Abstract
Genetic and biochemical analyses of the Gag protein of HIV-1 indicate a crucial role for this protein in several functions related to viral replication, including viral assembly. It has been suggested that Gag may fulfill some of the functions by recruiting host cellular protein(s). In our effort to identify structural and functional homologies between Gag and cellular cytoskeletal and secretory proteins involved in transport, we observed that HIV-1 Gag contains a unique PGQM motif in the capsid region. This motif was initially noted in the regulatory domain of synexin the membrane fusion protein of Xenopus laevis. To evaluate the functional significance of the highly conserved PGQM motif, we introduced alanine (A) in place of individual residues of the PGQM and deleted the motif altogether in a Gag expression plasmid and in an HIV-1 proviral DNA. The proviral DNA containing mutations in the PGQM motif showed altered expression, assembly, and release of viral particles in comparison to parental (NL4-3) DNA. When tested in multiple- and single-round replication assays, the mutant viruses exhibited distinct replication phenotypes; the viruses containing the A for the G and Q residues failed to replicate, whereas A in place of the P and M residues did not inhibit viral replication. Deletion of the tetrapeptide also resulted in the inhibition of replication. These results suggest that the PGQM motif may play an important role in the infection process of HIV-1 by facilitating protein–protein interactions between viral and/or viral and cellular proteins.
Keywords: Gag precursor, capsid, homology, viral assembly, infectivity
Retroviruses are broadly divided into two groups, based on the organizational complexity of the viral genome. The genomes of simple retroviruses encompass genes gag, pol, and env, which are essential for the productive replication of the virus. Complex retroviruses, on the other hand, contain additional coding sequences located both in the middle and at the 3′ end of the genome (1, 2). HIV-1, a member of the complex retrovirus family, contains six auxiliary genes (vif, vpr, tat, rev, vpu, and nef) in addition to gag, pol, and env (2). Of all the proteins encoded by HIV-1, Gag has been shown to be the most versatile in terms of the role it plays in the replication of HIV and other retroviruses. Its four major functions are (i) to assist in the assembly of virus particles; (ii) to help transduce the Gag-pol polyprotein precursor into the virus particle as a source of viral enzymatic functions; (iii) to incorporate viral genomic RNA into the virus particles; and (iv) to incorporate the Env protein into the viral membrane (3, 4). Furthermore, it has been shown that the expression of Gag in the absence of other viral proteins and RNA genome results in the assembly and release of virus-like particles (5, 6). This finding has stimulated enormous interest in the use of Gag as a model system to understand the processes associated with viral morphogenesis.
The Gag precursor protein is synthesized in the free ribosomes contained in the cytoplasm. Viral genomic RNA serves as the messenger in this process, and the addition of the myristyl residue at the N terminus of the precursor and the surrounding basic residues helps to transport the protein to the cell membrane (7). Biochemical studies have identified a prominent role for protein–protein and protein–nucleic acid interactions in the assembly and infection process of HIV-1 (1, 5, 7). These observations have led to the speculation that host cellular protein(s) may contribute to the functions carried out by Gag (3, 5). In this regard, an interaction between HIV-1 Gag and cyclophilin has been detected by using several approaches, including a yeast-based, two-hybrid system (8–10); the incorporation of cyclophilin A (CyPA) has also been demonstrated in the virus particles (11–15). These findings have prompted a search for additional host cellular proteins and/or specific structural/functional features present in viral protein that may participate in Gag-mediated functions. Toward this goal, we have analyzed the predicted amino acid sequence of the Gag precursor for possible structural and functional homologies to cytoskeletal and secretory proteins. Through this comparative approach, we have identified a PGQM motif in the capsid (CA) region of HIV-1 Gag that is present in multiple copies within the synexin molecule. Synexin is a calcium and GTP sensor in the exocytotic process and belongs to the annexin gene family. The PGQM motif in synexin occurs in the regulatory region of the N-terminal domain of the protein, which contains sites for phosphorylation, proteolysis, and protein–protein interaction (16). Because conceptual parallels are often invoked between vesicle fusion and viral assembly and release, we have focused on the role of the synexin/CA parallel at the level of the shared PGQM motif in HIV-1 infection and replication. Mutational analysis of the PGQM motif has provided us with strong evidence for its functional significance in HIV-1 replication.
MATERIALS AND METHODS
Generation of Gag Expression Plasmid and HIV-1 Proviral DNA Containing Alterations in the PGQM Motif.
