Abstract
We have taken a comprehensive approach to the generation of novel DNA binding zinc finger domains of defined specificity. Herein we describe the generation and characterization of a family of zinc finger domains developed for the recognition of each of the 16 possible 3-bp DNA binding sites having the sequence 5′-GNN-3′. Phage display libraries of zinc finger proteins were created and selected under conditions that favor enrichment of sequence-specific proteins. Zinc finger domains recognizing a number of sequences required refinement by site-directed mutagenesis that was guided by both phage selection data and structural information. In many cases, residues not expected to make base-specific contacts had effects on specificity. A number of these domains demonstrate exquisite specificity and discriminate between sequences that differ by a single base with >100-fold loss in affinity. We conclude that the three helical positions −1, 3, and 6 of a zinc finger domain are insufficient to allow for the fine specificity of the DNA binding domain to be predicted. These domains are functionally modular and may be recombined with one another to create polydactyl proteins capable of binding 18-bp sequences with subnanomolar affinity. The family of zinc finger domains described here is sufficient for the construction of 17 million novel proteins that bind the 5′-(GNN)6-3′ family of DNA sequences. These materials and methods should allow for the rapid construction of novel gene switches and provide the basis for a universal system for gene control.
The paradigm that the primary mechanism for governing the expression of genes involves protein switches that bind DNA in a sequence specific manner was established in 1967 (1). Since that time diverse structural families of DNA binding proteins have been described. Despite this wealth of structural diversity, the Cys2-His2 zinc finger motif constitutes the most frequently used nucleic acid binding motif in eukaryotes. This observation is as true for yeast as it is for humans. The Cys2-His2 zinc finger motif, identified first in the DNA and RNA binding transcription factor TFIIIA (2), is perhaps the ideal structural scaffold on which a sequence-specific protein might be constructed. A single zinc finger domain consists of approximately 30 aa with a simple ββα fold stabilized by hydrophobic interactions and the chelation of a single zinc ion (2, 3). Presentation of the α-helix of this domain into the major groove of DNA allows for sequence-specific base contacts. Each zinc finger domain typically recognizes 3 bp of DNA (4–7), though variation in helical presentation can allow for recognition of a more extended site (8–11). In contrast to most transcription factors that rely on dimerization of protein domains for extending protein-DNA contacts to longer DNA sequences or addresses, simple covalent tandem repeats of the zinc finger domain allow for the recognition of longer asymmetric sequences of DNA by this motif.
Recognition of these unique properties led us to propose and perform experiments aimed at creating what might be a universal system for the control of gene expression. In recent experiments we have described polydactyl zinc finger proteins that contain six zinc finger domains and bind 18 bp of contiguous DNA sequence (12). Recognition of 18 bp of DNA is sufficient to describe a unique DNA address within all known genomes, a requirement for our proposal for using polydactyl proteins as highly specific gene switches. Indeed, we have demonstrated control of both gene activation and repression by using these polydactyl proteins in a model system (12).
Because each zinc finger domain typically binds 3 bp of sequence, a complete recognition alphabet requires the characterization of 64 domains. Existing information, which could guide the construction of these domains, has come from three types of studies: structure determination (4–11, 13, 14), site-directed mutagenesis (15–20), and phage-display selections (21–27). All have contributed significantly to our understanding of zinc finger/DNA recognition, but each has its limitations. Structural studies have identified a diverse spectrum of protein/DNA interactions but do not explain whether alternative interactions might be more optimal. Further, while interactions that allow for sequence specific recognition are observed, little information is provided on how alternate sequences are excluded from binding. These questions have been partially addressed by mutagenesis of existing proteins, but the data are always limited by the number of mutants that can be characterized. Phage display and selection of randomized libraries overcomes certain numerical limitations, but providing the appropriate selective pressure to ensure that both specificity and affinity drive the selection is difficult. Experimental studies from several laboratories (21–26), including our own (27), have demonstrated that it is possible to design or select a few members of this recognition alphabet. However, the specificity and affinity of these domains for their target DNA was rarely investigated in a rigorous fashion in these early studies.
In this work we have taken a more systematic approach. We describe the selection by phage display, refinement by site-directed mutagenesis, and rigorous characterization of 16 zinc finger domains representing the 5′-GNN-3′ subset of this 64-member recognition code. We demonstrate that the identity of the residues at the three helical positions −1, 3, and 6 of a zinc finger domain are typically insufficient to describe in detail the specificity of the domain. While current zinc finger recognition codes attempt to define the specificity of the domain based on the residue identity at helical positions −1, 3, and 6, our results suggest that the predictive value of this code is limited.
MATERIALS AND METHODS
Selection by Phage Display.
