Abstract
The PU.1 interaction partner (Pip) is a member of the interferon regulatory factor family that regulates gene expression through heterodimerization with the ETS transcription factor PU.1. Binding of Pip alone to DNA is weak, and usually it is recruited by phosphorylated PU.1 to form a strong ternary complex with specific DNA sequences. An approach combining sequence homology analysis, secondary structure predictions, and a precise mutational strategy has been used to determine critical residues within the Pip heterodimerization domain that contribute to ternary complex formation. We have delimited the Pip interaction domain to residues 245–422 by using deletion analysis. Site-directed mutagenesis of conserved polar amino acids within two predicted α-helices contained in this region, and which are highly conserved in the IRF family, confirmed the importance of these residues for Pip–PU.1 interaction with DNA as well as for trans-activation activity. Our results suggest the existence of a functional epitope essential for heterodimerization between Pip and PU.1 and possibly, in general, between interferon regulatory factor family members and their partners.
Gene expression is regulated by a sequence of protein–protein and protein–DNA interactions. Sequence specific protein–DNA interactions characterize binding of tissue and cell-specific transcription factors, which are further refined by protein–protein interactions with other coactivators/corepressors and the basal transcription machinery (1). Further level of gene regulation is accomplished by posttranslational modifications, such as phosphorylation (2, 3). PU.1 is an ETS family member implicated in the developmental regulation of cells in the hematopoietic system and in the regulation of multiple genes in B and myeloid cells (4–6). PU.1 can be phosphorylated at multiple Ser residues including the Pro, Glu, Ser, and Thr rich (PEST) domain (7). Phosphorylation of PU.1 enables recruitment of the PU.1 interaction partner (Pip), also known as NF-EM5, LSIRF, IRF-4, or ICSAT (7–12).
Pip is a lymphoid restricted member of the interferon regulatory factor (IRF) family of transcription factors, implicated in the regulation of gene expression in B cells through cell-type-specific enhancers (Eκ3′, Eλ2–4, and Eλ3–1) (9). Pip is a weak DNA-binding protein and a poor transcriptional activator (9, 11, 12). However, the binding of Pip to DNA in vitro is enhanced in the presence of phosphorylated PU.1, and PU.1–Pip interaction results in a synergistic activation of reporters containing adjacent PU.1- and Pip-binding sites (9, 13). Pip also represses transcription of interferon-stimulated response element (ISRE) reporter constructs when induced by IRF-1 (12, 13). The generation of knockout mice lacking Pip shows a severe deficiency in B cell function, suggesting that Pip is probably involved in the regulation of genes implicated in late B cell differentiation (14).
We designed a combined approach that used sequence homology studies, secondary structure predictions, and a detailed mutational analysis to determine residues within the Pip interaction domain (ID) that are essential for ternary complex (TC) formation with PU.1 and DNA. Deletion analysis demonstrates that residues 245–422 of Pip are absolutely necessary for its interaction with PU.1. Modification of polar amino acids within two conserved putative α-helices (spanning residues 300–335) abrogates protein–protein interaction between PU.1 and Pip in vitro and have a detrimental effect on the transcriptional activity of the complex in transient transfection experiments.
MATERIALS AND METHODS
Plasmids.
The Pip cDNA was amplified by using reverse transcription–PCR from mouse spleen mRNA using SuperScript reverse transcriptase (GIBCO/BRL) and 5′-TTGCTGCCCTCAGCTAAGAG-3′ and 5′-GCCCTGTCAGAGTATTTCTTC-3′ as 5′ and 3′ primers, respectively. Internal deletions were prepared by digestion with appropriate restriction enzymes or by overlapping PCR fragments. Point mutations were generated by PCR using primers with partial degeneracies at the site of interest. The hemagglutinin (HA) epitope tag sequence was amplified by using PCR (15) and fused to the C-terminal end of wild-type (wt) and ΔPip. All cDNAs were cloned into pcDNA3 (Invitrogen). Double-stranded oligonucleotides used in electrophoretic mobility-shift assay (see below) were inserted into the BglII site of a TK-pGL3 reporter plasmid (Promega). The nucleotide sequence of all constructs was confirmed by sequencing. Detailed information on the generation of the plasmids will be provided on request.
Sequence-Homology Analysis and Secondary-Structure Predictions.