HIV-1 proviral DNA, designated NL4-3 (17), was used for the experiments. For the introduction of the A in place of individual amino acid residues in the PGQM motif, as well as for the deletion of this motif entirely, PCR-based methods were utilized (18) involving the primers shown in Table 1. The PCR amplification was initially carried out by using two separate reactions: Sph-UP (+) and a minus-strand primer composed the site to be mutated in the first reaction, whereas the second reaction involved Apa-DS (−) and a plus-strand primer corresponding to the site to be mutated. The DNAs derived from both reactions were mixed and amplified by using Sph-UP (+) and Apa-DS (−) primers. On amplification, the DNA was digested, using the restriction enzymes SphI and ApaI, and ligated to the proviral DNA, which was previously cleaved with the appropriate enzymes. The ligated DNA was used to transform Escherichia coli cells, and the plasmid DNA isolated from the bacterial colonies was screened by using the SphI and ApaI enzymes. The mutated codon corresponding to the residue of interest was verified by DNA sequence analysis. The large-scale preparation of plasmid DNA was carried out as described (19). A Gag expression plasmid was constructed in which the Gag coding sequences were cloned into pCDNA3 vector (20). To enable detection, sequences corresponding to Flag epitope (DYKDDDDK) were added in-frame to the Gag.
Table 1.
Primers used for the mutagenesis of PGQM motif
| Primer Designation | Sequence 5′ → 3′ | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sph-UP | + | GGA | GCC | ACC | CCA | CAA | GAT | TT | ||
| Apa-DS | − | TCC | ACA | TTT | CCA | ACA | GCC | CT | ||
| P93 | + | GGG | CCT | ATT | GCA | GCA | GGC | CAG | ATG | AGA |
| − | TCT | CAT | CTG | GCC | TGG | TGC | AAT | AGG | CCC | |
| G94 | + | GGG | CCT | ATT | GCA | GCA | GGC | CAG | ATG | AGA |
| − | TCT | CAT | CTG | GCC | TGC | TGC | AAT | AGG | CCC | |
| Q95 | + | ATT | GCA | CCA | GGC | GCG | ATG | AGA | GAA | CCA |
| − | TGG | TTC | TCT | CAT | CGC | GCC | TGG | TGC | AAT | |
| M96 | + | GCA | CCA | GGC | CAG | GCG | AGA | GAA | CCA | AGG |
| − | CTT | TGG | TTC | TCT | CGC | CTG | GCC | TGG | TGC | |
| ΔPGQM | + | CTT | ATT | GCA | AGA | GAA | CCA | AGG | GGA | AGT |
| − | TGG | TTC | TCT | TGC | AAT | AGG | CCC | TGC | ATG | |
In Vitro Transcription/Translation and Radioimmunoprecipitation Analysis.
The coupled T7 transcription/translation system (Promega) was used. For expression studies involving Gag, the recombinant vaccinia virus vTF7–3 that expresses T7 RNA polymerase in infected cells was used (20). Immunoblot analysis was carried out by using procedures as described (20). Regarding HIV-1 proviral DNA studies, 48 hours after transfection of RD cells in 100 mm Petri dishes (21), cells were labeled for 18 hours with 8 ml of methionine-free RPMI 1640 medium containing 2% dialyzed fetal calf serum and 50 uCi/ml (1 Ci = 37 GBq) [35S]methionine. The medium was removed and filtered through a 0.45 μm filter. The labeled cells were solubilized in 5 ml of PBS-TD buffer (PBS containing 0.5% Triton X-100 and 1% deoxycholate). The labeled HIV-1 proteins in the medium and cells were immunoprecipitated with HIV-1 antibody-positive serum, separated in 11% SDS/polyacrylamide gels, dried, and exposed to x-ray film (22).
Transfection.
Proviral DNA (10 μg) was transfected into human rhabdomyosarcoma (RD) cells or HeLa cells by using the calcium phosphate coprecipitation method (21). The virus particles released in the culture supernatant were collected at the end of 48 hours and quantitated by either reverse transcriptase- or HIV-1 p24 antigen-capture assay using the commercial kit from Organon Teknika–Cappel.
Virus Infectivity Assays.
Three separate procedures were performed to monitor viral replication and infectivity.
Viral replication in CEM 50 cells.
