Abstract
The histone variant H2A.Z is evolutionarily conserved and plays an essential role in mice, Drosophila, and Tetrahymena. The essential function of H2A.Z is unknown, with some studies suggesting a role in transcriptional repression and others in activation. Here we show that Caenorhabditis elegans HTZ-1/H2A.Z and the remodeling complex MYS-1/ESA1–SSL-1/SWR1 synergize with the FoxA transcription factor PHA-4 to coordinate temporal gene expression during foregut development. We observe dramatic genetic interactions between pha-4 and htz-1, mys-1, and ssl-1. A survey of transcription factors reveals that this interaction is specific, and thus pha-4 is acutely sensitive to reductions in these three proteins. Using a nuclear spot assay to visualize HTZ-1 in living embryos as organogenesis proceeds, we show that HTZ-1 is recruited to foregut promoters at the time of transcriptional onset, and this recruitment requires PHA-4. Loss of htz-1 by RNAi is lethal and leads to delayed expression of a subset of foregut genes. Thus, the effects of PHA-4 on temporal regulation can be explained in part by recruitment of HTZ-1 to target promoters. We suggest PHA-4 and HTZ-1 coordinate temporal gene expression by modulating the chromatin environment.
Synopsis
During development, a single fertilized egg gives rise to the different cell types within an embryo. These different cell types are characterized by the different genes that they express. A critical question in biology is how embryonic cells activate genes at the appropriate time and place to generate the different cell types. In this paper, the authors explore temporal regulation of gene expression during formation of an organ, namely the Caenorhabditis elegans foregut. They have discovered that foregut genes require a variant of the canonical H2A histone for the onset of transcription. This variant, called H2A.Z, or htz-1 in C. elegans, promotes transcription by modifying how DNA is packaged within cells. Their data suggest that a key regulator of foregut development, the transcription factor PHA-4, recruits HTZ-1 to pharyngeal promoters, and this association contributes to their timely activation.
Introduction
In eukaryotes, DNA and histones are assembled into nucleosomes, which present a daunting barrier to the transcriptional apparatus. The transcriptional machinery uses several approaches to augment or disrupt the repressive activity of chromatin, one of which is the exchange of canonical histones for histone variants. There are multiple variants of the core histone H2A, including MacroH2A, which is involved in mammalian X inactivation, H2A.X for DNA repair, and H2A.Z, whose function is linked to transcription [1]. Early studies with H2A.Z in Tetrahymena demonstrated that it was associated with the transcriptionally active macronucleus [2,3]. In yeast, H2A.Z has also been implicated in gene activation. H2A.Z antagonizes telomeric silencing and synergizes with the SWI/SNF remodeling complex to activate a subset of yeast genes [4]. In yeast, global chromatin binding studies have revealed that H2A.Z preferentially occupies promoter regions [5–8]. The function of this occupancy is currently being debated as some studies have found no correlation between H2A.Z occupancy and transcription rate [8], whereas others have suggested a strong inverse correlation between the presence of H2A.Z and the rate of transcription [5–7]. Despite these differences, the general consensus is that yeast H2A.Z configures chromatin to poise genes for transcriptional activation.
In metazoans, H2A.Z has been implicated in transcriptional repression more than activation. H2A.Z behaves genetically like a Polycomb group gene in Drosophila and is required for the stable association of Polycomb with polytene chromosomes [9]. In both mice and flies, H2A.Z localizes to heterochromatin, as well as euchromatin, and can associate with the heterochromatin protein HP1 [10,11]. One possibility is that H2A.Z enrichment in heterochromatin might prevent this domain from spreading. For example, H2A.Z is enriched in the 5′ insulator of the chicken β-globin locus that flanks a heterochromatic region [12]. However, this explanation is hard to resolve with a Polycomb-like function in Drosophila. These alternative roles may explain why vertebrate H2A.Z has diverged from yeast H2A.Z (61% similarity [7]) despite the general conservation of this histone variant in all eukaryotes [13]. Thus, the role of H2A.Z in metazoans remains unclear.
H2A.Z has not been studied previously in Caenorhabditis elegans, but the complex that assembles H2A.Z into chromatin in other organisms, the Esa1-Swr1 complex in yeast or the human SRCAP complex [13], was identified in C. elegans in a screen for regulators of vulval development [14]. Inactivation of Esa1–Swr1 homologs in C. elegans enhance mutations in known repressor complexes such as NuRD and Rb to de-repress the lag-2/Delta ligand during vulval development [14,15]. These observations raise the question of whether yeast and metazoans have evolved divergent functions for H2A.Z. One difficulty addressing this question is the paucity of direct H2A.Z target genes in metazoans, and the lack of knowledge of how and when H2A.Z is recruited to specific regions of DNA. Here we address these issues by examining the role of H2A.Z during C. elegans foregut (pharynx) development and the interplay of H2A.Z with the pharynx selector gene pha-4.
PHA-4 belongs to the FoxA family of transcription factors, which are critical to form the digestive tract during embryogenesis and also regulate metabolism during post-embryonic life [16,17]. In C. elegans, pha-4 is required to specify cells of the pharynx during the earliest stages of organogenesis, and is also important later for organ differentiation, morphogenesis, and function [18,19]. This global role reflects the broad range of PHA-4 target genes. PHA-4 directly activates foregut genes expressed both early and late during foregut development and is required during post-embryonic life as well [18,20–22]. Thus, a key question is how diverse transcriptional responses are orchestrated by a single transcription factor. Previous studies have shown that the affinity of PHA-4 for its DNA binding site is one input for temporal regulation: promoters with high-affinity sites are competent to be expressed early whereas those with low-affinity sites are typically activated at later times [18,19]. A second strategy is combinatorial regulation of foregut target promoters. In C. elegans, association of PHA-4 with a target is rarely sufficient for expression, and additional factors contribute towards the precise temporal or cell type activation of pharyngeal genes [19,20]. Similarly, in vertebrates, expression in the liver critically depends on FoxA proteins that function in combination with additional liver transcription factors [17]. Here we explore a third input involved in the timely activation of pharyngeal genes: co-factors that are recruited to pharyngeal promoters.
FoxA proteins are thought to modify the chromatin environment of their target genes [17]. Gualdi et al. proposed that FoxA proteins function as competence factors that bind to promoters before they are transcriptionally active [23]. This association is thought to decompact nucleosomal DNA, which allows additional transcription factors to find their binding sites, leading to transcriptional activation [24]. The mechanism by which FoxA proteins modulate chromatin is still murky. Studies in vitro have largely focused on how FoxA itself can interact with nucleosomal DNA, and have not addressed the contribution of additional components. Here we explore the role of the histone variant HTZ-1/H2A.Z for pharyngeal gene activation, recruitment by PHA-4, and timing of expression.
Results
Requirement of ssl-1, mys-1, and htz-1 for Pharyngeal Development
To find genes involved in pharynx development, we had previously created a temperature-sensitive configuration of pha-4 that would allow us to examine suppressors and enhancers of compromised pha-4 [25]. The pha-4(ts) strain contains the pha-4(zu225) allele, which carries a premature stop codon and renders pha-4 mRNA subject to degradation by the nonsense-mediated decay pathway [25–27]. We took advantage of a temperature-sensitive allele of the nonsense-mediated decay pathway component, smg-1(cc546ts) [28], to modulate the accumulation of truncated PHA-4 by varying the temperature. At 15 °C, SMG-1 is active and mRNA derived from pha-4(zu225) is degraded, which results in Pha-4 lethality at the first larval stage (L1). At the permissive temperature of 24 °C, SMG-1 is compromised and pha-4 mRNA is stabilized, which allows truncated PHA-4 to accumulate and worms to survive. At 20 °C, an intermediate level of truncated PHA-4 accumulates, producing a less severe pharyngeal phenotype that is nonetheless lethal. The pha-4(ts) strain provided a sensitive means to examine genetic interactions between pha-4 and other loci, in particular the ability to test for enhanced lethality at permissive temperature or suppressed lethality at intermediate or restrictive temperatures.