Sequences for IRF family members were obtained from GenBank, and sequence alignments were performed with geneworks (IntelliGenetics). Secondary-structure predictions were obtained by using a consensus of four methods to minimize bias of individual methods: Levin (16), dpm (17), sopma (18, 19), and Gibrat (20).
Electrophoretic Mobility-Shift Assay.
Proteins were synthesized by using a coupled in vitro transcription-translation rabbit reticulocyte lysate system (Promega) following the manufacturer’s directions. Translation efficiency was estimated by parallel reactions in the presence of [35S]Met and SDS/PAGE. Protein–DNA complexing was performed at room temperature for 20 min in 20 mM Hepes (pH 7.9), 75 mM KCl, 0.5 mM EDTA, 1 mM DTT, 0.5 μg of poly(dI,dC), 5% glycerol, and 32P-labeled probe and then resolved in a 5% nondenaturing polyacrylamide gel with 0.25× Tris-borate buffer at 12.5 V/cm. For antibody supershifts, 1 μg of anti-HA antibody (Boehringer Mannheim) was added after the initial incubation, and the reactions were further incubated for 20 min before electrophoresis. Sense sequence of oligonucleotides used in this study are as follows: λ1B, 5′-gaaaaagagaaataaaaGGAAgtGAAAcccaag-3′; κE3′, 5′-gatccctttgaGGAActGAAAacagaacct-3′; ISG15, 5′-gatcctcgGGAAaggGAAAccgaaactgaagcca-3′ (capital letters indicate the PU.1 and the Pip core binding sites).
Transient Transfections.
NIH 3T3 cells were grown in DMEM supplemented with 10% newborn calf serum. Typically, 300 ng of a four-copy κE3′-luciferase reporter plasmid and 50 ng of expression vectors were cotransfected in triplicate in 24-well plates and luciferase activity was determined approximately 36 hr after transfection (21, 22). Samples were corrected by protein concentration estimated by a Bradford standard microassay. Transfection experiments were performed at least three times.
RESULTS
Putative Structural Motifs Within the ID of Pip Are Conserved Between IRF Family Members.
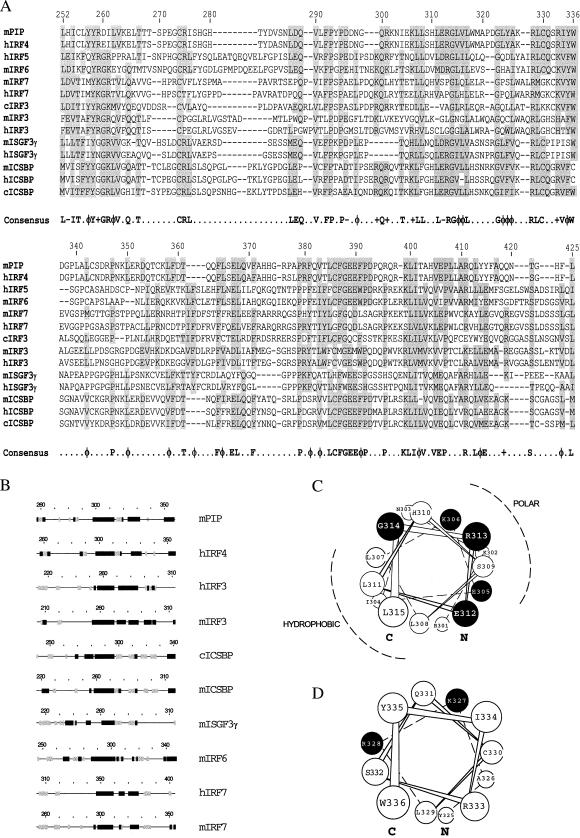
The IRF family shares a modular structure with a highly conserved DBD and a less conserved ID. The degree of identity between different members of the family is quite variable within the ID and is generally <50% (23). Within this domain, several areas are well conserved in all members analyzed, and a consensus sequence can be found (Fig. 1A). A strong conservation of structural motifs also is predicted in a region spanning residues 260–360 in Pip (Fig. 1B). A conserved amphipathic α-helix encompassing residues 300–314 in mPip (Fig. 1C) was noticed in all family members analyzed (Fig. 1B). Another conserved α-helix was predicted to exist between residues 325 and 335 in mPip (Fig. 1D), although the extent of the helical structure remains variable in the family.