An equivalent amount of virus (250,000 cpm reverse transcriptase activity or 10 ng of HIV-1 p24 equivalent) was used to infect CEM cells for viral replication studies. The cells were incubated with virus inoculum for 24 hours, washed, and resuspended in medium. The culture supernatant from infected cells was monitored periodically for virus production (20). The infected cultures also were inspected routinely for the appearance of syncytia.
Single-cycle replication assay.
Proviral DNA was cleaved at the NheI recognition site, and a hygromycin (Hygr) gene under the control of SV40 early promoter was inserted in the env, disrupting its expression. Cotransfection of the modified proviral DNA and the murine amphotropic env expression plasmid into cells resulted in the generation of virus particles capable of only a single round of replication. The virus particles released into the culture medium were centrifuged and suspended in medium, and an aliquot was used to infect HeLa cells in the presence of DEAE-dextran. The infected cells were washed 48 hours after infection and placed in medium containing hygromycin. At the end of 14 days, colonies of cells resistant to hygromycin were stained and counted.
Multinuclear activation of a galactosidase indicator (MAGI) assay.
The infectivity of the virus particles was also tested by using a MAGI assay as described (23). This assay determines the extent of infection in a cell by measuring the induction of an endogenous β-galactosidase gene under the transcriptional control of HIV-1 long terminal repeat.
RESULTS
Identification of the PGQM Motif in HIV-1 Gag.
HIV-1 Gag is synthesized as a precursor protein Pr55, which contains 500 amino acid residues. There are five HIV-1 protease cleavage recognition sequences within the precursor leading to the generation of MA (p17), CA (p24), p2, NC (p7), p1, and p6. In our effort to search for the structural and functional homologies of Gag, we initially turned our attention to host cellular cytoskeletal and secretory proteins. Our earlier work revealed the presence of the PGQM motif in the synexin molecule of Xenopus laevis, and we were able to determine that the number of copies of this motif varies in a tissue- and developmental stage-specific manner (16). Furthermore, a synthetic peptide, containing six copies of the PGQM motif, has been shown to interact with multiple cellular proteins (24). This observation prompted us to search for the occurrence of the PGQM motif in HIV proteins by using a fine pattern algorithm. This search revealed the presence of the PGQM motif in the predicted amino acid sequence of HIV-1 Gag and Env. Furthermore, a search of the SwissProt and GenBank databases revealed that the PGQM motif has also been found in several other cellular and viral proteins (unpublished data).
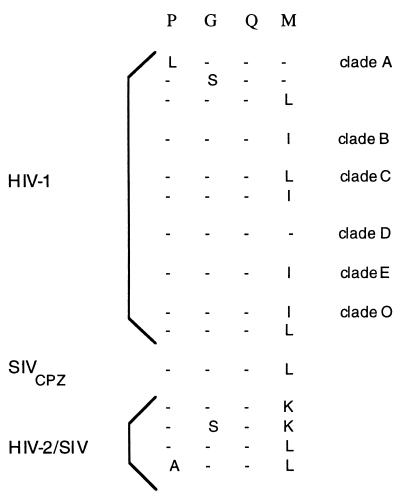
Having identified the PGQM motif in HIV-1 Gag, we wished to determine whether this motif is present in divergent HIV-1 isolates. A comparison of the predicted amino acid sequences of Gag (25) showed that the PGQM motif is highly conserved among the HIV-1 isolates of different clades (Fig. 1). No differences were found among the G and Q residues of the various isolates, and variations were observed among the P and M residues. In all members of the HIV-1 group, the PGQM motif is located in the capsid (CA amino acid residues 93–96). Interestingly, x-ray crystallographic and NMR structural analysis of partial capsid (1–151 residues) show that the PGQM motif is present in the exposed loop between helices 4 and 5 (4).
Figure 1.
The conservation of PGQM motif among HIV-1 isolates of different clades. The predicted amino acid sequences of Gag were according to Myers et al. (25). The designation of the clades is indicated on the right.
In the course of the analysis of plasma virions for intrapatient variability of Gag coding sequences, Yoshimura et al. (26) noted an additional copy of PGQM motif (amino acids 7–10) in the capsid protein. Interestingly, PGQM motif is present in SIVCPZ GAB, a virus isolated from a chimpanzee and rat leukemia virus. The PGQM noted in the Env of HIV-1 from Thailand is not conserved in other HIV-1 isolates (27). HIV-2/SIV isolates contain a modified PGQK sequence in the capsid protein (Fig. 1).