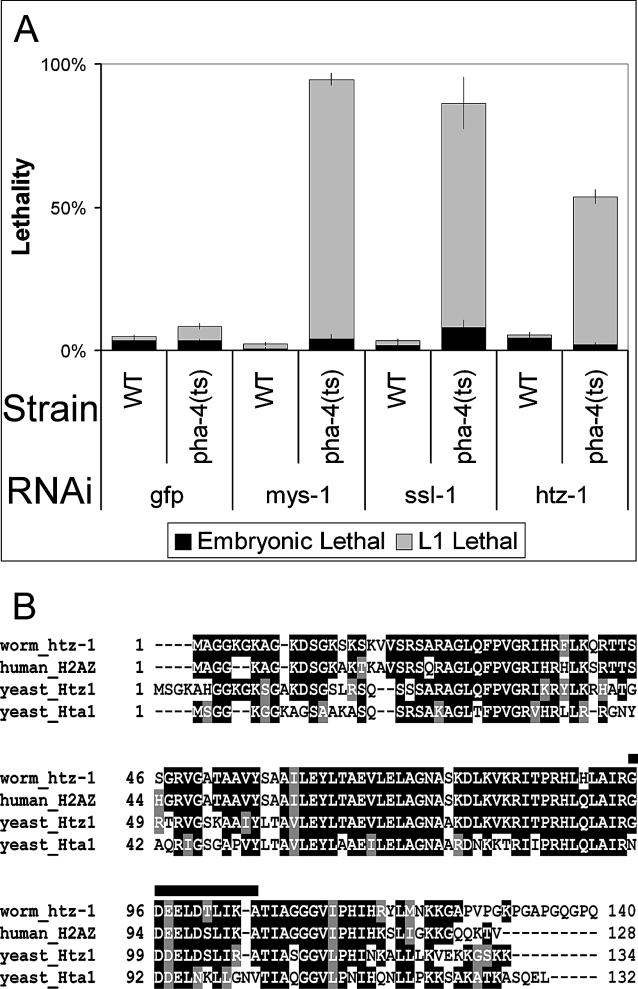
To investigate whether C. elegans homologs of the Esa1-Swr1 complex (known as mys-1 and ssl-1 [14]), were involved in pharynx development, we inactivated each of these genes using RNAi either alone or in combination with pha-4(ts). Disrupting these genes by feeding worms bacteria expressing dsRNA [29] had no effect on viability (Figure 1A), and most animals developed into sterile adults, as had been observed previously [14,30,31]. Likewise, pha-4(ts) with control GFP(RNAi) was largely viable at permissive temperature (24 °C) (Figure 1A). However, when pha-4(ts) was combined with either mys-1(RNAi) or ssl-1(RNAi), we observed a dramatic enhancement of lethality at L1 (Figure 1A). htz-1(RNAi) alone had no effect on viability [30], whereas we observed enhanced L1 lethality for pha-4(ts); htz-1(RNAi) animals (Figure 1A and 1B, and Figure S1), consistent with the idea that SSL-1/Swr1 likely interacts with HTZ-1 in C. elegans, as it does in other organisms [32,33]. Importantly, htz-1 has little sequence identity at the nucleotide level with H2A genes that could lead to “off-target” effects [34]. Moreover, we observed enhancement of pha-4(ts) lethality with double-stranded RNA (dsRNA) for the 3′UTR of htz-1, which lacks sequence identity with H2A genes (unpublished data). These data reveal that loss of htz-1 is synthetically lethal with pha-4(ts).
Figure 1. Enhancement of pha-4(ts) by mys-1, ssl-1, and htz-1 .
(A) Feeding dsRNA to wild-type (WT) or pha-4(ts) [18,25] worms at the permissive temperature of 24 °C. WT worms generate viable progeny with mys-1, ssl-1 or htz-1 RNAi. In the pha-4(ts) background, L1 arrest increased with mys-1, ssl-1, or htz-1 RNAi compared to control GFP(RNAi) (grey bars). Embryonic lethality remained unchanged (black bars). Effectiveness of RNAi feeding was manifest through viable, but sterile, progeny for mys-1 and ssl-1 [14], as well as repeated enhancement of L1 lethality for pha-4(ts), performed in parallel. n = 100 worms/plate, three plates per column. Error bars indicate the standard deviation.
(B) Alignment of C. elegans htz-1 (R08C7.3) with human H2A.Z, yeast Htz1, and one of the core H2A genes from yeast, Hta1. Extended acid patch region essential for H2A.Z function is indicated by the bar [50,60,67].
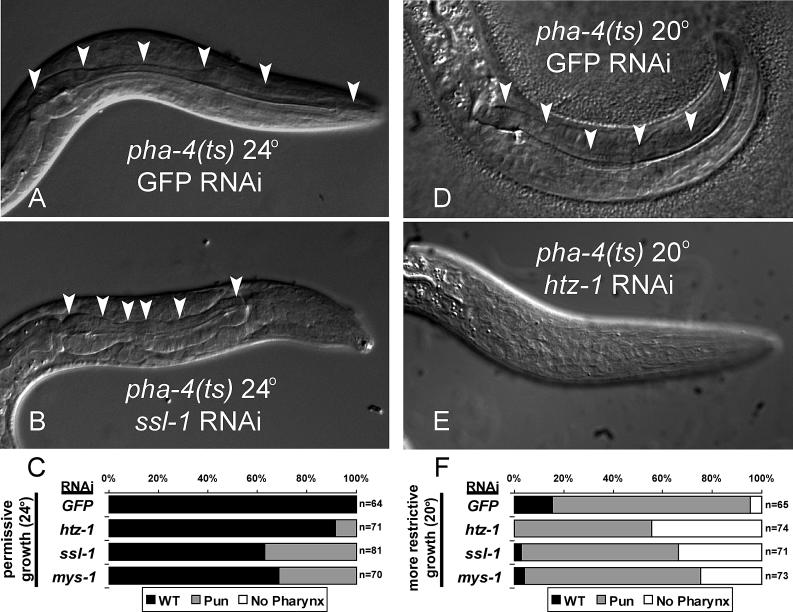
pha-4(ts); mys-1(RNAi), pha-4(ts); ssl-1(RNAi), and pha-4(ts); htz-1(RNAi) larvae raised at both the permissive temperature (24 °C) and the intermediate temperature (20 °C) exhibited pronounced defects in pharynx development. At permissive temperature, RNAi against either mys-1, ssl-1, or htz-1 generated Pun (pharynx unattached) pha-4(ts) larvae not observed with the pha-4(ts); GFP(RNAi) control (Figure 2A–2C). At intermediate temperature, RNAi against mys-1, ssl-1, or htz-1 increased the percentage of L1 larvae lacking any detectable pharynx (Figure 2D–2F). For example, 44.6% of pha-4(ts); htz-1(RNAi) animals had no detectable pharynx (n = 74, Figure 2E and 2F), similar to pha-4 null alleles [35]. By comparison, only 4.6% of pha-4(ts); GFP(RNAi) control larvae lacked an obvious pharynx (n = 65, Figure 2D and 2F). Other tissues appeared normal in pha-4(ts); mys-1(RNAi), pha-4(ts); ssl-1(RNAi), or pha-4(ts); htz-1(RNAi) larvae, as viewed under the light microscope. These data indicate that enhancement of pha-4 lethality by mys-1, ssl-1, or htz-1 reflects, at least in part, a defect in pharyngeal development.