Construction of zinc-finger libraries by PCR overlap extension was essentially as described (27). Growth and precipitation of phage were as described (28, 29), except that ER2537 cells (New England Biolabs) were used to propagate the phage and 90 μM ZnCl2 was added to the growth media. Precipitated phage were resuspended in zinc buffer A (ZBA; 10 mM Tris, pH 7.5/90 mM KCl/1 mM MgCl2/90 μM ZnCl2)/1% BSA/5 mM DTT. Binding reactions [500 μl: ZBA/5 mM DTT/1% Blotto (50 mM Tris⋅HCl, pH 7.4/100 mM NaCl/5% nonfat dry milk, Bio-Rad)/competitor oligonucleotides/4 μg sheared herring sperm DNA (Sigma)/100 μl filtered phage (1013 colony-forming units)] were incubated for 30 min at room temperature, before the addition of 72 nM biotinylated hairpin target oligonucleotide. Incubation continued for 3.5 hr with constant gentle mixing. Streptavidin-coated magnetic beads (50 μl; Dynal) were washed twice with 500 μl ZBA/1% BSA, then blocked with 500 μl of ZBA/5% Blotto/antibody-displaying (irrelevant) phage (≈1012 colony-forming units) for ≈4 hr at room temperature. At the end of the binding period, the blocking solution was replaced by the binding reaction and incubated 1 hr at room temperature. The beads were washed 10 times over a 1-hr period with 500 μl of ZBA/5 mM DTT/2% Tween 20, then once without Tween 20. Bound phage were eluted 30 min with 10 μg/μl of trypsin.
Hairpin target oligonucleotides had the sequence 5′-Biotin-GGACGCN′N′N′CGCGGGTTTTCCCGCGNNNGCGTCC-3′, where NNN was the 3-nt finger-2 target sequence and N′N′N′ its complement. A similar nonbiotinylated oligonucleotide, in which the target sequence was TGG (compTGG), was included at 7.2 nM in every round of selection to select against contaminating parental phage. Two pools of nonbiotinylated oligonucleotides also were used as competitors: one containing all 64 possible 3-nt targets sequences (compNNN), the other containing all of the GNN target sequences except for the current selection target (compGNN). These pools typically were used as follows: round 1, no compNNN or compGNN; round 2, 7.2 nM compGNN; round 3, 10.8 nM compGNN; round 4, 1.8 μM compNNN, 25 nM compGNN; round 5, 2.7 μM compNNN, 90 nM compGNN; round 6, 2.7 μM compNNN, 250 nM compGNN; round 7, 3.6 μM compNNN, 250 nM compGNN.
Multitarget Specificity Assays.
The fragment of pComb3H (28, 30) phagemid RF DNA containing the zinc-finger coding sequence was subcloned into a modified pMAL-c2 (New England Biolabs) bacterial expression vector and transformed into XL1-Blue (Stratagene). Freeze/thaw extracts containing the overexpressed maltose binding protein-zinc finger fusion proteins were prepared from isopropyl β-d-thiogalactoside-induced cultures by using the Protein Fusion and Purification System (New England Biolabs). In 96-well ELISA plates, 0.2 μg of streptavidin (Pierce) was applied to each well for 1 hr at 37°C, then washed twice with water. Biotinylated target oligonucleotide (0.025 μg) was applied similarly. ZBA/3% BSA was applied for blocking, but the wells were not washed after incubation. All subsequent incubations were at room temperature. Eight 2-fold serial dilutions of the extracts were applied in binding buffer (ZBA/1% BSA/5 mM DTT/0.12 μg/μl sheared herring sperm DNA). The samples were incubated 1 hr, followed by 10 washes with water. Mouse anti-maltose binding protein mAb (Sigma) in ZBA/1% BSA was applied to the wells for 30 min, followed by 10 washes with water. Goat anti-mouse IgG mAb conjugated to alkaline phosphatase (Sigma) was applied to the wells for 30 min, followed by 10 washes with water. Alkaline phosphatase substrate (Sigma) was applied, and the OD405 was quantitated with softmax 2.35 (Molecular Devices).
Gel Mobility Shift Assays.
Fusion proteins were purified to >90% homogeneity by using the Protein Fusion and Purification System (New England Biolabs), except that ZBA/5 mM DTT was used as the column buffer. Protein purity and concentration were determined from Coomassie blue-stained 15% SDS/PAGE gels by comparison to BSA standards. Target oligonucleotides were labeled at their 5′ or 3′ ends with [32P] and gel purified. Eleven 3-fold serial dilutions of protein were incubated in 20 μl of binding reactions (1× binding buffer/10% glycerol/≈1 pM target oligonucleotide) for 3 hr at room temperature, then resolved on a 5% polyacrlyamide gel in 0.5× TBE buffer (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3). Quantitation of dried gels was performed by using a PhosphorImager and imagequant software (Molecular Dynamics), and the KD was determined by Scatchard analysis.
RESULTS AND DISCUSSION
Library Construction and Selection.
As in our previous studies (27), we have used the murine Cys2-His2 zinc finger protein Zif268 for construction of phage-display libraries. Zif268 is structurally the most well-characterized of the zinc finger proteins (4, 5, 31). DNA recognition in each of the three zinc finger domains of this protein is mediated by residues in the N terminus of the α-helix contacting primarily 3 nt on a single strand of the DNA. The operator binding site for this three-finger protein is 5′-GCGTGGGCG-′3 (finger-2 subsite is underlined). Structural studies of Zif268 and other related zinc finger-DNA complexes (6–11, 13, 14) have shown that residues from primarily three positions on the α-helix (−1, 3, and 6) are involved in specific base contacts. Typically, the residue at position −1 of the α-helix contacts the 3′ base of that finger’s subsite while positions 3 and 6 contact the middle base and the 5′ base, respectively.