Figure 1.
mPip ID shares regions of extensive sequence homology and conserved secondary structure motifs with other IRF family members. (A) Sequence of the ID of different IRFs were aligned by using the geneworks program. Amino acid numbers for the mPip protein are represented on top. Regions of similarity (allowing for conservative substitutions) are shaded. A consensus sequence can be deduced, which is indicated at the bottom. Symbols are as follows: −, acidic; φ, hydrophobic; +, basic. (B) Secondary-structure predictions were obtained for the full-length sequences of IRF family members by using a consensus of four different methods. α-helical structures, β-sheets, and coiled regions are represented by a solid or a stripped box and a line, respectively. Predicted structures have been aligned with respect to the conserved G314 of mPip. c, chicken; h, human; m, mouse. (C and D) Schematic representation of α-helices 300–314 and 325–335 in mPip, respectively. C and N indicate the C and the N terminus. ●, Conserved charged residues with a potential role in protein–protein interaction and that have been further modified by site-directed mutagenesis.
Definition of a Minimal Functional Pip by Using Deletion Analysis.
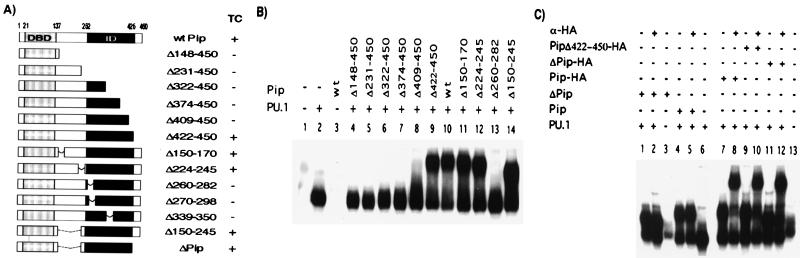
The observation that conserved areas are present within the ID raises the possibility that these subdomains may be involved in protein–protein interactions. Therefore, we mapped regions within the ID of Pip necessary for TC formation by using deletion analysis (Fig. 2A). No TC was detected between mutants Δ148–450, Δ231–450, Δ322–450, Δ374–450, and Δ409–450 and PU.1 on the λ1B probe in our experimental conditions, whereas a deletion of the most C-terminal 28 aa (Pip Δ422–450) still permitted the interaction (Fig. 2B). Internal deletions of residues 150–170, 224–245, or 150–245 within the hinge region between the DBD and ID, generated mutant proteins still able to form a TC with PU.1-DNA. However, a deletion of residues 260–282 completely abrogated the ability of Pip to form a TC (Fig. 2B). The internal and C-terminal deletions were further combined to give a minimal construct, hereafter referred to as ΔPip, containing residues 1–149 and 245–422 that still retained the ability to bind PU.1–DNA (Fig. 2C, lane 1). The TC formed between PU.1, the λlB element, and HA-tagged wt or Pip mutants could be supershifted by the addition of anti-HA tag antibody (Fig. 2C, lanes 8, 10, and 12), demonstrating that it contained Pip proteins. Control reactions showed that this antibody did not react with the untagged proteins (lanes 2 and 5). Our results clearly delineate the Pip ID to residues 245–422.
Figure 2.
Serial deletions delineate the boundaries of the mPip ID. (A) Schematic representation of the C-terminal and internal deletion mutants used in this study. The solid box indicates the Pip ID. On the right, the ability of the different mutants to form TC. Equal amounts of proteins prepared by in vitro transcription/translation were assayed for interaction with PU.1 and λ1B probe. On the left (−), nonprogrammed reticulocyte lysate was used as control for nonspecific binding. (C) Pip and the deletion mutants ΔPip and Δ422–450 were tagged with a HA epitope to demonstrate the presence of Pip protein in the complex, in the presence of an anti-HA antibody.
Site-Directed Mutagenesis of Pip Reveals Crucial Residues Required for Protein–Protein Interactions.
By using random mutagenesis, we observed that mutations that impaired the interaction of Pip with PU.1 and DNA accumulated in the region between residues 296 and 336 and, mostly, conserved charged residues within this region were affected (data not shown). The PEST region of PU.1 is required for interaction with Pip, and it has been suggested that the acidic residues within this region could interact via a charge–charge basis with a basic region in Pip (8). Accordingly, α-helices 300–314 and 325–335 have a net positive charge and could paticipate in the contact surface between Pip and PU.1. Therefore, we focused on the charged residues within these helices for further mutation analysis.