Figure 2. mys-1, ssl-1, and htz-1 Enhance Pharyngeal Defects of pha-4(ts) .
(A–C) Feeding dsRNA to pha-4(ts) worms at the permissive temperature of 24 °C.
(A) A pha-4(ts); GFP(RNAi) L1 with a wild-type pharynx (arrowheads).
(B) A pha-4(ts); ssl-1(RNAi) L1 with an unattached pharynx (arrowheads).
(C) Quantitation of pha-4(ts) animals exhibiting a normal pharynx (WT), an unattached or incomplete pharynx (Pun), or no detectable pharynx. htz-1, ssl-1, or mys-1 RNAi significantly increased the number of Pun pha-4(ts) animals (htz-1: p=0.0290; ssl-1 and mys-1: p < 0.0001, Fisher exact test).
(D–F) Feeding dsRNA to pha-4(ts) animals at the intermediate temperature of 20 °C.
(D) A pha-4(ts); GFP(RNAi) L1 at the intermediate temperature of 20 °C with a morphologically wild-type pharynx (arrowheads).
(E) A pha-4(ts); htz-1(RNAi) worm at 20 °C missing a detectable pharynx.
(F) Quantitation of pha-4(ts) animals exhibiting a normal pharynx (WT), an unattached or incomplete pharynx (Pun), or no detectable pharynx. htz-1, ssl-1, or mys-1 RNAi significantly increased the number of worms with no detectable pharynx (htz-1 and ssl-1: p < 0.0001; mys-1: p = 0.0015; Fisher exact test). WT worms with reduced htz-1, ssl-1, or mys-1 activity had a wild-type pharynx at 24 °C and 20 °C (unpublished data). RNAi was conducted by feeding dsRNA [29].
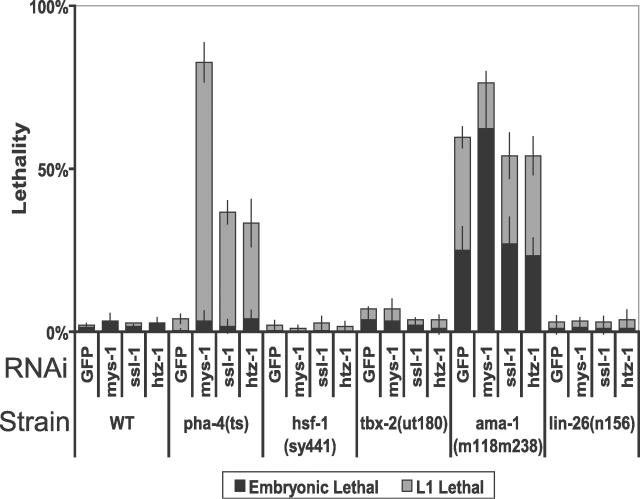
The synergy of ssl-1, mys-1, and htz-1 with pha-4 was specific since we did not observe enhancement by ssl-1, mys-1, or htz-1 with strains carrying other compromised transcription factors (Figure 3). First we examined the heat-shock factor hsf-1, since H2A.Z binds some heat-shock promoters in Drosophila and yeast [5,10]. Normally, hsf-1(sy441) worms exhibit a temperature-sensitive developmental arrest at 25 °C and are viable at 15 °C [36]. We observed a low level of L1 lethality for hsf-1(sy441); GFP(RNAi) at the intermediate temperature of 20 °C. However, lethality was not enhanced by ssl-1, mys-1, or htz-1 RNAi with hsf-1(sy441). Null alleles of tbx-2 and lin-26 are each lethal [37,38], whereas tbx-2(ut180) or lin-26(n156) mutants survive with neuronal or vulval defects, respectively [39]. We did not detect increased lethality when ssl-1, mys-1, or htz-1 was inactivated in combination with tbx-2(ut180) or lin-26(n156). Unlike pha-4(ts), percent lethality for these mutants remained similar to the level observed with GFP(RNAi). Finally, we examined ama-1, which encodes the large subunit of C. elegans RNA polymerase II [40]. ama-1(m118m238) worms were sick either as single mutants or in combination with control GFP(RNAi), and segregated many embryonic and L1-arrested animals (Figure 3). Strikingly, neither ssl-1 nor htz-1 RNAi enhanced the lethality associated with ama-1(m118m238). However, embryonic lethality was significantly increased with mys-1(RNAi), suggesting that mys-1 may have additional roles that are independent of ssl-1 and htz-1. Thus, strains exhibiting comparable baseline lethality to pha-4(ts) (i.e., hsf-1, tbx-2, and lin-26) or significantly higher baseline lethality (ama-1) did not exhibit enhanced lethality with either htz-1 or ssl-1. We conclude that pha-4 is exceptionally sensitive to reduction of mys-1, ssl-1, or htz-1.
Figure 3. Specificity of mys-1, ssl-1, and htz-1 Synergy with pha-4(ts) .
The indicated worm strains were fed dsRNA for GFP (negative control) or mys-1, ssl-1, or htz-1 at 20 °C (or 24 °C for WT and pha-4(ts)). Lethal embryos (black bars) or lethal L1 progeny (grey bars) were scored for each strain. Effectiveness of RNAi feeding was manifest through viable, but sterile, progeny for mys-1 and ssl-1 [14], as well as repeated enhancement of L1 lethality for pha-4(ts). n =1 00 worms/plate, three plates per column. Error bars indicate the standard deviation.
As a second test for specificity, we examined a temperature-sensitive configuration of unc-54, which mimicked the pha-4(ts) strain. Like pha-4(ts), RNA derived from unc-54(r293) is subject to degradation by the nonsense-mediated decay pathway [27,28]. Combining smg-1(cc546ts) with unc-54 leads to stabilization of unc-54 mRNA at 24 °C and degradation at 15 °C [25,26]. Inactivation of ssl-1, mys-1, or htz-1 with unc-54(ts) did not enhance the Unc-54 phenotype at permissive temperature or suppress unc-54 at intermediate temperature (Figure S2). These results demonstrate that enhancement of pha-4(ts) by ssl-1, mys-1, and htz-1 does not reflect indirect effects on the nonsense-mediated decay pathway. These data implicate roles for chromatin remodeling and histone exchange in the positive regulation of pharyngeal development.
Association of HTZ-1/H2A.Z with Pharyngeal Promoters
We wondered what was the basis for the synthetic interaction between htz-1 and pha-4. One intriguing possibility was that HTZ-1 might be recruited to pharyngeal promoters, which depend on PHA-4 for activation [18]. To address this idea, we used a nuclear spot assay [41,42] to visualize HTZ-1 association with target promoters in living embryos. We chose the nuclear spot assay because it allowed us to examine living embryos at precise times in development. We tagged the amino terminus of HTZ-1 with YFP and placed this chimera under control of the htz-1 promoter. We also created a LacI::CFP construct [43] driven by the htz-1 promoter. These two constructs were used to create transgenic worms that expressed both YFP::HTZ-1 and LacI::CFP from an extrachromosomal array. Because there were no mutations available, we could not assess whether our YFP::HTZ-1 reporter had rescuing activity. However, we believe the construct is functional since (1) we placed YFP at the equivalent location to a functional HA::Htz1 reporter in yeast [44], (2) the YFP::HTZ-1 localized with chromosomes in mitotic cells (Figure S3), and (3) YFP::HTZ-1 was distributed non-homogenously in interphase nuclei, similar to reports for endogenous H2A.Z in other organisms [10,11].