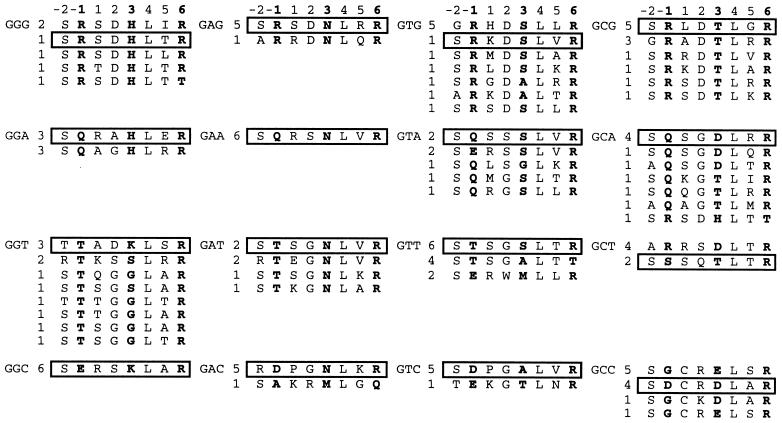
To select a family of zinc finger domains recognizing the 5′-GNN-3′ subset of sequences, we constructed two highly diverse zinc finger libraries in the phage-display vector pComb3H (28, 30). Both libraries involved randomization of residues within the α-helix of finger 2 of C7, a variant of Zif268 (27). The NNK library was constructed by randomization of positions −1, 1, 2, 3, 5, and 6 by using a condon doping strategy that allows for all amino acid combinations within 32 condons. The VNS library was constructed by randomization of positions −2, −1, 1, 2, 3, 5, and 6, which precludes Tyr, Phe, Cys, and all stop condons in its 24-codon set. The libraries consisted of 4.4 × 109 and 3.5 × 109 members, respectively, each capable of recognizing sequences of the 5′-GCGNNNGCG-3′ type. The size of the NNK library ensured that it could be surveyed with 99% confidence while the VNS library was highly diverse but somewhat incomplete. These libraries are, however, significantly larger than previously reported zinc finger libraries (21–27). Seven rounds of selection were performed on the zinc finger displaying-phage with each of the 16 5′-GCGGNNGCG-3′ biotinylated hairpin DNAs targets by using a solution binding protocol. Stringency was increased in each round by the addition of competitor DNA. Sheared DNA was provided for selection against phage that bound nonspecifically to DNA. Stringent selective pressure for sequence specificity was obtained by providing DNA of the 5′-GCGNNNGCG-3′ type as specific competitors (see Materials and Methods). Excess DNA of the 5′-GCGGNNGCG-3′ type was added to provide even more stringent selection against binding to DNAs with single or double base changes as compared with the biotinylated target. Phage binding to the single biotinylated DNA target sequence were recovered by using streptavidin-coated beads. In some cases the selection process was repeated. The finger-2 recognition helices of several randomly chosen seventh-round clones are shown in Fig. 1.
Figure 1.
The finger-2 recognition helices of randomly chosen clones from the seventh round of selection. The selection target site is shown to the left of each set, followed by the frequency with which each sequence was observed. The helix position of each amino acid is shown at the top, with positions −1, 3, and 6 shown in bold. Boxed sequences were studied in detail.
Striking conservation of all three of the primary DNA contact positions (−1, 3, and 6) was observed for virtually all the clones of a given target. Although many of these residues were observed previously at these positions after selections with much less complete libraries, the extent of conservation observed here represents a dramatic improvement over earlier studies (21–25, 27). Typically, phage selections have shown a consensus selection in only one or two of these positions. The greatest sequence variation occurred at the residues in positions 1 and 5, which do not make base contacts in the Zif268/DNA structure and were expected not to contribute significantly to recognition (4, 5). Variation in positions 1 and 5 also implied that the conservation in the other positions was the result of their interaction with the DNA and not simply the fortuitous amplification of a single clone caused by other reasons. Conservation of residue identity at position 2 also was observed. The conservation of position −2 is somewhat artifactual; the NNK library had this residue fixed as serine. This residue makes contacts with the DNA backbone in the Zif268 structure. Both libraries contained an invariant leucine at position 4, a critical residue in the hydrophobic core that stabilizes folding of this domain.
