same number in A; lane 9, 5 μl each of proteins used in lanes 2 and 5; lane 10, 8.5 μl each of proteins used in lanes 2 and 5; lane X, partially purified VITF-X; lane B, buffer control.
When the same partially purified proteins were tested in the transcription complementation assay, only those from bacteria coexpressing the A8R and A23R ORFs exhibited stimulatory activity regardless of which one had the histidine tag (Fig. 2B). Moreover, transcription activity was not detected when the two ORFs were expressed separately and the proteins were mixed together (Fig. 2B). This result provided further support for the idea that the two polypeptides were subunits of one transcription factor.
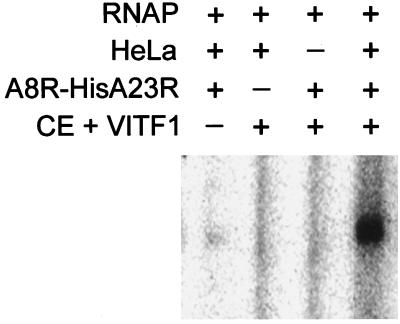
Previous studies had established that intermediate transcription factor activity could be reconstituted by mixing fractions containing vaccinia virus-encoded RNA polymerase, capping enzyme, VITF-1, VITF-2 (cell factor), and VITF-X. The coexpressed recombinant proteins also reconstituted intermediate transcription activity in this semidefined system (Fig. 3), thereby distinguishing it from previously identified components. Taken together, these data suggest that the A8R and A23R ORFs encode subunits of an intermediate transcription factor that we named VITF-3.
Figure 3.
Recombinant A8R–A23R complex complements other components required for intermediate transcription. The following components were added to a transcription assay: vaccinia virus RNA polymerase (RNAP), extract of uninfected HeLa cells (HeLa), capping enzyme plus VITF-1-containing fraction from infected HeLa cells (CE + VITF-1), and the A8R–HisA23R complex purified from E. coli. Transcripts were analyzed as described in the legend to Fig. 1.
Copurification of the A8R- and A23R-Encoded Proteins.
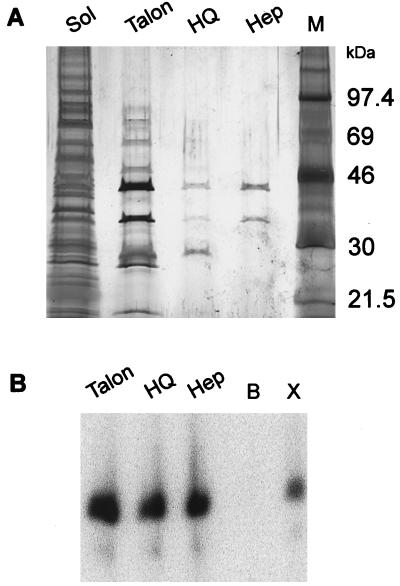
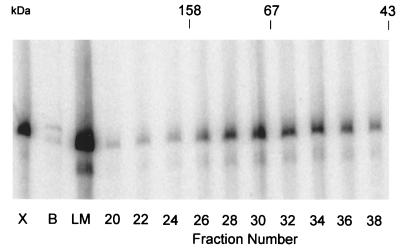
After metal-affinity chromatography, the A8R and A23R polypeptides copurified with VITF-3 transcription factor activity on HQ and heparin columns (Fig. 4). After the final purification step, the 45- and 34-kDa proteins were the major species detected by silver staining. In addition, VITF-3 peak activity eluted from a Sephacryl S300 size-exclusion column just ahead of a 67-kDa marker (Fig. 5), consistent with a heterodimer of 34- and 45-kDa subunits and similar to the size estimate for VITF-X (22).
Figure 4.
Association of 45- and 34-kDa subunits with VITF-3 activity during purification. Soluble proteins from E. coli coexpressing the A8R and HisA23R proteins were purified by column chromatography. At each chromatographic step, fractions were tested for intermediate transcription factor activity and active fractions were pooled. (A) Total soluble protein (Sol) and pooled active fractions from Talon, HQ, and heparin (Hep) columns were analyzed by SDS/PAGE and silver staining. The positions of protein markers (M) are shown on the right. (B) Pooled fractions in A were tested for intermediate transcription factor activity as described in the legend to Fig. 1. Additional lanes: B, buffer control; X, VITF-X.