Generation of Gag Expression Vector and HIV-1 Proviral DNA Containing Alterations in the PGQM Motif.
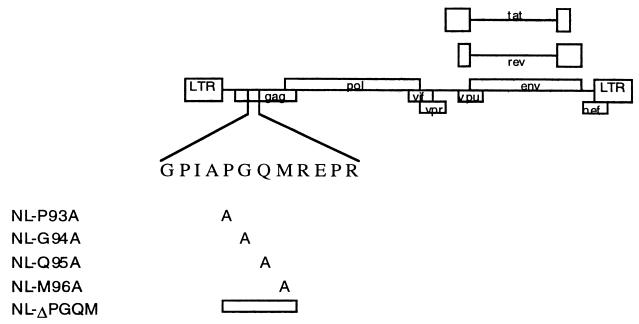
To assess the functional significance of the PGQM motif in HIV-1 replication, we have generated HIV-1 proviral DNA (NL4-3) containing changes in the PGQM sequence by using site-specific mutagenesis methods. The first group of mutants was generated by inserting A for individual amino acids in the PGQM motif. The second group consists of mutant forms of DNA in which the PGQM motif is completely removed (Fig. 2). The genetic changes were introduced into the proviral DNA through PCR, by using the primers listed in Table 1. The PCR-amplified DNA was cleaved with SphI and ApaI and ligated to the proviral DNA, as these enzymes have unique cleavage sites. To generate Gag expression vector, the respective proviral DNA was used as a template to amplify Gag coding sequences, and the DNA fragment was cloned into the plasmid vector. The resulting recombinant plasmids were examined for integrity by using DNA sequence analysis.
Figure 2.
Generation of HIV-1 proviral DNA with alteration for individual residues in PGQM motif. Schematic representation of full-length HIV-1 proviral DNA and the changes in the PGQM motif are indicated.
The PGQM Motif Is Not Required for Virion Assembly.
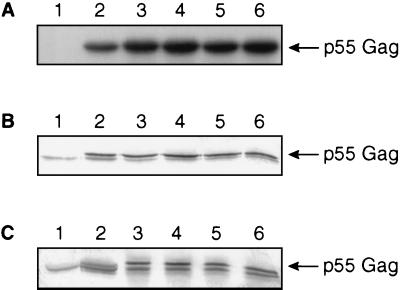
The expression of Gag was verified by using an in vitro T7 expression system. The in vitro-translated proteins were immunoprecipitated with antibodies to Flag epitope. Similar expression level was noted for each of the Gag plasmid (Fig. 3A). To determine the ability of mutant Gag to form virus-like particles, a vaccinia virus T7 RNA polymerase expression system (vTF7-3) was used. vTF7-3-infected HeLa cells were transfected with Gag plasmid. Immunoblot analysis of Gag was performed using cell lysate and culture supernatant with Flag antibodies. Results as shown in Fig. 3 B indicated the presence of p55 Gag in cell lysate but none in mock-transfected cells. The analysis of culture supernatant also showed similar results (Fig. 3C), indicating that Gag containing alterations in the PGQM motif retain the ability to form virus-like particle as Gag without alterations.
Figure 3.
Analysis of Gag protein. (A) Radioimmunoprecipitation assay analysis of in vitro-transcribed and -translated Gag. Antiserum to Flag epitope was used. Lane 1, pCDNA3 vector; lane 2, P93A; lane 3, G94A; lane 4, Q95A; lane 5, M96A; lane 6; ΔPGQM. (B) Cell lysate. (C) Culture supernatant of HeLa cells transfected with Gag plasmid. Immunoblot analysis was used. Details as above.