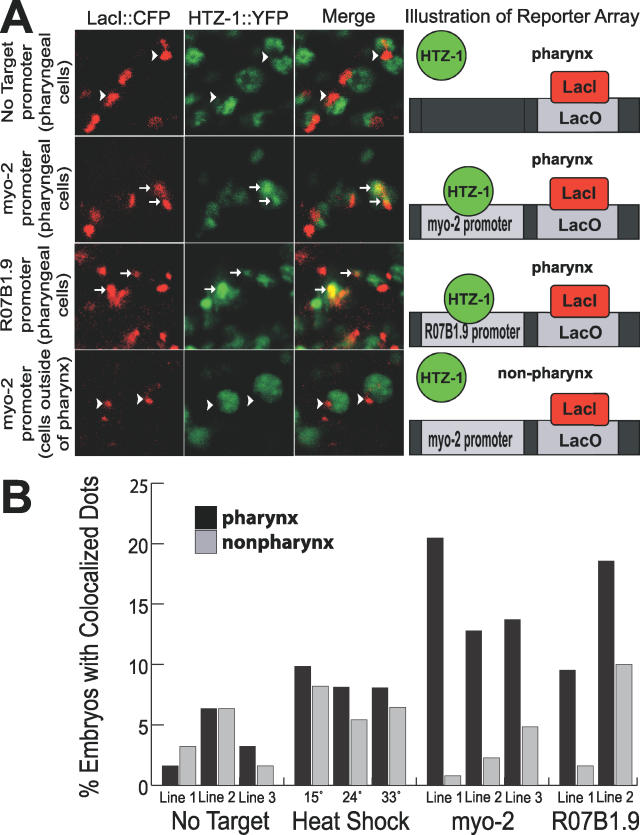
To observe association, we introduced a second extrachromosomal array (the reporter array) into the strain bearing YFP::HTZ-1 and LacI::CFP. The reporter array contained multiple copies of LacO operator sequences and a target promoter of interest. Three independent lines were generated in which the reporter array contained no target promoter. One line was generated with a reporter array containing the divergent heat-shock promoter for the hsp-16.1 and hsp-16.48 genes [45]. Three lines were generated in which the reporter array contained the promoter sequences for pharyngeally expressed myo-2 [46], and two lines were created with a reporter array containing the pharyngeal promoter for R07B1.9 [19]. Both myo-2 and R07B1.9 are PHA-4 target genes [18,19,21]. A single 0.7-μ confocal section was taken blindly through the center of an embryo (and pharynx) of an equal number of comma, 1.5-fold, 2-fold, and 3-fold stage embryos (see [47] for embryonic stages). This section typically bisected the nucleus of about 20 pharyngeal cells. Binding of LacI::CFP to LacO enabled us to localize the position of the extrachromosomal reporter array within the nucleus. Association of YFP::HTZ-1 with the reporter array was visualized as a bright dot co-localized with LacI::CFP. We scored the proportion of embryos with co-localized dots. To quantify our data, we counted the number of embryos with dots, rather than individual cells, because we could not always distinguish between individual cells when cells were packed closely together.
We observed YFP::HTZ-1 associated with pharyngeal promoters in developing embryos. In our negative control, YFP::HTZ-1 rarely bound to reporter arrays when no promoter was present (Figure 4A, row 1, and 4B). However, YFP::HTZ-1 localized to reporter arrays carrying promoter sequences for either myo-2 or R07B1.9, both of which are selectively expressed in the pharynx [18,46] (Figure 4A, rows 2 and 3, and 4B). This association was significantly higher in the three myo-2 promoter lines (p < 0.0001) and two R07B1.9 lines (p = 0.0014) compared to the no-target promoter controls. Strikingly, the no-target reporter arrays typically excluded YFP::HTZ-1 (Figure 4A, row 1), in contrast to the enrichment seen for reporter arrays with pharyngeal promoters. These data reveal that YFP::HTZ-1 associates with pharyngeal promoters in vivo.
Figure 4. Association of YFP::HTZ-1 with Pharyngeal Promoters.
(A) Extrachromosomal target arrays were visualized by LacI::CFP (red) bound to the Lac operator (LacO). YFP::HTZ-1 (green) was excluded (arrowheads) from arrays with no-target promoter (row 1), but associated with target arrays containing promoters for pharyngeal genes myo-2 or R07B1.9 in pharyngeal cells (rows 2 and 3). Merge is yellow. YFP::HTZ-1 was excluded from arrays containing the myo-2 promoter in non-pharyngeal cells (row 4). Cartoons depict interpretation of data.
(B) Percentage of embryos containing one or more co-localized LacI::CFP and YFP::HTZ-1 dots in the pharynx (black) or outside of the pharynx (grey). Association was significantly higher in the three myo-2 promoter lines (p < 0.0001) and two R07B1.9 lines (p = 0.0014). No significant difference in association was found when comparing the no-target lines to the heat-shock promoter target at 15 °C, 24 °C, or heat-shock at 33 °C for 30 min (p > 0.095). More than 60 embryos were scored for each no-target, heat-shock, and R07B1.9 line. More than 120 embryos were scored for each myo-2 line. YFP::HTZ-1 association was predominantly pharyngeal for myo-2 (p < 0.0001) and R07B1.9 (p = 0.0407) target arrays, but not control arrays (p = 1.00). An equivalent number of images were taken at the 1.5-, 2-, and 3-fold stages of embryogenesis for each line. The p-values were calculated using Fisher exact test.
H2A.Z has been implicated in both transcriptional activation and repression, depending on the organism and the assay. Therefore, we wanted to determine whether HTZ-1 associated with active or repressed promoters in C. elegans. First, we compared association with pharyngeal promoters within pharyngeal versus non-pharyngeal cells. We observed a dramatic enrichment of embryos with spots within pharyngeal cells, despite the larger number of non-pharyngeal nuclei present in each embryo. (We estimate a maximum of 5–10 pharyngeal muscle nuclei versus 50–100 non-pharyngeal nuclei per optical section, Figure 4A, row 4, and 4B). YFP::HTZ-1 association was significantly pharyngeal for myo-2 (p < 0.0001) and R07B1.9 (p = 0.0407) (Figure 4B). These data indicate that pharyngeal promoters preferentially associate with YFP::HTZ-1 within pharyngeal cells. Thus, we observed increased YFP::HTZ-1 association in cells that express myo-2 and R07B1.9, and rare association in cells in which these genes are permanently silent. These data suggest HTZ-1/H2A.Z is not involved in constitutive repression of pharyngeal genes.
By comparison, the no-target reporter array lines had a low level of YFP::HTZ-1 spots, and these were evenly distributed in both pharyngeal and non-pharyngeal cells (Figure 4B; p = 1.0000, three lines). We also observed an even distribution of pharyngeal and non-pharyngeal spots when we examined a line containing the divergent heat-shock promoters in the reporter array [48]. There was no significant increase in spots for the heat-shock promoter array compared to the no-target control, and YFP::HTZ-1 association did not change at different temperatures (15 °C, 24 °C, or after heat shock at 33 °C for 30 min; p > 0.095). Thus, not all promoters are enriched for HTZ-1, and pharyngeal cells do not promote non-specific association of HTZ-1 with reporter arrays.