Figure 5.
Gel filtration of the native recombinant transcription factor. Partially purified A8R–HisA23R complex from E. coli was loaded on a Sephacryl S300 column. Samples, 5 μl of the loading material (LM), or 17 μl of numbered fractions, were assayed for intermediate transcription factor activity. VITF-X (X) was used as a positive control and buffer alone (B) as a negative control. The elution positions of standards used to calibrate the column (aldolase, 158 kDa; serum albumin, 67 kDa; ovalbumin, 43 kDa) are indicated at the top.
Early Expression of the A8R and A23R Genes.
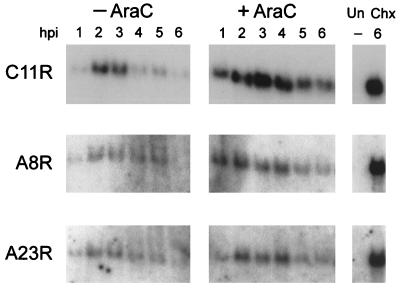
Previous studies had shown that VITF-X could be isolated from cells infected with vaccinia virus in the presence of AraC, an inhibitor of DNA replication. To serve as an intermediate transcription factor, the A8R and A23R genes would have to be expressed early in infection. Northern blotting was used to measure the accumulation of A8R and A23R transcripts under standard conditions and in the presence of an inhibitor of DNA replication. Neither the A8R nor the A23R RNA was detectable with specific 32P-labeled DNA probes in uninfected cells, but discrete RNAs of the expected length were present in infected cells (Fig. 6). The two RNAs were detected within 1 h after infection and were most abundant between 2 and 5 h. Moreover, the RNAs were synthesized in the presence of AraC and were greatly overexpressed in the presence of cycloheximide, an inhibitor of protein synthesis. These properties are characteristic of early transcripts, as shown for the C11R RNA encoding the vaccinia virus growth factor (Fig. 6).
Figure 6.
Northern blot analysis of mRNAs from cells infected with vaccinia virus. HeLa cells were infected with vaccinia virus in the absence (−) or presence (+) of AraC and harvested from 1 to 6 h later. In addition, uninfected HeLa cells (Un) and HeLa cells infected in the presence of cycloheximide (Chx) for 6 h were harvested. RNAs were resolved by electrophoresis on a denaturing agarose gel, transferred to a nylon membrane, and hybridized with 32P-labeled DNA probes specific for the C11R, A8R, and A23R ORFs. Autoradiograms are shown.
DISCUSSION
The vaccinia virus intermediate transcription factor provisionally named VITF-X was initially discovered by fractionation of infected cell extracts and partially purified (22), whereas VITF-3 was identified by expression of the candidate vaccinia virus ORFs A8R and A23R. Several lines of evidence indicated that VITF-3 is VITF-X. Most importantly, VITF-3 replaced VITF-X for in vitro transcription of a template containing an intermediate promoter. In addition, both were expressed in the presence of the DNA replication inhibitor AraC and have similar native molecular masses. Furthermore, highly purified preparations of VITF-X contained polypeptides of ≈35 and 45 kDa as prominent components (22). The additional polypeptides detected in VITF-X preparations are likely to be contaminants.
Our data indicated that the A8R and A23R polypeptides are subunits of VITF-3. Coexpression of the two ORFs in E. coli was required to obtain an active transcription factor; activity was not obtained by simply mixing the individually expressed soluble proteins. In addition, a histidine tag on either protein allowed the other coexpressed protein to bind to a metal-affinity column. Moreover, the two polypeptides copurified on successive chromatography columns and the gel filtration properties were consistent with a heterodimer.
The proposed heterodimeric structure of VITF-3 is reminiscent of that of VETF (7), and one of the VETF subunit genes (11) is located adjacent to the A8R gene. Nevertheless, there is no sequence similarity between the subunits of these two transcription factors. VETF has a DNA-dependent ATPase activity (28) and binds specifically to early promoters (29, 30). Neither the A8R nor the A23R ORF contains an ATPase or any other recognizable functional motif, and specific DNA binding has not been demonstrated for VITF-3 thus far. Sequence comparisons indicate the presence of homologs of A8R and A23R in a distantly related vertebrate poxvirus (31) but not in unrelated viruses or organisms.