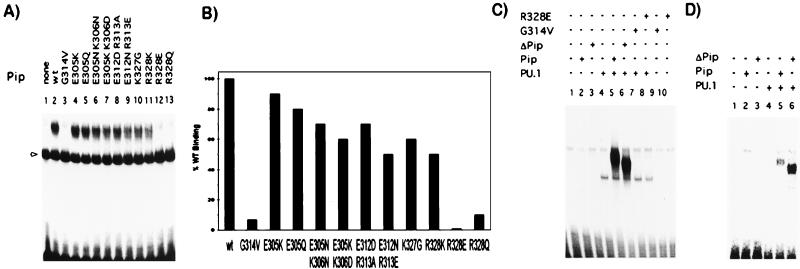
Within the first putative α-helix (residues 300–314), mutation of amino acids E305, K306, E312, or R313 to another polar or oppositely charged residue (E305K, E305Q, E305N-K306N, E305K-K306D, E312D-R313A, or E312N-R313E) had small-to-moderate effects on the ability of Pip to interact with PU.1 and DNA (Fig. 3A, lanes 4–9). Quantitation of the band intensities showed a ≈50% inhibition of binding with mutant E312N–R313E (Fig. 3B). In the second region (residues 325–335), mutations of two positively charged residues K327 and R328 were examined. Mutant K327G showed 60% of TC formation as compared with wt Pip (Fig. 3B). Amino acid R328 is absolutely conserved at equivalent positions in all IRF IDs (see Fig. 1A). A conservative mutation of this residue to K (R328K) decreased the amount of TC to 50%, indicating that a positive charge at this position is not sufficient for maximal interaction. Importantly, when residue R328 was changed to the opposite (R328E) or neutral charge (R328Q), no TC was detected (Fig. 3A). G314 is another amino acid absolutely conserved in all of the IRF members, and substitution by V (G314V) completely abrogated PU.1–Pip–DNA interaction under our experimental conditions (Fig. 3A).
Figure 3.
Site-directed mutagenesis identifies residues essential for TC formation. The effect of specific point mutations within the Pip ID was analyzed by gel retardation assay on the λ1B (A), kE3′ (C), and ISG15 (D) probes. Proteins were prepared by in vitro transcription/translation and equal amounts of PU.1 and wt Pip or mutants were incubated with DNA probes as indicated. Binding of PU.1 as a monomer in A is indicated by an arrow. (B) The amount of TC formed by the different Pip mutants was quantitated by using a PhosphorImager and is represented as a percentage relative to wt Pip. The results of a representative experiment are shown.
We next addressed how ternary complex formation was affected by using different response elements, such as the κE3′ or the ISG15. We observed that Pip mutations that abrogated PU.1–Pip interaction in the λ1B element, such as G314V and R328E, also disrupted TC formation in the κE3′ or the ISG15 elements, whereas the ΔPip mutant could still interact efficiently with PU.1 on these probes (Fig. 3 C and D; data not shown). The pattern of binding observed for the other point mutants on the κE3′ or the ISG15 elements was similar to that observed on the λ1B probe (data not shown). A weaker binding activity to the ISG15 element was noticed, which could probably be caused by the presence in this element of a longer spacer between the PU.1 and Pip binding sites with respect to the κE3′ or λ1B elements (3 vs. 2 bp, respectively).
Mutations Within the ID of Pip Affect Its Ability to Activate Transcription.
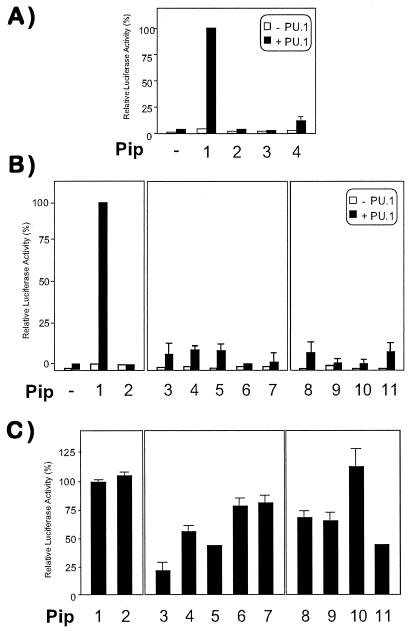
Transient transfection experiments were next performed to understand the functional significance of mutations in the Pip ID. When NIH 3T3 cells were transfected with PU.1 or Pip alone, a very modest transcriptional activation of a κE3′-driven luciferase reporter construct was observed (Fig. 4A). However, cotransfection of PU.1 and Pip expression vectors rendered a strong synergistic trans-activation, as previously described (Fig. 4A) (8, 9, 13). When the effect of internal deletions was analyzed, we observed that mutants with impaired TC-formation ability had also lost their trans-activation potential (Fig. 4A). However, the minimal construct ΔPip behaved as a poor transcriptional activator (Fig. 4A), although it could still form a strong TC with PU.1 and DNA.