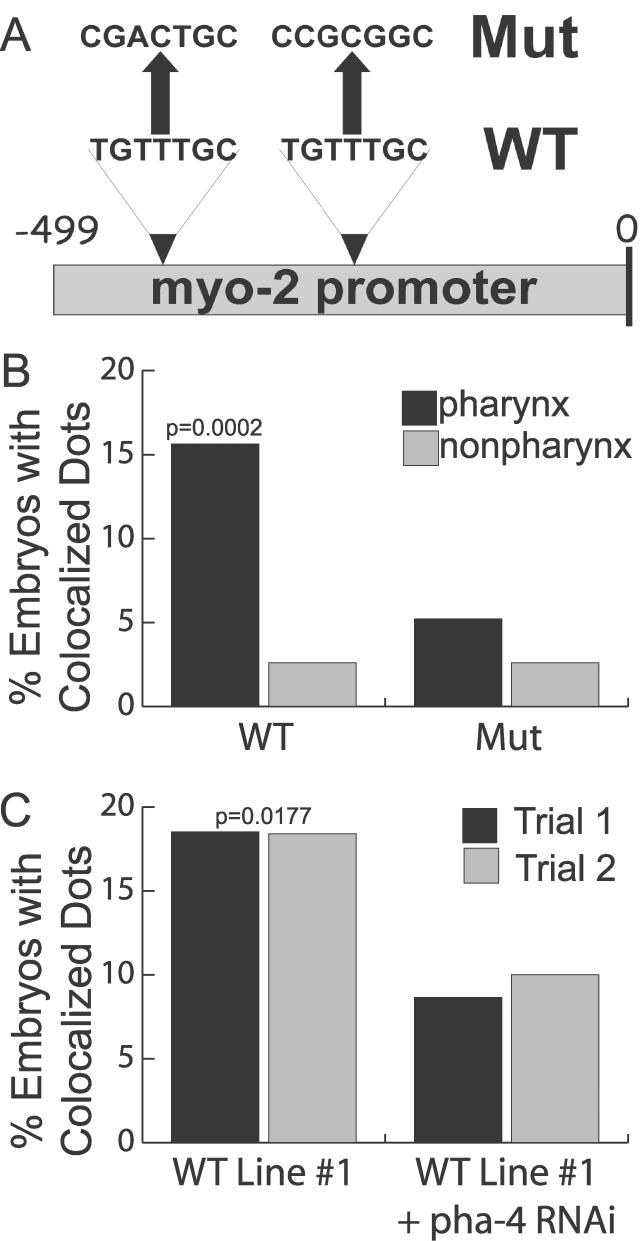
Our second test to explore roles in activation versus repression was to examine whether PHA-4 was required for YFP::HTZ-1 recruitment to pharyngeal promoters. We created three independent lines with reporter arrays containing a myo-2 promoter with mutated PHA-4 binding sites (Figure 5A) [18]. Each of these lines showed a significant decrease in the percent of embryos with YFP::HTZ-1 occupancy on the reporter array in the pharynx (Figure 5B). This level resembled the degree of association observed in non-pharyngeal cells or lines bearing no-target reporter arrays (Figure 4B versus Figure 5B). Similarly, inactivation of pha-4 by RNAi decreased the occupancy of YFP::HTZ-1 on myo-2 sequences by one half (Figure 5C). These results revealed a requirement for pha-4 for robust association of YFP::HTZ-1 with the myo-2 promoter. The dependency on pha-4 for YFP::HTZ-1 association supports the genetic synergy between pha-4 and htz-1 to promote pharyngeal development.
Figure 5. Association of YFP::HTZ-1 Depends on pha-4 Activity.
(A) Two PHA-4 binding sites within the myo-2 promoter were mutated in the myo-2 Mut construct [18].
(B) YFP::HTZ-1 association in the pharynx (black) and outside of the pharynx (grey). n = 384 embryos for three wild-type (WT) lines and 192 embryos for three mutant (Mut) lines. p-value indicates a significant difference for pharyngeal association of YFP::HTZ-1 in WT versus Mut.
(C) Association of YFP::HTZ-1 with the wild-type myo-2 promoter (line #1) decreased in pha-4(RNAi) embryos within pharyngeal cells. An equivalent number of images were taken at the 1.5-, 2-, and 3-fold stages of embryogenesis and >80 embryos were imaged for each trial. p-values calculated using Fisher's exact test.
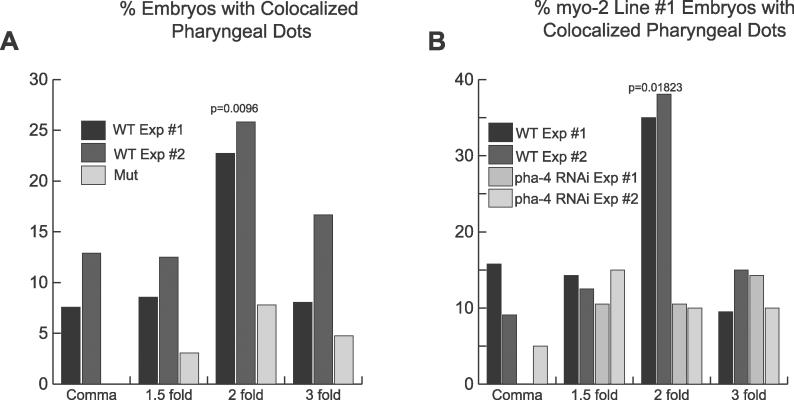
Our third test to examine roles in activation versus repression was to determine when YFP::HTZ-1 associated with pharyngeal promoters. We examined embryos from the three myo-2 target lines spanning 4 h of embryogenesis (comma, 1.5-, 2-, or 3-fold stages). At the 2-fold stage, the association of YFP::HTZ-1 nearly tripled above what was observed at the comma and 1.5-fold stages (Figure 6A and 6B). This peak required pha-4 activity since it was lost when pha-4 was inactivated by RNAi (Figure 6B) or when PHA-4 binding sites were mutated within the myo-2 promoter (Figure 6A). YFP::HTZ-1 association with myo-2 decreased to background levels after the gene was fully active at the 3-fold stage. Thus, YFP::HTZ-1 was enriched at the myo-2 promoter at the developmental stage when myo-2 transcription initiates [49].
Figure 6. YFP::HTZ-1 Association Peaks at the Onset of myo-2 Expression.
(A) Association of YFP::HTZ-1 with the myo-2 promoter in the pharynx during the comma, 1.5-, 2-, and 3-fold stages of development (two trials: black, dark grey). YFP::HTZ-1 association in the pharynx does not peak at the 2-fold stage for a myo-2 promoter with both PHA-4 binding sites mutated (Mut, light grey). n > 60 embryos imaged at each stage, for each experiment. Multiple lines were used for each trial.
(B) Pharyngeal YFP::HTZ-1 association with myo-2 target array #1 decreased at the 2-fold stage when pha-4 activity was reduced by RNAi. n > 20 embryos imaged at each stage for each of the two wild-type (WT) and two pha-4(RNAi) experiments.
The p-values for (A) and (B) indicate the significance of the WT peak at the 2-fold stage as calculated by a repeated measures analysis of variance (ANOVA).
HTZ-1 Influences the Onset of Pharyngeal Gene Expression
PHA-4 influences the timing of expression of genes within the pharynx, [18,19], raising the question of whether htz-1 might also contribute to temporal regulation. We removed htz-1 mRNA by microinjection of htz-1 dsRNA and examined the onset of R07B1.9::GFP and myo-2::GFP expression (Figure 7A–7C). Microinjection was used over RNAi feeding since microinjection typically gives the strongest RNAi phenotype. For example, microinjection of dsRNA for ssl-1 causes embryonic arrest, whereas feeding worms bacteria expressing dsRNA generates viable, sterile animals(Figure S4) [14].