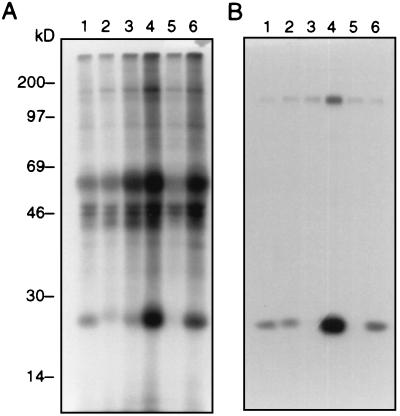
The HIV-1 proviral DNAs containing changes in the PGQM sequence were transfected into RD cells to determine its importance for the synthesis of viral proteins and viral assembly. The cells were metabolically labeled for 18 hours with [35S]methionine 48 hours after transfection. The culture supernatant and the cell lysates were analyzed separately to obtain a viral protein profile. The parental proviral DNA NL4-3 was included as a positive control in the assays described. On clearance by centrifugation, the cell lysate was subjected to immunoprecipitation by using antisera obtained from HIV-1-infected individuals. The labeled HIV-1 proteins in the medium and in cells were analyzed by using SDS/PAGE as described in Materials and Methods. The characteristic viral proteins detected in the cell lysates are shown in Fig. 4. The results indicate that there is a major difference in the synthesis of viral proteins in the cells transfected with PGQM mutants in comparison to parental NL4-3. The cells transfected with NL-P93A, NL-G94A, and NL-ΔPGQM mutants showed lower levels of Gag precursor Pr55 (Fig. 4, lanes 1, 2, and 5) compared with wild-type NL4-3 (lane 6) in cells. The mutant NL-Q95A (lane 3) showed Pr55 levels comparable to NL4-3, and NL-M96A registered a high amount of Pr55 (Fig. 4A). In the extracellular medium, although comparable level of gp120 protein was observed with all of the mutants, only a low level of p24 was noted in NL-Q95A- and NL-ΔPGQM-transfected cultures (lanes 3 and 5, respectively). Similar results also were observed when we analyzed the levels of p24 in the medium 48 hours after transfection (data not shown). Taken together, these data indicate that changes in the PGQM motif exerted an effect on the synthesis and the overall amount of virus particle released. It was also noted that processing of precursor proteins was similar in wild-type and mutant viruses.
Figure 4.
Radioimmunoprecipitation assay analysis of viral proteins. RD cells were transfected with 10 μg of HIV-1 proviral DNA by using the calcium phosphate precipitation method. Cells were labeled with [35S]methionine for 12 hours. Both cells and culture supernatants were processed as described in Materials and Methods. (A) Cells. (B) Culture supernatant. Lane 1, NL-P93-A; lane 2, NL-G94A; lane 3, NL-Q95A; lane 4, NL-M96A; lane 5, NL-ΔPGQM; lane 6, NL4-3. Immunoprecipitation with normal sera did not show reactivity with viral proteins (data not shown).
The Effect of the PGQM Motif on Virus Infectivity.
To determine whether the PGQM motif present in the CA has a role in the events associated with virus infectivity, we have carried out multiple- and single-round replication assays. For the assay involving multiple rounds of infections, an equivalent amount of viral particles based on p24 antigen values was incubated with CEM cells. The supernatant of the infected cultures was monitored for up to 15 days for the release of virus particles by p24 antigen assay. The cells infected with NL4-3-derived virus showed large syncytia at the end of 4 days after infection. By day 9 postinfection, syncytia also were noted in cells infected with NL-P93A and NL-M96A. These results corroborate well with the release of virus particles in the medium (Table 2). The level of replication among the viruses derived from proviral DNA containing residue A in place of P and M was similar to that of the wild-type NL4-3 virus. On the other hand, residue A substitution for G and Q completely eliminated virus replication. The deletion mutant, in which PGQM motif is entirely removed, also failed to replicate.
Table 2.
Effect of alterations in PGQM motif on virus replication
| Designation of virus | Days after infection
|
|||
|---|---|---|---|---|
| 4 | 7 | 10 | 13 | |
| NL4-3 | 8.3 | 2500 | 2590 | 322 |
| NL-P93A | 4.4 | 553 | 2160 | 630 |
| NL-G94A | 0 | 0 | 0 | 0 |
| NL-Q95A | 0 | 0 | 0 | 0 |
| NL-M96A | 7.3 | 1340 | 1730 | 309 |
| NL-ΔPGQM | 0 | 0 | 0 | 0 |
Values given are ng/ml HIV-1 p24 antigen in culture supernatant.
To precisely quantitate the effect of changes to the PGQM motif on infectivity, a single-round replication assay was carried out as described (20, 28). HIV-1 proviral DNA containing mutations in the PGQM motif was cleaved by using the NheI restriction enzyme, and a hygromycin gene cassette under the control of SV40 early promoter was introduced. This modification resulted in the elimination of env gene expression. Cotransfection of modified proviral DNA with an amphotropic env expression plasmid (29) into cells resulted in the release of virus particles capable of initiating infection. However, the viruses generated were replication-defective, and the cells containing proviral DNA conferred resistance to hygromycin when grown in the selective medium. In this assay, each Hygr colony of HeLa cells represented an infection event. As observed with the multiple-round replication assay, both the mutant viruses containing residue A in place of G and Q residues, and the viruses whose DNA lacked the PGQM motif, failed to initiate infection (Table 3). The mutant viruses containing residue A in place of P and M registered ≈50% and 25% inhibition in comparison to the control. The MAGI assay showed similar results (data not shown).