Impressive amino acid conservation was observed for recognition of the same nucleotide in different targets. For example, Asn in position 3 (Asn3) was virtually always selected to recognize adenine in the middle position, whether in the context of GAG, GAA, GAT, or GAC. Gln−1 and Arg−1 were always selected to recognize adenine or guanine, respectively, in the 3′ position regardless of context. Amide side chain-based recognition of adenine by Gln or Asn is well documented in structural studies as is the Arg guanidinium side chain to guanine contact with a 3′ or 5′ guanine (6, 7, 10). More often, however, two or three amino acids were selected for nucleotide recognition. His3 or Lys3 (and to a lesser extent, Gly3) were selected for the recognition of a middle guanine. Ser3 and Ala3 were selected to recognize a middle thymine. Thr3, Asp3, and Glu3 were selected to recognize a middle cytosine. Asp and Glu also were selected in position −1 to recognize a 3′ cytosine, while Thr−1 and Ser−1 were selected to recognize a 3′ thymine.
Characterization of Finger-2 Proteins.
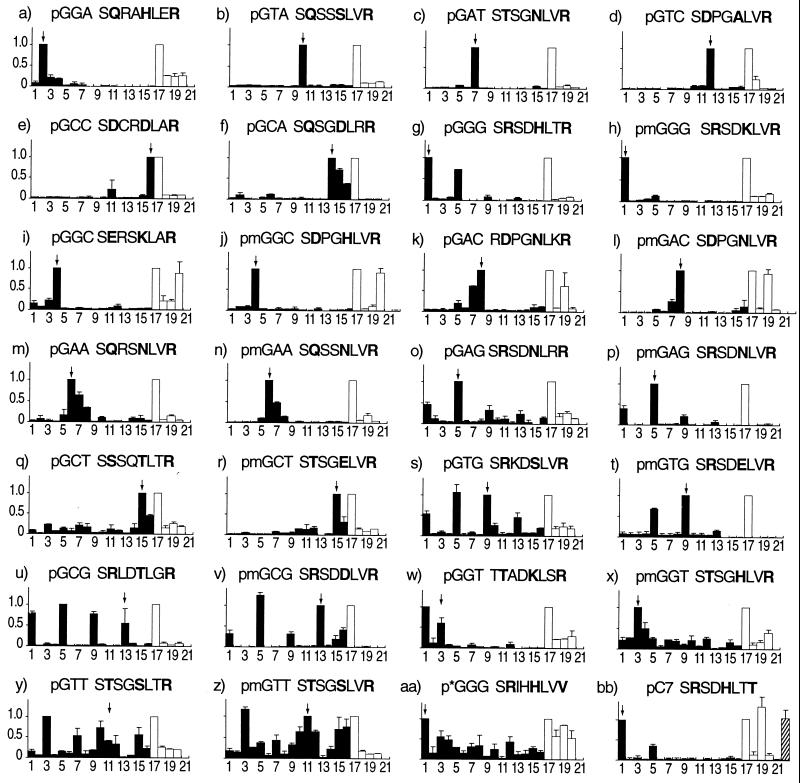
Selected Zif268 variants were subcloned into a bacterial expression vector, and the proteins were overexpressed (finger-2 proteins, hereafter referred to by the subsite for which they were panned). It is important to study soluble proteins rather than phage fusions because it is known that the two may differ significantly in their binding characteristics (32). The specificity profiles of representative clones are shown in Fig. 2. The proteins were tested for their ability to recognize each of the 16 5′-GNN-3′ finger-2 subsites by using a multitarget ELISA assay (Fig. 2, filled bars). This assay provided an extremely rigorous test for specificity because there were always six “nonspecific” sites that differed from the “specific” site by only a single nucleotide out of a 9-nt target. Many of the phage-selected finger-2 proteins showed exquisite specificity (for example, Fig. 2 a–e), while others demonstrated varying degrees of crossreactivity (Fig. 2 f, g, i, k, m, o, q, and s). Proteins pGCG, pGGT, and pGTT (Fig. 2 u, w, and y) actually bound better to subsites other than those for which they were selected.
Figure 2.
Multitarget ELISA titration assay for binding specificity. At the top of each graph is the DNA finger-2 target site for which each protein was selected or designed, and the recognition helix of that protein (positions −2 to 6). Helix positions −1, 3, and 6 are in bold. Proteins modified by site-directed mutagenesis have the prefix “m” before their DNA target. Columns 1–16 (filled bars) represent target oligos with different finger-2 subsites: 1 = GGG; 2 = GGA; 3 = GGT; 4 = GGC; 5 = GAG; 6 = GAA; 7 = GAT; 8 = GAC; 9 = GTG; 10 = GTA; 11 = GTT; 12 = GTC; 13 = GCG; 14 = GCA; 15 = GCT; 16 = GCC. Columns 17–20 (empty bars) represent oligonucleotide pools with a unique 5′ nucleotide in their finger-2 subsite: 17 = GNN; 18 = ANN; 19 = TNN; 20 = CNN. (j) Column 22 = CGC. (i) Column 22 = TAC. (bb) Column 22 = TGG. All data are background subtracted (column 21 = no target oligo). The height of each bar represents the average normalized titer from two independent experiments, with the highest signal normalized to the greatest value in columns 1–16 and 17–20. Error bars represent the deviation from the average. An arrow indicates the position of the cognate target oligonucleotide.