Figure 4.
Trans-activation activity of Pip is affected by mutations in putative α-helices 300–314 and 325–335. NIH 3T3 cells were cotransfected with 300 ng of a κE3′-driven luciferase reporter alone (open columns) or in the presence of 50 ng of PU.1 (filled columns), together with 50 ng of wt or Pip mutants expression vectors, respectively (A and B). Synergistic transcriptional activation observed when both PU.1 and wt Pip were cotransfected was taken as 100%. (C) The ability of PU.1 and wt Pip (50 ng each) to induce luciferase activity was measured in the presence of 50 ng of cotransfected mutant Pip expression vector. Relative luciferase activity obtained in the absence of mutant Pip (column 1) was used as control. (A) Column 1, wt; column 2, Δ374–450; column 3, Δ409–450; column 4: ΔPip. (B and C) Column 1: wt; column 2: K94E; column 3, E305K; column 4, E305N K306N; column 5, E305K K306D; column 6, E312N R313E; column 7, G314V; column 8, K327G; column 9, R328K; column 10, R328E; column 11, R328Q.
To determine whether protein–protein interaction was sufficient for maximum transcriptional activity of the heterodimer, we designed a point mutant within the DBD (K94E) of Pip that abolished its DNA-binding activity. K94 is conserved in all IRF members analyzed except the vIRF, and in IRF1 it participates in DNA binding by forming one hydrogen bond with the phosphate backbone (24). This mutant did not form TC with PU.1 (not shown) and failed to activate the luciferase reporter either in the absence or in the presence of PU.1 (Fig. 4B), suggesting that both proteins need to bind DNA to form a strong complex in vitro and for optimal transcriptional activity of the PU.1–Pip complex. When mutations of polar residues at the predicted α-helix 300–314 were analyzed, significantly reduced trans-activation capability of the PU.1–Pip complex was observed (Fig. 4B), whereas the same mutations affected only partially the ability of Pip to form TC (see Fig. 3A). Within the second α-helix, mutations in residues K327 and R328 had also a negative effect on the transcriptional activity of the PU.1–Pip complex (Fig. 4B). The transcriptional activity was not restored when higher amounts of mutant Pip expression vectors were cotransfected, indicating that the lack of transactivation was not caused by a reduced expression of the different mutant proteins (data not shown).
We next examined the possibility that these mutants exert a dominant-negative effect over wt Pip protein. NIH 3T3 cells were cotransfected with equal amounts of PU.1 and Pip expression vectors, in the absence or in the presence of an equimolar concentration of the different Pip mutants. K94E did not interfere with the PU.1–Pip-induced trans-activation of the reporter (Fig. 4C). However, a strong inhibition of luciferase activity (>80%) was observed in the presence of the E305K mutant. This strong dominant-negative activity of E305K was partially reversed by a second mutation, K306E. Mutations at amino acids E312, R313, or G314 did not result in a significant dominant negative activity of the Pip mutant. Mutations within the second helix had only a partial or no dominant-negative effect over the wt Pip (Fig. 4C). The lack of effect with mutant R328E was not surprising given its inability to form ternary complex in vitro with PU.1 (see Fig. 3). Altogether, these data provide evidence that the proper arrangement of polar side chains within regions 300–314 and 325–335 and a conserved structural topology are crucial for optimal protein–protein interactions and, more importantly, for the transcriptional activity of the Pip–PU.1 heterodimer.
DISCUSSION
It is well established that heterodimerization between different transcription factors is a required step for optimal binding to specific DNA sequences and subsequent transcriptional activation of genes nearby. A good example is represented by PU.1 and its interaction partner Pip. In this study we have delineated the minimal protein-protein interaction interface between Pip and PU.1 to residues 245–422 of Pip. Our results indicate that both proteins contact DNA, because a Pip mutant that lacks DNA-binding activity (K94E) is unable to interact with PU.1 and trans-activate through the κE3′ element. However, a minimal Pip construct (ΔPip) that lacks the intervening region, probably containing a putative Pip trans-activation domain, can still form TC but renders a transcriptionally inactive heterodimer (Fig. 4A). This minimal construct is particularly interesting in that we can separate the TC formation and the trans-activation activities within the Pip molecule. However, further studies will be required to rule out that the lack of transcriptional activity is not caused by a poor stability of this protein in transfected cells.