Table 3.
Effect in alterations in PGQM motif on virus replication involving a single round
| Proviral DNA | Titers, CFU/ml | Inhibition, % |
|---|---|---|
| ED84 (NL4-3) | 1792 | 0 |
| NL-P93A | 768 | 42 |
| NL-G94A | 0 | 100 |
| NL-Q95A | 0 | 100 |
| NL-M96A | 1376 | 24 |
| NL-ΔPGQM | 0 | 100 |
No hygromycin-resistant colonies were observed for any of the proviral clones when the transcomplementation was performed without pSV-A-MLV-env, pSV-A-MLV-env by itself, or mock-transfected. Extent of inhibition was calculated in comparison to pED84 control proviral DNA. CFU, colony-forming units.
DISCUSSION
In the studies presented here, we have identified a motif, designated PGQM, in the predicted amino acid sequence of HIV-1 Gag. Comparative analysis of several HIV-1 and HIV-2/SIV isolates further showed that this motif is conserved, with only minor variations involving mostly conservative changes (25). This suggests that PGQM motif may play a significant role in the replication cycle of HIV-1. The biological studies carried out with the PGQM mutant viruses showed a dramatic effect in virus replication by using multiple- and single-round replication. The multiple-round assay involved CEM cells and spreading infection. This assay, although useful for monitoring viral replication, cannot measure the extent of the infection in precise, quantitative terms. Hence, a single-round replication assay was utilized, involving pseudotyped virus particles containing A-MuLV env (29). Both assays showed that the introduction of A in place of the G and Q residues completely terminated viral replication. Similar results also were observed for mutant viruses lacking the PGQM motif. Analysis of virus particles further showed that the processing of the precursor proteins of the mutant viruses is not grossly altered in comparison to the parental NL4-3, despite a reduced level of viral particles released into the medium. Comparative analysis of the residues in the PGQM motif among diverse HIV-1 isolates showed the conservation of G94 and Q95. Alterations noted with respect to M are L and I, which are conservative substitutions. Insertion of A for P and M are well tolerated, and the replication phenotype is similar to that of the wild type. The biological experiments were carried out multiple times to confirm the viral phenotypes.
The mechanism(s) underlying the lack of viral replication in mutant viruses involving G and Q residues is not clear. The observation noted in the mutant, where A was introduced for P93, is in agreement with the data published by Luban and coworkers (30–32). The mutant P93A virus has been shown to incorporate CyPA, and it also exhibited replication phenotype (4). Aberham et al. (33) isolated spontaneous mutants resistant to cyclosporin A, and sequence analysis of the resistant virus showed alterations in two codons (A92E and G94D). This mutant also incorporated CyPA into the virus particles. Hence, it is likely that the failure of mutant G94A in our studies may not be caused by a lack of incorporation of CyPA into virus particles and may involve some other steps associated with virus infection. Viruses lacking the PGQM motif failed to replicate, as was shown earlier for the deletion mutant involving residues 90–93 (34). The decreased infectivity may not be caused by the presence of less Env protein, as is evident from the radioimmunoprecipitation analysis of viral proteins. It is possible that the PGQM motif may stabilize Gag and/or help recruit proteins binding to Gag for transport to the cell membrane. It should be mentioned that HIV-1 isolates belonging to Group O have been shown to be less susceptible to inhibition by cyclosporin A, as opposed to Group M isolates. Hence, the conserved nature of PGQM motif in Group O and the biological data reported here may suggest an as-yet-unidentified role for this motif in HIV-1 replication.