Attempts were made to improve binding specificity by modifying the recognition helix by using site-directed mutagenesis. Data from our selections and structural information guided mutant design. More than 100 mutant proteins were characterized in an effort to expand our understanding of the rules of recognition. Only the best example for each subsite is shown in Fig. 2 h, j, l, n, p, r, t, v, x, and z. Although helix positions 1 and 5 are not expected to play a direct role in DNA recognition, the best improvements in specificity always involved modifications in these positions. These residues have been observed to make phosphate backbone contacts, which contribute to affinity in a nonsequence-specific manner. Removal of nonspecific contacts increases the importance of the specific contacts to the overall stability of the complex, thereby enhancing specificity. For example, the specificity of proteins pGAC, pGAA, and pGAG (Fig. 2 k, m, and o) were improved simply by replacing atypical, charged residues in positions 1 and 5 with smaller, uncharged residues. Protein pGTT (Fig. 2y) also was improved by a change in position 5 (Fig. 2z), although several attempts at selection and mutagenesis failed to identify a protein that could bind subsite GTT without crossreaction.
Another class of modifications involved changes to both binding and nonbinding residues. The crossreactivity of protein pGGG for the finger-2 subsite GAG (Fig. 2g) was abolished by the modifications His3–Lys and Thr5–Val (Fig. 2h). It is interesting to note that His3 was unanimously selected during panning to recognize the middle guanine, although Lys3 provided better discrimination of A and G. This finding suggests that panning conditions for this protein may have favored selection by a parameter such as affinity over that of specificity. Indeed, the affinity of protein pmGGG for subsite GGG is 15-fold less than that of pGGG (Table 1). In the Zif268 structure, His3 donates a hydrogen bond to the N7 of the middle guanine (4, 5). This bond also could be made with N7 of adenine, and in fact, Zif268 does not discriminate between G and A in this position (31). Although this reasoning explains the observed crossreactivity of protein pGGG, His3 was found to specify only a middle guanine in proteins pGGA, pmGGC, and pmGGT (Fig. 2 a, j, and x), even though Lys3 was selected during panning for proteins pGGC and pGGT. It should be noted that Lys3 also is found in finger 2 of YY1 and finger 1 of TFIIIA where both fingers recognize binding sites with a middle G (9, 11, 13). The ability of Lys3 to provide discrimination against adenine recognition at this position had not previously been suggested and is not evident from the structures of these proteins. In a TFIIIA structure this residue is involved in contact with a phosphate, not a base (9). The multiple crossreactivities of protein pGTG (Fig. 2s) were similarly attenuated by modifications Lys1–Ser and Ser3–Glu (Fig. 2t), resulting in a 5-fold loss in affinity (Table 1). The Ser3–Glu modification of pmGTG (Fig. 2t) was largely accidental; the intention had been to create a protein that could recognize the subsite GCG. Glu3 has been shown to be very specific for cytosine in binding site selection studies of Zif268 (31). No structural studies show an interaction of Glu3 with the middle thymine, and Glu3 was never selected to recognize a middle thymine in our study or any others (21–27). Despite this paucity of predictive data, the Ser3–Glu modification favored the recognition of a middle thymine over cytosine (compare Fig. 2 s and t). These examples illustrate the limitations of relying on previous structures and selection data to understand the structural elements underlying specificity. It also should be emphasized that improvements by modifications involving positions 1 and 5 could not have been predicted by existing “recognition codes” (20, 33–35), which typically consider only positions −1, 2, 3, and 6. Only by the combination of selection and site-directed mutagenesis can we begin to fully understand the intricacies of zinc finger/DNA recognition.
Table 1.
Affinities of finger-2 proteins
| Protein1 | Finger-2 helix2 | Finger-2 subsite3 | KD, nM4 | KD. Prot/KD. Zif268 |
|---|---|---|---|---|
| pGGG | SRSDHLTR | GGG | 0.4 | 0.04 |
| pmGGG | SRSDKLVR | GGG | 6 | 0.6 |
| GTG | >1,400 | |||
| pGGA | SQRAHLER | GGA | 3 | 0.3 |
| pmGGT | STSGHLVR | GGT | 15 | 1.5 |
| GGC | >2,400 | |||
| pmGGC | SDPGHLVR | GGC | 40 | 4.0 |
| pmGAG | SRSDNLVR | GAG | 1 | 0.1 |
| GGG | 45 | 4.5 | ||
| pmGAA | SQSSNLVR | GAA | 0.5 | 0.05 |
| pGAT | STSGNLVR | GAT | 3 | 0.3 |
| pmGAC | SDPGNLVR | GAC | 3 | 0.3 |
| GCC | 90 | 9.0 | ||
| pGTG | SRKDSLVR | GTG | 3 | 0.3 |
| pmGTG | SRSDELVR | GTG | 15 | 1.5 |
| GAG | 30 | 3.0 | ||
| pGTA | SQSSSLVR | GTA | 25 | 2.5 |
| GTG | >1,000 | |||
| pmGTT | STSGSLVR | GTT | 5 | 0.5 |
| pGTC | SDPGALVR | GTC | 40 | 4.0 |
| GCC | >4,400 | |||
| pmGCG | SRSDDLVR | GCG | 9 | 0.9 |
| GAG | 6 | 0.6 | ||
| pGCA | SQSGDLRR | GCA | 2 | 0.2 |
| GCT | 10 | 1 | ||
| pmGCT | STSGELVR | GCT | 65 | 6.5 |
| pGCC | SDCRDLAR | GCC | 80 | 8.0 |
| C7 | SRSDHLTT | TGG | 0.5 | 0.05 |
| Zif268 | SRSDHLTT | TGG | 10 | 1 |
Protein designations are as in Fig. 2.