Analysis by random mutagenesis of the minimal Pip ID indicates that a region spanning residues 296–336 accumulates loss-of-function mutations, as determined by their inability to form a TC with PU.1 and DNA in gel-shift experiments. The importance of this region is further supported by three factors: (i) secondary-structure prediction analysis indicates the probable existence of two α-helical structures in this region (amino acids 300–314 and 325–335); (ii) homology studies show a high degree of conservation in this region within the IRF family; and (iii) this structural domain contains a conserved hydrophobic motif located between both helical structures: GφφL(X)3–5 Gφφφ(X)1–3RL (φ indicates a hydrophobic residue, and X any amino acid), in which G and R are invariable in all of the members analyzed.
How can we explain the effects of the point mutations in the general context of protein–protein interactions between PU.1 and Pip? The nature of the mutations introduced by site-directed mutagenesis was intended to be incompatible with the function of the side chain in the interaction surface. Therefore, we first substituted charged residues to alter specific shape or charge interactions, while the structural integrity of the helices would be maintained, as indicated by secondary structure predictions (data not shown). In the first helix, substitution of a E residue by a K or N inverts or cancels the charge and also increases or reduces the side-chain length, respectively. Similarly, the modification of K residues by D, N, or Q will invert or reduce the charge and reduce the side-chain length. In the second helix, modification of the charge and/or the side-chain length also are expected when R328 is substituted by a K, E, or Q. The role of charged side chains in both helices could be to participate in intermolecular contacts with PU.1. Of special interest is the effect of R328 mutations in TC formation as well as in the biological function of the complex. R residues are present in the recognition site of many enzymes (25). The guanidinium group of R is able to form multiple hydrogen bonds with a phosphate group, and the charged interaction is much stronger than that generated with just one positive charge in the side chain, as in K residues. Our evidence suggests that R328 could play a critical role during TC formation by interacting with PU.1 through the phosphoryl group of S148. It could also interact with carboxyl groups, quite abundant in the side chains of acidic residues (E and D) in the PEST domain (8). Mutation R328K supports this hypothesis, because K residues do not form hydrogen bonds with phosphate groups as efficiently as R residues do, which in turn could be translated into weaker TC (as shown in Fig. 2). This mutant is not able to induce luciferase expression in transfection experiments, further demonstrating also a critical role for R328 in the trans-activation capability of the complex. Substitution of R328 by an E or Q residue completely abrogates TC formation and has a detrimental effect on transfection experiments.
A second type of mutation is represented by G314V, which completely abrogates the PU.1–Pip interaction as measured by TC formation and transfection experiments. Because G is a flexible residue, its location at the end of a helical structure may play a critical role in the positioning of α-helices 300–314 and 325–335 with respect to each other. A mutation that replaces the proton moiety in G314 by a bulky branched hydrophobic side chain (G314V) may interfere with the proper packing of those two conserved helices, as previously described in the Drosophila Antennapedia homeodomain and other structures that contain a helix–turn–helix motif or an arrangement of two closely positioned helices (26). Alternatively, G residues are known to terminate α-helices, therefore its substitution for a V residue may result in the extension of the helix (27). Our results suggest an important structural role for G314, possibly in the packing of the two α-helices. In the second helix, substitution of the K327 by a G will reduce the side chain to only a hydrogen residue, and it will possibly terminate the α-helix at this position, as indicated by secondary-structure predictions (data not shown), making it shorter than in wt Pip (28, 29). Although the ability of this mutant to form a TC is only partially impaired, this modification has a dramatic effect on the transcriptional activity of the complex.
Our results suggest that residues with a modest contribution to TC formation, as analyzed by gel retardation assay, play a major role in the transcriptional activity of the complex, further supporting the importance of helices 300–314 and 325–335. Furthermore, our data indicate that specific mutations within the Pip interaction domain can interfere with the biological function of the PU.1–wt Pip heterodimer and function as dominant-negative mutants. For instance, mutant E305K has a strong dominant-negative activity, most probably by competing with wt Pip for complex formation, resulting in an ineffective trans-activation complex. As noted, the PEST domain in PU.1 is very rich in negatively charged residues (8), and E305K substitution will increase the total positive charge in Pip. Interestingly, cotransfection of PipK94E mutant did not affect the transcriptional activity of the PU.1–wt Pip (Fig. 4C), further confirming the necessity of Pip binding to DNA for its optimal activity in intact cells.