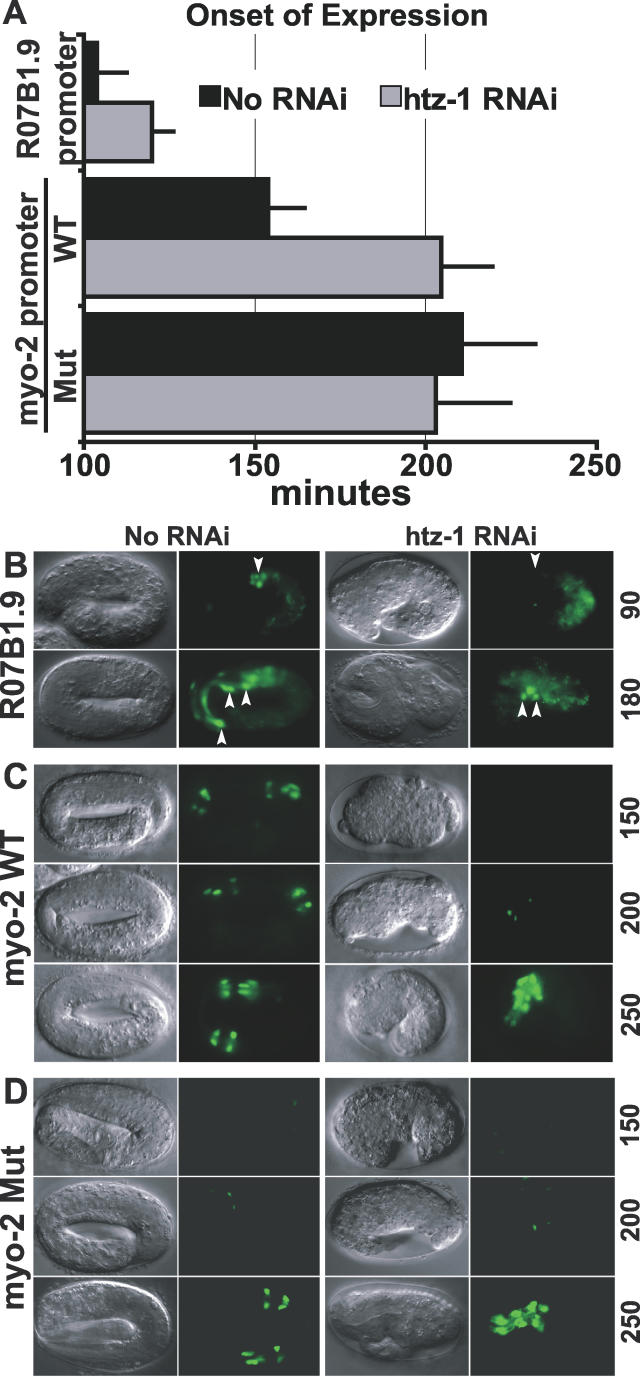
Figure 7. htz-1 Influences the Onset of Late Pharyngeal Gene Expression.
(A) Onset of R07B1.9::GFP and myo-2::GFP after the comma stage for wild-type (black) or htz-1(RNAi) (grey) embryos. Activation of the R07B1.9 reporter was delayed after injection of htz-1 dsRNA (p < 0.0001, Student t-test). No RNAi: n = 18, htz-1 RNAi: n = 12 embryos. myo-2 activation was scored in embryos carrying either a wild-type myo-2::GFP reporter (WT) or a myo-2::GFP reporter with both PHA-4 binding sites mutated (Mut) [18]. Activation of the WT reporter was delayed after injection of htz-1 dsRNA (p < 0.0001, Student t-test), whereas onset of the Mut reporter was unchanged (p = 0.2068). No RNAi: n = 15 WT, n = 33 Mut Embryos; htz-1 RNAi: n = 9 WT, n = 21 Mut embryos. Error bars indicate the standard deviation.
(B) Expression of R07B1.9::GFP at 90 and 180 min after the comma stage with htz-1 RNAi or without (No RNAi). Pharyngeal R07B1.9::GFP is indicated by arrowheads.
(C) Expression of myo-2::GFP (WT) at 150, 200, and 250 min after the comma stage with htz-1 RNAi or without (No RNAi).
(D) Expression of myo-2::GFP lacking PHA-4 binding sites [18] at 150, 200, 250 min after the comma stage with htz-1 RNAi or without (No RNAi).
Microinjection of htz-1(RNAi) caused highly penetrant, late-embryonic arrest (Figure 7B–7D), which phenocopied embryonic arrest from ssl-1(RNAi) microinjection (Figure S4). We focused on events after the comma stage of embryogenesis because htz-1(RNAi) embryos appeared to develop identically to the wild type up to the comma stage, based on morphological criteria (Figure S5). After the 2-fold stage, embryos failed to elongate (Figures 7B–7D). In terminal embryos, the pharynx and intestine appeared differentiated, but misshapen morphologically (Figure S4).
R07B1.9::GFP was first activated 104 min after the comma stage in the wild type, whereas activation was delayed to 120 min in htz-1(RNAi) embryos (Figure 7A and 7B). Likewise, myo-2::GFP activation was delayed from 154 min after the comma stage in the wild type to 204 min in htz-1(RNAi) embryos (Figure 7A and 7C). The delay of myo-2::GFP expression with htz-1(RNAi) was similar to that seen when PHA-4 binding sites were mutated within the myo-2 promoter (Figure 7A and 7D) [18]. htz-1 was not essential to repress R07B1.9 or myo-2 expression at early stages of pharyngeal development (Figure 7B, 90 min, and 7C, 150 min), nor was it important to achieve the ultimate strong expression during the terminal stages of development (Figure 7B, 180 min, and 7C, 250 min). These data suggest that htz-1 is critical for the timely activation of R07B1.9 and myo-2. We note that these findings do not rule out a role for H2A.Z in transcriptional repression or synergy with other transcription factors in other contexts.
We observed no difference in GFP expression between wild-type and htz-1(RNAi) embryos for a myo-2::GFP reporter bearing mutated PHA-4 binding sites (Figure 7A and 7D, 200 min). This result indicates that the delay in myo-2 activation after htz-1(RNAi) does not reflect non-specific effects of htz-1(RNAi) on embryonic development. In summary, we can account for the effects of PHA-4 on timing of myo-2 activation [18] by its ability to recruit HTZ-1.
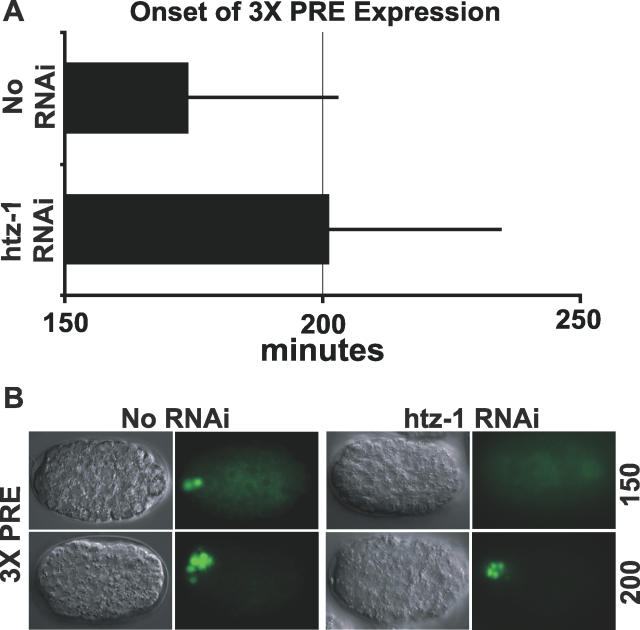
We also examined the effects of htz-1 on a GFP reporter driven by three copies of a high-affinity PHA-4 binding site (TGTTTGC) upstream of a minimal Δpes-10 promoter (3xPRE::GFP) [19]. This reporter reproducibly activates pharyngeal expression early, at the beginning of pharynx primordium formation [19]. Activation of 3xPRE::GFP was delayed from 174 min past the two-cell stage of embryogenesis in wild-type embryos to 201 min in htz-1(RNAi) embryos (Figure 8A and 8B). These data reveal that htz-1 is important for the timely activation of genes within the pharynx at early as well as late stages of development.