The transcription of intermediate genes in vitro requires at least four proteins—namely, RNA polymerase (9), capping enzyme (19, 32), VITF-1 [which also serves as an RNA polymerase subunit (20)], an unidentified cellular protein called VITF-2 (21), and VITF-3. Except for VITF-3, the viral proteins have roles in the biosynthesis of all three temporal classes of mRNAs. Thus, VITF-3 may be the viral factor chiefly responsible for regulating the transcription of intermediate genes. Intermediate mRNAs are synthesized for only a short period of time before they are superceded by the transcription of late genes at ≈6 h after infection (33). The subunits that constitute VITF-3 are encoded by early genes, and their mRNAs were most abundant between 2 and 5 h after infection and were largely gone by 6 h. Therefore, there can be little additional VITF-3 made at late times. Whether intermediate transcription ceases because VITF-3 is unstable, is out competed by more abundant late transcription factors, or for other reasons is not yet known. The stability of the A8R and A23R proteins could not be determined because the amounts made were too low for detection by antibodies that readily detected overexpressed recombinant proteins (unpublished data).
Why poxviruses have an intermediate class of genes that requires unique transcription factors is puzzling. The early genes encode many different types of proteins that have roles in gene expression, DNA replication, and host defenses. The numerous late genes are highly expressed and encode the major structural proteins and other components of virus particles. By contrast, only five vaccinia virus genes have been shown to have intermediate promoters. Three of these genes encode late transcription factors (15), one an RNA helicase (14, 34), and the other a phosphoprotein that has both early and intermediate promoters (14, 35). If these and other intermediate genes also had early promoters, there would seem to be no need for intermediate transcription factors. However, the existence of three classes of genes suggests that there are regulatory advantages to having such a complex system. For example, because a single virus particle is sufficient for a productive infection, there might be a problem if late proteins were synthesized as soon as viral DNA uncoating occurred. In this case, structural proteins might sequester the DNA before synthesis of sufficient genome copies for continued exponential replication and gene expression. Thus, one important advantage of the poxvirus cascade regulatory system may be to delay the production of late proteins.
Acknowledgments
We thank Tatiana Senkevich for suggestions regarding candidate intermediate transcription factors, Norman Cooper for preparing virus and cells, Andrea Weisberg for help with the figures, and Tom Kristie for comments on the manuscript. Support for this work was provided by National Institute of Allergy and Infectious Diseases intramural funding. B.M. is a coinventor on U.S. patents 5,135,855 and 5,550,035 concerning recombinant vaccinia virus/bacteriophage T7 expression systems.
ABBREVIATIONS
- VETF
vaccinia virus early transcription factor
- VITF
vaccinia virus intermediate transcription factor
- AraC
cytosine arabinonucleoside
References
- 1.Moss B. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Vol. 2. Philadelphia: Lippincott-Raven; 1996. pp. 2637–2671. [Google Scholar]
- 2.Moss B, Salzman N P. J Virol. 1968;2:1016–1027. doi: 10.1128/jvi.2.10.1016-1027.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pennington T H. J Gen Virol. 1974;25:433–444. doi: 10.1099/0022-1317-25-3-433. [DOI] [PubMed] [Google Scholar]
- 4.Davison A J, Moss B. J Mol Biol. 1989;210:771–784. doi: 10.1016/0022-2836(89)90108-3. [DOI] [PubMed] [Google Scholar]
- 5.Davison A J, Moss B. J Mol Biol. 1989;210:749–769. doi: 10.1016/0022-2836(89)90107-1. [DOI] [PubMed] [Google Scholar]