Helix positions −1, 3, and 6 are shown in bold.
Altered nucleotides are underlined.
Values represent at least two independent experiments. The SE was ± 50%.
From the combined selection and mutagenesis data it emerged that specific recognition of many nucleotides could be best accomplished by using motifs, rather than a single amino acid. For example, the best specification of a 3′ guanine was achieved by using the combination of Arg−1, Ser1, and Asp2 (the RSD motif). By using Val5 and Arg6 to specify a 5′ guanine, recognition of subsites GGG, GAG, GTG, and GCG could be accomplished by using a common helix structure (SRSD-X-LVR) differing only in the position 3 residue (Lys3 for GGG, Asn3 for GAG, Glu3 for GTG, and Asp3 for GCG). Similarly, 3′ thymine was specified by using Thr−1, Ser1, and Gly2 in the final clones (the TSG motif). This finding is in stark contrast to the prediction of the code that Asn−1 and Gln−1 best recognize 3′ thymine (34, 35). Further, a 3′ cytosine could be specified by using Asp−1, Pro1, and Gly2 (the DPG motif) except when the subsite was GCC; Pro1 was not tolerated by this subsite. Specification of a 3′ adenine was with Gln−1, Ser1, and Ser2 in two clones (QSS motif). Residues at positions 1 and 2 of the motifs were studied for each of the 3′ bases and found to provide optimal specificity for a given 3′ base as described here (data not shown).
The multitarget ELISA assays were designed with the assumption that all of the proteins preferred guanine in the 5′ position because all proteins contained Arg6, and this residue is known from structural studies to contact guanine at this position (4–11, 13). This interaction was demonstrated here by using the 5′ binding site signature assay (ref. 34; Fig. 2, empty bars). Each protein was applied to pools of 16 oligonucleotide targets in which the 5′ nucleotide of the finger-2 subsite was fixed as G, A, T, or C (Fig. 2, columns 17, 18, 19, and 20, respectively) and the middle and 3′ nucleotides were randomized. All proteins (Fig. 2 a–z) preferred the GNN pool with essentially no crossreactivity. As a control we studied p*GGG that contains Val6. This recognition helix was reported in another selection study (22). As seen in Fig. 2aa, Val does not specify a single base at this position. The crossreactivity of proteins pGGC and pGAC (Fig. 2 i–l) is an artifact as shown by the lack of binding to subsites CGC (Fig. 2j, column 22) and TAC (Fig. 2l, column 22). Target oligonucleotides with a finger-2 subsite of CCC or TCC were found to create a perfect GGC or GAC subsite, respectively, on their complementary strand.
The results of the multitarget ELISA assay were confirmed by affinity studies of purified proteins (Table 1). In cases where crossreactivity was minimal in the ELISA assay, a single nucleotide mismatch typically resulted in a greater than 100-fold loss in affinity. This degree of specificity had yet to be demonstrated with zinc finger proteins. In general, proteins selected or designed to bind subsites with G or A in the middle and 3′ position had the highest affinity, followed by those that had only one G or A in the middle or 3′ position, followed by those that contained only T or C. The former group typically bound their targets with a higher affinity than Zif268 (10 nM), the latter with somewhat lower affinity, and almost all of the proteins had an affinity lower than that of the parental C7 protein. Proteins pGTC, pmGCT, and pGCC had the lowest affinities (40, 65, and 80 nM, respectively) and yet were among the most specific (Fig. 2 d, r, and e, respectively) suggesting that specificity can result not only from specific protein-DNA contacts, but also from interactions that exclude all but the correct nucleotide and common backbone interactions.
Position 2 and Target Site Overlap.