Figure 8. htz-1 Influences the Onset of Early PHA-4–Dependent Expression.
(A) Onset of 3XPRE::GFP after the two-cell stage for wild-type (black) or htz-1(RNAi) (grey) embryos. 3XPRE::GFP is a reporter construct with three copies of a high-affinity PHA-4 response element upstream of the Δpes-10 promoter that reproducibly activates early pharyngeal expression [19]. Activation of the 3XPRE::GFP reporter is delayed after injection of htz-1 dsRNA (p = 0.0289, Student t-test). No RNAi: n = 23, htz-1 RNAi: n = 9 embryos. Error bars indicate the standard deviation.
(B) Expression of 3XPRE::GFP at 150 and 200 min after the two-cell stage with htz-1 RNAi or without (No RNAi).
Discussion
Developmental events such as organogenesis are sensitive to precise temporal regulation, to integrate the processes of cell-fate specification, differentiation, and morphogenesis. Our studies with C. elegans have revealed three inputs that control temporal gene expression during pharyngeal organogenesis. The first is PHA-4 and its affinity for target promoters. PHA-4 functions globally to activate both early- and late-expressed pharyngeal genes. The affinity of PHA-4 for its DNA binding site contributes to the timing of expression of these targets. High-affinity binding sites promote expression during early organogenesis, whereas low-affinity sites are typically restricted to later development [18,19]. The second input for temporal control are cis-regulatory elements that function combinatorially with PHA-4. Enhancer and repressor elements have been discovered that promote either early or late pharyngeal gene expression, in combination with PHA-4 [19]. The third input, described here, is the histone variant HTZ-1. The data we present reveal that PHA-4 is required to recruit HTZ-1 to a subset of pharyngeal promoters, and that HTZ-1 is required for the timely onset of transcription. Thus, activation of pharyngeal genes by PHA-4 may depend on its ability to recruit HTZ-1.
Roles for MYS-1, SSL-1, and HTZ-1
Our studies have revealed that pha-4 synergizes with mys-1/TIP60–ssl-1/swr1 and htz-1/H2A.Z to promote activation of pharyngeal gene expression during organogenesis. Partial inactivation of pha-4 in combination with htz-1, mys-1, or ssl-1 RNAi resulted in a specific, synergistic lethality. YFP::HTZ-1 associated with pharyngeal gene promoters. This association was particularly striking at the onset of transcription and required PHA-4. htz-1 was required for the late onset of expression for the pharyngeal genes R07B1.9 and myo-2, and for the early onset of expression of 3xPRE. For myo-2, the association and transcriptional effects of HTZ-1 depended on PHA-4 binding sites and pha-4 activity. Together, these data suggest that HTZ-1 promotes the timely activation of pharyngeal genes in response to PHA-4 (Figure 9).
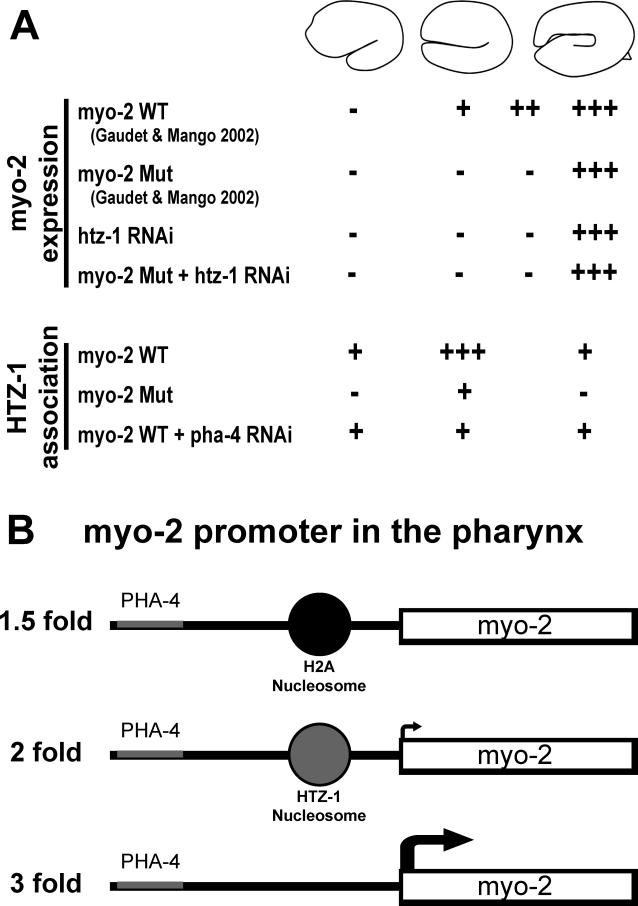
Figure 9. Summary and Model: HTZ-1 Synergizes with PHA-4 to Establish the Foregut.
(A) Summary of the data presented in this paper and in [18]. myo-2 expression initiates at the 2-fold stage, and onset at this stage requires pha-4 and htz-1 because mutation of PHA-4 binding sites (Mut) or htz-1 RNAi lead to a delay in myo-2 activation. HTZ-1 association with the myo-2 promoter peaks at the onset of myo-2 transcription, and this association requires pha-4.
(B) Model to explain how HTZ-1 synergizes with PHA-4. PHA-4 association with the myo-2 promoter leads to exchange of H2A-containing nucleosomes for one or more nucleosomes carrying HTZ-1/H2A.Z at the 2-fold stage. Based on data from other organisms [1], we propose MYS-1 and/or SSL-1 function in a complex that performs the exchange reaction. HTZ-1–containing nucleosomes promote transcriptional activation by the 2-fold stage.
How broad is the influence of HTZ-1 for transcriptional activation? We note that loss of htz-1 function did not enhance unc-54, hsf-1, tbx-2, ama-1, or lin-26. Thus, some transcription appears relatively insensitive to loss of htz-1. Intriguingly, a genome-wide survey of Htz1 occupancy in yeast revealed a correlation between Htz1 and four transcription factors, one of which was the Fox homolog Fkh1 [5]. In Xenopus, endogenous H2A.Z is highly expressed in the notochord [50], a tissue that depends on the pha-4 orthologue FoxA2 for establishment [51–53]. These observations suggest that certain transcription factors, including forkhead proteins, may be acutely dependent on H2A.Z.
The occupancy of HTZ-1 at pharyngeal promoters was particularly striking at the onset of transcription, but we also observed a small percentage of embryos with HTZ-1 association at other stages. One possibility is that there are two pools of HTZ-1: one that is constitutively associated with genes for an unknown function, and a second pool that is recruited transiently to promoters to facilitate their activation. The idea of distinct pools of H2A.Z is supported by studies in yeast that have examined H2A.Z location, acetylation, and Swr1 dependency [6,54–57]. Alternatively, it may be that the low, constitutive association reflects non-specific association with DNA or ambiguities with the nuclear spot assay. Future studies will be needed to tease these possibilities apart.