- 6.Baldick C J, Keck J G, Moss B. J Virol. 1992;66:4710–4719. doi: 10.1128/jvi.66.8.4710-4719.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broyles S S, Yuen L, Shuman S, Moss B. J Biol Chem. 1988;263:10754–10760. [PubMed] [Google Scholar]
- 8.Wright C F, Moss B. J Virol. 1989;63:4224–4233. doi: 10.1128/jvi.63.10.4224-4233.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vos J C, Sasker M, Stunnenberg H G. Cell. 1991;65:105–114. doi: 10.1016/0092-8674(91)90412-r. [DOI] [PubMed] [Google Scholar]
- 10.Moss B, Ahn B-Y, Amegadzie B Y, Gershon P D, Keck J G. J Biol Chem. 1991;266:1355–1358. [PubMed] [Google Scholar]
- 11.Gershon P D, Moss B. Proc Natl Acad Sci USA. 1990;87:4401–4405. doi: 10.1073/pnas.87.11.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broyles S S, Fesler B S. J Virol. 1990;64:1523–1529. doi: 10.1128/jvi.64.4.1523-1529.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahn B-Y, Moss B. Proc Natl Acad Sci USA. 1992;89:3536–3540. doi: 10.1073/pnas.89.8.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vos J C, Stunnenberg H G. EMBO J. 1988;7:3487–3492. doi: 10.1002/j.1460-2075.1988.tb03224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keck J G, Baldick C J, Moss B. Cell. 1990;61:801–809. doi: 10.1016/0092-8674(90)90190-p. [DOI] [PubMed] [Google Scholar]
- 16.Kovacs G R, Moss B. J Virol. 1996;70:6796–6802. doi: 10.1128/jvi.70.10.6796-6802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunasinghe S K, Hubbs A E, Wright C F. J Biol Chem. 1998;273:27524–27530. doi: 10.1074/jbc.273.42.27524. [DOI] [PubMed] [Google Scholar]
- 18.Zhu M, Moore T, Broyles S S. J Virol. 1998;72:3893–3899. doi: 10.1128/jvi.72.5.3893-3899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vos J C, Sasker M, Stunnenberg H G. EMBO J. 1991;10:2553–2558. doi: 10.1002/j.1460-2075.1991.tb07795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosales R, Harris N, Ahn B-Y, Moss B. J Biol Chem. 1994;269:14260–14267. [PubMed] [Google Scholar]
- 21.Rosales R, Sutter G, Moss B. Proc Natl Acad Sci USA. 1994;91:3794–3798. doi: 10.1073/pnas.91.9.3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanz P, Moss B. J Virol. 1998;72:6880–6883. doi: 10.1128/jvi.72.8.6880-6883.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ward G A, Stover C K, Moss B, Fuerst T R. Proc Natl Acad Sci USA. 1995;92:6773–6777. doi: 10.1073/pnas.92.15.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexander W A, Moss B, Fuerst T R. J Virol. 1992;66:2934–2942. doi: 10.1128/jvi.66.5.2934-2942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. ; 517–563. [DOI] [PubMed] [Google Scholar]
- 26.Antoine G, Scheiflinger F, Dorner F, Falkner F G. Virology. 1998;244:365–396. doi: 10.1006/viro.1998.9123. [DOI] [PubMed] [Google Scholar]
- 27.Janknecht R, De Martynoff G, Lou J, Hipskind R A, Nordheim A, Stunnenberg H G. Proc Natl Acad Sci USA. 1991;88:8972–8976. doi: 10.1073/pnas.88.20.8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Broyles S S, Moss B. J Biol Chem. 1988;263:10761–10765. [PubMed] [Google Scholar]
- 29.Broyles S S, Li J, Moss B. J Biol Chem. 1991;266:15539–15544. [PubMed] [Google Scholar]
- 30.Cassetti M C, Moss B. Proc Natl Acad Sci USA. 1996;93:7540–7545. doi: 10.1073/pnas.93.15.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Senkevich T G, Koonin E V, Bugert J J, Darai G, Moss B. Virology. 1997;233:19–42. doi: 10.1006/viro.1997.8607. [DOI] [PubMed] [Google Scholar]
- 32.Harris N, Rosales R, Moss B. Proc Natl Acad Sci USA. 1993;90:2860–2864. doi: 10.1073/pnas.90.7.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baldick C J, Jr, Moss B. J Virol. 1993;67:3515–3527. doi: 10.1128/jvi.67.6.3515-3527.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shuman S. Proc Natl Acad Sci USA. 1992;89:10935–10939. doi: 10.1073/pnas.89.22.10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rochester S C, Traktman P. J Virol. 1998;72:2917–2926. doi: 10.1128/jvi.72.4.2917-2926.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]