The random mutagenesis studies revealed that TC formation is also disturbed by few mutations in hydrophobic residues in the region spanning residues 300–335. Four of these mutations involved a change from a hydrophobic residue to a P, possibly disrupting the integrity of the putative helices and consequently TC formation (data not shown). We also observe a conserved hydrophobic motif GφφL(X)3–5 Gφφφ(X)1–3RL between α-helices 300–314 and 325–335. These residues may be important in the recognition by Pip of PU.1 or other modulators representing a functional epitope, as previously reported for other transcriptional activators (30–33). They may also be essential for proper folding of the ID and their contribution will be important as well for TC formation and transactivation. A combination of the functional epitope with the charged and polar residues modulate PU.1–Pip interaction by adding specificity to the interaction.
Previous deletion analysis suggests that a region spanning residues 410–439 is important for Pip binding to DNA. An inhibitory function has been proposed for this region, because removal of residues 410–439 greatly reduces TC formation and enables Pip to bind to the λ1B site (13). Our results confirm the importance of this region, because deletion of residues 409–450 abrogates TC formation. A consensus α-helix is predicted to exist between residues 399–414 (13), therefore deletion of residues 409–450 (this study) would disrupt this putative helix and possibly drastically affect the tertiary fold of the ID. Furthermore, deletion of residues 422–450 is still able to interact with PU.1 and DNA and still contains this helical motif (Fig. 2B). We note that this helix contains several conserved hydrophobic residues, that could be implicated in structural hydrophobic interactions with another helix in the ID (like α-helix 300–314) (13), therefore contributing to complete domain stabilization.
In summary, we have gained insight into functional regions of Pip necessary for optimal interaction with PU.1. Our combined approach of sequence-homology analysis, secondary-structure prediction, and extensive mutational analysis has disclosed the existence of critical residues within the Pip ID. We show that ternary-complex formation between PU.1, Pip, and DNA requires both DNA–protein and protein–protein interactions, and alteration of either the DNA-binding activity or the protein–protein ID has detrimental consequences on the biological activity of the complex. Whether the residues identified in this study account for a tight protein–protein interaction or play another role, such as influencing the rate of association/dissociation, specificity of interaction, or stability of the protein is under further investigation. Structural studies will improve our understanding on the binding interface between Pip and PU.1 and reveal important knowledge on the intermolecular contacts occurring between both proteins.
Acknowledgments
We thank C. Geourjon and G. Deleage for providing secondary-structure predictions and J. Piedrafita for critical reading of the manuscript. This work was supported by National Institutes of Health and Department of Defense grants to N.A.-M. and R.A.M.
ABBREVIATIONS
- PEST
Pro, Glu, Ser and Thr-rich domain
- DBD
DNA binding domain
- ID
interaction domain
- wt
wild type
- TC
ternary complex
- Pip
PU.1 interaction partner
- IRF
interferon regulatory factor
- HA
hemagglutinin
References
- 1.Ptashne M, Gann A. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T, Karin M. Cell. 1992;70:375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- 3.Karin M. Curr Opin Cell Biol. 1994;6:415–424. doi: 10.1016/0955-0674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 4.Scott E W, Simon M C, Anastasi J, Singh H. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 5.McKercher S R, Torbett B E, Anderson K L, Henkel G W, Vestal D J, Baribault H, Klemsz M, Feeney A J, Wu G E, Paige C J, Maki R A. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 6.Henderson A, Calame K. Annu Rev Immunol. 1998;16:163–200. doi: 10.1146/annurev.immunol.16.1.163. [DOI] [PubMed] [Google Scholar]