In the life cycle of HIV-1, as in other retroviruses, the Gag protein (the precursor and its cleavage products mediated by protease) carries out multiple functions. These include assembly of the virus , encapsidation of virion RNA, and uncoating of the core to facilitate reverse transcription. On synthesis, the Gag precursor is targeted to the cell membrane for virus budding by an ill-defined mechanism (5). Although a role for cellular proteins has been suggested, there is no information available regarding this. Recent studies using a yeast-based, two-hybrid system have determined that CyPA interacts with HIV-1 Gag, which results in the incorporation of CyPA into virus particles. The mutational analysis of the proline-rich domain in CA revealed that G89 and P90 are critical for the incorporation of CyPA. In the absence of CyPA, the infectivity level of the virus is affected, despite the fact that virus particles are formed. This suggests that CyPA binding is not required for the transport of Gag to the cell membrane. Considering the presence of PGQM motif in Gag, it is tempting to speculate that cellular proteins binding to the PGQM sequence may have a role in this process.
The PGQM motif was initially recognized in the predicted amino acid sequence of the synexin molecule of X. laevis (16). The human synexin contains a modified PGQM sequence as a single copy. Another feature of the synexin molecule of X. laevis is that it contains multiple copies of PGQM, the maximum being six, and the pattern is characteristic of the tissue of origin as well as the developmental stage. As no structural or functional information is available regarding the PGQM motif, Srivastava et al. (24) initiated studies with a synthetic peptide representing six copies of PGQM. Incubation of the peptide with cellular extract, followed by UV cross-linking, showed that this peptide interacts with three different cellular proteins. Based on these studies, it was speculated that cellular proteins interacting with PGQM motif may play an important role in the functions of synexin molecule (24). The database analysis carried out in our study revealed that, in addition to HIV-1 Gag, proteins of cellular and viral origin harbor this motif, implicating a possible functional role. The identification of PGQM motif in the predicted Gag sequence of rat leukemia virus and in the VP1 capsid protein of autonomous parvovirus minute virus of mice (35) provides further support in this regard.
In a recent study of the high-resolution structure of the HIV-1 capsid (36–38), it is indicated that the PGQM motif is located in the exposed loop between helices IV and V. Specifically, this loop is bounded by P85 and P99, which contains residues for binding to CyPA (amino acids 87–92), as well as a type II tight turn (residues A92–Q95). The contact between residues 92 and 95 is made through a hydrogen bond. Given this, the introduction of A for the G and Q residues may disrupt the type II turn, and thus may also interfere with certain essential functions contributing to the replication process, such as the uncoating of the core.
Undoubtedly, a systematic analysis of the events associated with virus infection will furnish further information on the role played by PGQM in viral replication. The PGQM mutant viruses may prove to be useful as unique agents for the dissection of the events before reverse transcription in HIV-1 infection. Furthermore, our results suggest potential strategies for developing anti-HIV-1 agents based on the loop containing the PGQM motif present in the capsid protein.
Acknowledgments
We thank Dr. Antonito Panganiban (University of Wisconsin, Madison, WI) for the gift of pED84. pSV-A-MLV-env was obtained from the AIDS Research and Reference Reagent Program of the National Institutes of Health. We also thank Mr. Stephen Whitney of Advanced BioScience Laboratories, Inc. for technical help, Mr. Sashi Reddy for sequence analysis, and Mr. Edward Frede of Thomas Jefferson University for his help in the preparation of this manuscript. This work was supported by funds from the National Institutes of Health (AI29306)(A.S.), a grant from the Commonwealth of Pennsylvania to the Biotechnology Foundation, Inc., and a grant from the Biomedical Research Support Committee and institutional funds (T.A.R.).