Asp2 always was coselected with Arg−1 in all proteins for which the target subsite was GNG. It is now understood that there are two reasons for this. From structural studies of Zif268 (4, 5), it is known that Asp2 of finger 2 makes a pair of buttressing hydrogen bonds with Arg−1 that stabilize the Arg−1/3′ guanine interaction, as well as some water-mediated contacts. However, the carboxylate of Asp2 also accepts a hydrogen bond from the N4 of a cytosine that is base paired to a 5′ guanine of the finger-1 subsite. Adenine base-paired to T in this position can make an analogous contact to that seen with cytosine. This interaction is particularly important because it extends the recognition subsite of finger 2 from three nucleotides (GNG) to four [GNG(G/T)] (15, 25, 26). This phenomenon is referred to as target site overlap and has three important ramifications. First, Asp2 was favored for selection by our library when the finger-2 subsite was GNG because our finger-1 subsite contained a 5′ guanine. Second, it may limit the utility of the libraries used in this study to selection on GNN or TNN finger-2 subsites because finger 3 of these libraries contains an Asp2, which may help specify the 5′ nucleotide of the finger-2 subsite to be G or T. In Zif268 and C7, which have Thr6 in finger 2, Asp2 of finger 3 enforces G or T recognition in the 5′ position (T/G)GG (Fig. 2bb). This interaction also may explain why previous phage display studies, which all used Zif268-based libraries, have found selection limited primarily to GNN recognition (21, 23–27). One of these studies stated that 5′G recognition is coded by Ser6 and Thr6 (34), yet all of the characterized finger 2 proteins here use Arg6 for exquisite 5′G recognition. Recognition of 5′G by Ser6 and Thr6 proteins is likely an artifact of target site overlap as seen in Zif268 and C7 and therefore is not a coded interaction.
Finally, target site overlap potentially limits the use of these zinc fingers as modular building blocks. From structural data it is known that there are some zinc fingers in which target site overlap is quite extensive, such as those in GLI (8) and YY1 (9), and others that are similar to Zif268 and display only modest overlap. In our final set of proteins, Asp2 is found in pmGGG, pmGAG, pmGTG, and pmGCG. The overlap potential of other residues found at position 2 is largely unknown; however, structural studies reveal that many other residues found at this position may participate in such cross-subsite contacts. Fingers containing Asp2 may limit modularity, because they would require that each GNG subsite be followed by a T or G.
CONCLUSIONS
We have demonstrated that many of the 16 possible GNN triplet sequences can be recognized with exquisite specificity by zinc finger domains. Optimized zinc finger domains can discriminate single base differences by greater than 100-fold loss in affinity. While many of the amino acids found in the optimized proteins at the key contact positions −1, 3, and 6 are those that are consistent with a simple code of recognition, we have discovered that optimal specific recognition is sensitive to the context in which these residues are presented. Residues at positions 1, 2, and 5 have been found to be critical for specific recognition. Further we demonstrate, that in contrast to the expectations of a simple recognition code, that sequence motifs at positions −1, 1, and 2 rather than the simple identity of the position 1 residue are required for highly specific recognition of the 3′ base. We believe these residues provide the proper stereochemical context for interactions of the helix both in terms of recognition of specific bases and in the exclusion of other bases, the net result being highly specific interactions. Thus our understanding of a recognition code is weak even when the recognition helix is constrained within the same zinc finger framework. We anticipate that attempts to apply a recognition code derived from the study of finger-2 variants of Zif268 will be limited as the effects of the zinc finger framework on helix presentation are not appreciated. One motivation for increasing our understanding of the recognition codes is to apply it to the many naturally occurring zinc finger proteins of unknown function. It is clear, however, that many more studies will be required to make this goal feasible.
Broad utility of the domains described here would be realized if they were modular in both their interactions with DNA and other zinc finger domains. This cooperativity could be achieved by working within the likely limitations imposed by target site overlap, namely that sequences of the 5′-(GNN)x-3′ type should be targeted. Indeed, we have now demonstrated the functional modularity of the zinc finger domains described here in the construction of polydactyl proteins that bind 18 bp of DNA with subnanomolar affinity (36). These polydactyl proteins have been used to activate and repress transcription driven by the human erbB-2 promoter in living cells. The family of zinc finger domains described here should be sufficient for the construction of 166 or 17 million novel proteins that bind the 5′-(GNN)6-3′ family of DNA sequences. Together, the materials and methods of these reports should allow for the rapid construction of novel gene switches and provide the basis for a universal system for gene control.
Acknowledgments
We thank Jayant Ghiara for his contributions and Jessica Saldana, Kris Bower, and Marikka Elia for their technical assistance. This study was supported in part by National Institutes of Health Grant GM 53910 to C.F.B. Postdoctoral fellowships were received by B.D. from the Deutsche Forschungsgemeinschaft, and by R.R.B. from the Swiss National Science Foundation and the Krebsliga beider Basel.