- 7.Pongubala J M, van Beveren C, Nagulapalli S, Klemsz M J, McKercher S R, Maki R A, Atchison M L. Science. 1993;259:1622–1625. doi: 10.1126/science.8456286. [DOI] [PubMed] [Google Scholar]
- 8.Pongubala J M, Nagulapalli S, Klemsz M J, McKercher S R, Maki R A, Atchison M L. Mol Cell Biol. 1992;12:368–378. doi: 10.1128/mcb.12.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisenbeis C F, Singh H, Storb U. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 10.Grossman A, Mittrucker H W, Nicholl J, Suzuki A, Chung S, Antonio L, Suggs S, Sutherland G R, Siderovski D P, Mak T W. Genomics. 1996;37:229–233. doi: 10.1006/geno.1996.0547. [DOI] [PubMed] [Google Scholar]
- 11.Matsuyama T, Grossman A, Mittrucker H W, Siderovski D P, Kiefer F, Kawakami T, Richardson C D, Taniguchi T, Yoshinaga S K, Mak T W. Nucleic Acids Res. 1995;23:2127–2136. doi: 10.1093/nar/23.12.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamagata T, Nishida J, Tanaka S, Sakai R, Mitani K, Yoshida M, Taniguchi T, Yazaki Y, Hirai H. Mol Cell Biol. 1996;16:1283–1294. doi: 10.1128/MCB.16.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brass A L, Kehrli E, Eisenbeis C F, Storb U, Singh H. Genes Dev. 1996;10:2335–2347. doi: 10.1101/gad.10.18.2335. [DOI] [PubMed] [Google Scholar]
- 14.Mittrucker H W, Matsuyama T, Grossman A, Kundig T M, Potter J, Shahinian A, Wakeham A, Patterson B, Ohashi P S, Mak T W. Science. 1997;275:540–543. [PubMed] [Google Scholar]
- 15.Field J, Nikawa J, Broek D, MacDonald B, Rodgers L, Wilson I A, Lerner R A, Wigler M. Mol Cell Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levin J M, Robson B, Garnier J. FEBS Lett. 1986;205:303–308. doi: 10.1016/0014-5793(86)80917-6. [DOI] [PubMed] [Google Scholar]
- 17.Deleage G, Roux B. Protein Eng. 1987;1:289–294. doi: 10.1093/protein/1.4.289. [DOI] [PubMed] [Google Scholar]
- 18.Geourjon C, Deleage G. Protein Eng. 1994;7:157–164. doi: 10.1093/protein/7.2.157. [DOI] [PubMed] [Google Scholar]
- 19.Geourjon C, Deleage G. Comput Appl Biosci. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 20.Gibrat J F, Garnier J, Robson B. J Mol Biol. 1987;198:425–443. doi: 10.1016/0022-2836(87)90292-0. [DOI] [PubMed] [Google Scholar]
- 21.Ausubel R, Brent R, Kingston R, Moore D, editors. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 22.Hauser C A, Westwick J K, Quilliam L A. Methods Enzymol. 1995;255:412–426. doi: 10.1016/s0076-6879(95)55043-7. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen H, Hiscott J, Pitha P M. Cytokine Growth Factor Rev. 1997;8:293–312. doi: 10.1016/s1359-6101(97)00019-1. [DOI] [PubMed] [Google Scholar]
- 24.Escalante C R, Yie J, Thanos D, Aggarwal A K. Nature (London) 1998;391:103–106. doi: 10.1038/34224. [DOI] [PubMed] [Google Scholar]
- 25.Riordan J F, McElvany K D, Borders C L., Jr Science. 1977;195:884–886. doi: 10.1126/science.190679. [DOI] [PubMed] [Google Scholar]
- 26.Harrison S C, Aggarwal A K. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- 27.Pabo C O, Sauer R T. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 28.Gunasekaran K, Nagarajaram H A, Ramakrishnan C, Balaram P. J Mol Biol. 1998;275:917–932. doi: 10.1006/jmbi.1997.1505. [DOI] [PubMed] [Google Scholar]
- 29.Parker M H, Hefford M A. Protein Eng. 1997;10:487–496. doi: 10.1093/protein/10.5.487. [DOI] [PubMed] [Google Scholar]
- 30.Radhakrishnan I, Perez-Alvarado G C, Parker D, Dyson H J, Montminy M R, Wright P E. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 31.Uesugi M, Nyanguile O, Lu H, Levine A J, Verdine G L. Science. 1997;277:1310–1313. doi: 10.1126/science.277.5330.1310. [DOI] [PubMed] [Google Scholar]
- 32.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. Nature (London) 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 33.McInerney E M, Rose D W, Flynn S E, Westin S, Mullen T-M, Krones A, Inostroza J, Nolte R T, Assa-Munt N, Milburn M V, et al. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]