ABBREVIATIONS
- CA
capsid
- CyPA
cyclophilin A
References
- 1.Coffin J M. In: Field’s Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippencott; 1996. pp. 1767–1847. [Google Scholar]
- 2.Levy J A. Microbiol Rev. 1993;57:183–289. doi: 10.1128/mr.57.1.183-289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wills J W, Craven R C. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 4.Luban J. Cell. 1996;87:1157–1159. doi: 10.1016/s0092-8674(00)81811-5. [DOI] [PubMed] [Google Scholar]
- 5.Hunter E. Semin Virol. 1994;5:71–83. [Google Scholar]
- 6.Katz R A, Skalka A M. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 7.Luciw P A. In: Field’s Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippencott; 1996. pp. 1881–1952. [Google Scholar]
- 8.Luban J, Bossolt K L, Franke E K, Kalpana G V, Goff S P. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 9.Colgan J, Yuan H E H, Franke E K, Luban J. J Virol. 1996;70:4299–4310. doi: 10.1128/jvi.70.7.4299-4310.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schutkowski M, Drewello M, Wollner S, Jakob M, Reimer U, Scherer G, Schierhorn A, Fischer G. FEBS Lett. 1996;394:289–294. doi: 10.1016/0014-5793(96)00972-6. [DOI] [PubMed] [Google Scholar]
- 11.Franke E K, Yuan H E H, Luban J. Nature (London) 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 12.Thali M, Bukovsky A A, Kondo E, Rosenwirth B, Walsh C T, Sodroski J, Gottlinger H G. Nature (London) 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- 13.Dorfman T, Gottlinger H G. J Virol. 1996;70:5751–5757. doi: 10.1128/jvi.70.9.5751-5757.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mlynar E, Bevec D, Billich A, Rosenwirth B, Steinkasserer A. J Gen Virol. 1997;78:825–835. doi: 10.1099/0022-1317-78-4-825. [DOI] [PubMed] [Google Scholar]
- 15.Franke E K, Luban J. Virology. 1996;222:279–282. doi: 10.1006/viro.1996.0421. [DOI] [PubMed] [Google Scholar]
- 16.Srivastava M, Zhang-Keck Z Y, Caohuy H, McPhie P, Pollard H B. Biochem J. 1996;316:729–735. doi: 10.1042/bj3160729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 20.Serio D, Rizvi T A, Cartas M, Kalyanaraman V S, Weber I T, Koprowski H, Srinivasan A. Proc Natl Acad Sci USA. 1997;94:3346–3351. doi: 10.1073/pnas.94.7.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graham F N, Van der Eb J. Virology. 1973;53:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 22.Nagashunmugam T, Velpandi A, Goldsmith C S, Zaki S R, Kalyanaraman V S, Srinivasan A. Proc Natl Acad Sci USA. 1992;89:4114–4118. doi: 10.1073/pnas.89.9.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimpton J, Emerman M. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srivastava M, Goping G, Caohury H, McPhie P, Pollard H B. Exp Cell Res. 1996;229:14–19. doi: 10.1006/excr.1996.0338. [DOI] [PubMed] [Google Scholar]
- 25.Myers G, Korber B, Wain-Hobson S, Smith R F. Human Retroviruses and AIDS. Los Alamos, NM: Los Alamos National Lab.; 1993. [Google Scholar]
- 26.Yoshimura F K, Diem K, Learn G H, Jr, Riddell S, Corey L. J Virol. 1996;70:8879–8887. doi: 10.1128/jvi.70.12.8879-8887.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu X F, Wang Z, Beyrer C, Celentano D D, Khamboonruang C, Allen E, Nelson K. J Virol. 1995;69:4649–4655. doi: 10.1128/jvi.69.8.4649-4655.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rizvi T A, Lew K A, Murphy E C, Schmidt R D. Virology. 1996;224:517–532. doi: 10.1006/viro.1996.0558. [DOI] [PubMed] [Google Scholar]
- 29.Page K A, Landau N, Littman D R. J Virol. 1990;64:5270–5276. doi: 10.1128/jvi.64.11.5270-5276.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Braaten D, Franke E K, Luban J. J Virol. 1996;70:3551–3560. doi: 10.1128/jvi.70.6.3551-3560.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braaten D, Franke E K, Luban J. J Virol. 1996;70:4220–4227. doi: 10.1128/jvi.70.7.4220-4227.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Braaten D, Aberham C, Franke E K, Yim L, Phares W, Luban J. J Virol. 1996;70:5170–5176. doi: 10.1128/jvi.70.8.5170-5176.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aberham C, Weber S, Phares W. J Virol. 1996;70:3536–3544. doi: 10.1128/jvi.70.6.3536-3544.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dorfman T, Bukovsky A, Ohagen A, Hoglund S, Gottlinger H G. J Virol. 1994;68:8180–8187. doi: 10.1128/jvi.68.12.8180-8187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahli R, McMaster G K, Hirt B. Nucleic Acids Res. 1985;13:3617–3633. doi: 10.1093/nar/13.10.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Momany C, Kovari L C, Prongay A J, Keller W, Gitti R K, Lee B M, Gorbalenya A E, Tong L, McClure J, Ehrlich L S, et al. Nat Struct Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 37.Gitti R K, Lee B M, Walker J, Summers M F, Yoo S, Sundquist W I. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 38.Gamble T R, Vajdos F, Yoo S, Worthylake D K, Houseweart M, Sundquist W I, Hill C P. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]