ABBREVIATION
- ZBA
zinc buffer A
References
- 1.Ptashne M. Nature (London) 1967;214:232–234. doi: 10.1038/214232a0. [DOI] [PubMed] [Google Scholar]
- 2.Miller J, McLachlan A D, Klug A. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee M S, Gippert G P, Soman K V, Case D A, Wright P E. Science. 1989;245:635–637. doi: 10.1126/science.2503871. [DOI] [PubMed] [Google Scholar]
- 4.Pavletich N P, Pabo C O. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 5.Elrod-Erickson M, Rould M A, Nekludova L, Pabo C O. Structure (London) 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- 6.Elrod-Erickson M, Benson T E, Pabo C O. Structure (London) 1998;6:451–464. doi: 10.1016/s0969-2126(98)00047-1. [DOI] [PubMed] [Google Scholar]
- 7.Kim C A, Berg J M. Nat Struct Biol. 1996;3:940–945. doi: 10.1038/nsb1196-940. [DOI] [PubMed] [Google Scholar]
- 8.Pavletich N P, Pabo C O. Science. 1993;261:1701–1707. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- 9.Houbaviy H B, Usheva A, Shenk T, Burley S K. Proc Natl Acad Sci USA. 1996;93:13577–13582. doi: 10.1073/pnas.93.24.13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fairall L, Schwabe J W R, Chapman L, Finch J T, Rhodes D. Nature (London) 1993;366:483–487. doi: 10.1038/366483a0. [DOI] [PubMed] [Google Scholar]
- 11.Wuttke D S, Foster M P, Case D A, Gottesfeld J M, Wright P E. J Mol Biol. 1997;273:183–206. doi: 10.1006/jmbi.1997.1291. [DOI] [PubMed] [Google Scholar]
- 12.Liu Q, Segal D J, Ghiara J B, Barbas III C F. Proc Natl Acad Sci USA. 1997;94:5525–5530. doi: 10.1073/pnas.94.11.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nolte R T, Conlin R M, Harrison S C, Brown R S. Proc Natl Acad Sci USA. 1998;95:2938–2943. doi: 10.1073/pnas.95.6.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narayan V A, Kriwacki R W, Caradonna J P. J Biol Chem. 1997;272:7801–7809. doi: 10.1074/jbc.272.12.7801. [DOI] [PubMed] [Google Scholar]
- 15.Isalan M, Choo Y, Klug A. Proc Natl Acad Sci USA. 1997;94:5617–5621. doi: 10.1073/pnas.94.11.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nardelli J, Gibson T J, Vesque C, Charnay P. Nature (London) 1991;349:175–178. doi: 10.1038/349175a0. [DOI] [PubMed] [Google Scholar]
- 17.Nardelli J, Gibson T, Charnay P. Nucleic Acids Res. 1992;20:4137–4144. doi: 10.1093/nar/20.16.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor W E, Suruki H K, Lin A H T, Naraghi-Arani P, Igarashi R Y, Younessian M, Katkus P, Vo N V. Biochemistry. 1995;34:3222–3230. doi: 10.1021/bi00010a011. [DOI] [PubMed] [Google Scholar]
- 19.Desjarlais J R, Berg J M. Proteins Struct Funct Genet. 1992;12:101–104. doi: 10.1002/prot.340120202. [DOI] [PubMed] [Google Scholar]
- 20.Desjarlais J R, Berg J M. Proc Natl Acad Sci USA. 1992;89:7345–7349. doi: 10.1073/pnas.89.16.7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choo Y, Klug A. Proc Natl Acad Sci USA. 1994;91:11163–11167. doi: 10.1073/pnas.91.23.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greisman H A, Pabo C O. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- 23.Rebar E J, Pabo C O. Science. 1994;263:671–673. doi: 10.1126/science.8303274. [DOI] [PubMed] [Google Scholar]
- 24.Jamieson A C, Kim S-H, Wells J A. Biochemistry. 1994;33:5689–5695. doi: 10.1021/bi00185a004. [DOI] [PubMed] [Google Scholar]
- 25.Jamieson A C, Wang H, Kim S-H. Proc Natl Acad Sci USA. 1996;93:12834–12839. doi: 10.1073/pnas.93.23.12834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isalan M, Klug A, Choo Y. Biochemistry. 1998;37:12026–12033. doi: 10.1021/bi981358z. [DOI] [PubMed] [Google Scholar]
- 27.Wu H, Yang W-P, Barbas III C F. Proc Natl Acad Sci USA. 1995;92:344–348. doi: 10.1073/pnas.92.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbas III C F, Kang A S, Lerner R A, Benkovic S J. Proc Natl Acad Sci USA. 1991;88:7978–7982. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barbas III C F, Lerner R A. Methods Companion Methods Enzymol. 1991;2:119–124. [Google Scholar]
- 30.Rader C, Barbas III C F. Curr Opin Biotechnol. 1997;8:503–508. doi: 10.1016/s0958-1669(97)80075-4. [DOI] [PubMed] [Google Scholar]
- 31.Swirnoff A H, Milbrandt J. Mol Cell Biol. 1995;15:2275–2287. doi: 10.1128/mcb.15.4.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crameri A, Cwirla S, Stemmer W P. Nat Med. 1996;2:100–102. doi: 10.1038/nm0196-100. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki M, Gerstein M, Yagi N. Nucleic Acids Res. 1994;22:3397–3405. doi: 10.1093/nar/22.16.3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choo Y, Klug A. Proc Natl Acad Sci USA. 1994;91:11168–11172. doi: 10.1073/pnas.91.23.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choo Y, Klug A. Curr Opin Struct Biol. 1997;7:117–125. doi: 10.1016/s0959-440x(97)80015-2. [DOI] [PubMed] [Google Scholar]
- 36.Beerli R R, Segal D J, Dreier B, Barbas C F., III Proc Natl Acad Sci USA. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]