FoxA and Chromatin Remodeling
FoxA transcription factors have been implicated in chromatin regulation. In vertebrates, FoxA2 associates with genes long before they are transcriptionally active and is thought to promote their transcriptional potentiation [17]. FoxA can decompact reconstituted H1-containing nucleosomal arrays in vitro [24], suggesting a mechanism by which FoxA could promote transcriptional activation in vivo. Our data suggest that H2A.Z contributes to FoxA modulation of chromatin. Previous studies implicated H2A.Z in decompaction of oligonucleosome arrays [58,59], and the crystal structure of H2A.Z revealed that a key interaction between H2A and H3 was disrupted in H2A.Z [60]. Thus, an interesting possibility is that H2A.Z contributes to decompaction of target promoters by FoxA proteins, perhaps by affecting the structure or stability of nucleosomes.
How many pharyngeal genes depend on HTZ-1? We examined the promoters of two pharyngeal genes, myo-2 and R07B1.9, in detail. Although HTZ-1 associated with the promoters of these two target genes late in development, the lack of a detectable pharynx in many pha-4(ts); htz-1(RNAi) larvae cannot be attributed to myo-2 and R07B1.9. Most likely, HTZ-1 works with PHA-4 at multiple targets and has synergistic effects on early as well as late pharyngeal genes (see, for example, 3xPRE). However, some pharyngeal genes may be independent of htz-1. For example, htz-1(RNAi) did not affect the timing of activation of pax-1 (Figure S6), a gene that is expressed in pharyngeal marginal and epithelial cells (J. Stevenson, A. Chisholm, and S. E. Mango, unpublished data). Nor did we observe YFP::HTZ-1 associated with pax-1 (n = 60 embryos; Figure S6). It will be intriguing to determine the characteristics of individual pharyngeal promoters that determine whether it is htz-1 dependent or not.
YFP::HTZ-1 association with reporter arrays containing the myo-2 promoter decreased at the 3-fold stage, when myo-2 expression is at its peak [49]. One speculative explanation of this result is that HTZ-1 is lost from the myo-2 promoter as transcription gets under way. In yeast, Htz1 is displaced from the Gal1 and Pho5 promoters after they become active [1]. Moreover, Htz1-containing nucleosomes are less stable than canonical, H2A-containing nucleosomes in vitro [5]. Thus, it is possible that prior to PHA-4 association, HTZ-1 is absent from myo-2 because this promoter is not primed for activation, but at the 3-fold stage, when the gene is transcribed, HTZ-1 is absent because nucleosomes containing HTZ-1 have been evicted. On the other hand, in non-pharyngeal cells, no PHA-4 is available to bind myo-2 or R07B1.9, and these genes do not recruit HTZ-1. This model suggests that pharyngeal genes pass through distinct states as they transition towards active transcription.
Materials and Methods
Strains and alleles.
C. elegans strains were maintained as described [61]. Strains include: SM190 smg-1(cc546ts)I; pha-4(zu225)V [18,25]; SM279 is PD8120 smg-1(cc546ts)I 5X out-crossed [25], PR1182 cha-1(p1182)IV [62], SM1353 cha-1(p1182)IV; pxEx214(HTZ-1promoter::YFP::HTZ-1 + HTZ-1promoter::CFP::LacI +pRF4), SM1358 cha-1(p1182)IV; pxEx214 pxEx219(cha-1 + LacO), SM1469 cha-1(p1182)IV; pxEx214 pxEx240(cha-1 + LacO), SM1470 cha-1(p1182)IV; pxEx214 pxEx241(cha-1 + LacO), SM1357 cha-1(p1182)IV; pxEx214; pxEx218(cha-1 + HS prom + LacO), SM1356 cha-1(p1182)IV; pxEx214; pxEx217(myo-2 prom + LacO + cha-1), SM1393 cha-1(p1182)IV; pxEx214; pxEx222(myo-2 prom + LacO + cha-1), SM1394 cha-1(p1182)IV; pxEx214; pxEx223(myo-2 prom + LacO + cha-1), SM1390 cha-1(p1182)IV; pxEx214; pxEx220(myo-2 mut prom + LacO + cha-1), SM1395 cha-1(p1182)IV; pxEx214; pxEx224(myo-2 mut prom + LacO + cha-1), SM1397 cha-1(p1182)IV; pxEx214; pxEx233(myo-2 mut prom + LacO + cha-1), SM1391 cha-1(p1182)IV; pxEx214; pxEx221(R07B1.9 prom + LacO + cha-1), SM1408 cha-1(p1182)IV; pxEx214; pxEx227(R07B1.9 prom + LacO + cha-1), SM1355 cha-1(p1182)IV; pxEx214; pxEx216(pax-1 prom + LacO + cha-1), SM1031 contains pxEx171(myo-2::GFP::His2B + pRF4), and SM1033 contains pxEx173(myo-2mut::GFP::His2B + pRF4) [18], SM202 pxIs2(pax-1::GFP +pRF4), SM1064 pxEx187(R07B1.9::GFP::His2B + pRF4) [20], SM1535 pxIs17(3XPRE::GFP::His2B + pRF4) [19], unc-54(ts) is PD8118 smg-1(cc546ts) unc-54(r293)I, PS3551 is hsf-1(sy441)I, JC1970 is tbx-2(ut180)III, DR685 was used to obtain dpy-13(e184) ama-1(m118m236)IV, and MT156 is lin-26(n156)II.
Injections.
SM1353 cha-1(p1182)IV; pxEx214(HTZ-1promoter::YFP::HTZ-1 + HTZ-1promoter::CFP::LacI +pRF4) was created by injecting PR1182 with pRF4 (50 ng/μl) [63] linearized with EcoR1, HTZ-1P::YFP::HTZ-1 PCR product (0.5 ng/μl), HTZ-1P::CFP::LacI PCR product (4 ng/μl), and sheared herring sperm DNA to 100 ng/μl [64]. Target strains were created by injecting SM1353 with the KpnI/SphI lacO fragment from pSV2-dhfr-8.32 [43], 2 ng/μl of cha-1 plasmid RM#527p linearized with ApaI, 10 ng/μl of linear target promoter, and sheared herring sperm DNA to 100 ng/μl. For myo-2, we used 511 base pairs of the endogenous promoter upstream of the start codon, the same length used in the myo-2p::GFPHis2B reporter. For R07B1.9, we used 1,000 base pairs of the endogenous promoter upstream of the start codon, the same length used in the R07B1.9p::GFPHis2B reporter. A total of 265 base pairs of the pax-1 endogenous promoter were used, the same length used in the pax-1::GFP reporter.
Identification of enhancers of the partial loss of pha-4 activity.
HT115 bacteria expressing dsRNA for GFP, mys-1, ssl-1, or htz-1 from [30] were used for feeding RNAi according to standard procedures [29]. RNAi was initiated at the L4 stage and progeny scored 2–3 d later.
Nuclear spot assay.
Images were acquired from live embryos using a LSM510 Meta confocal microscope. A multi-track setting was used to acquire CFP and YFP from a 0.7-μ section taken blindly from the center of the embryo (and pharynx), as determined by differential interference contrast (DIC) microscopy. Positives must have both YFP::HTZ-1 and subnuclear CFP::LacI foci. YFP::HTZ-1 foci are unevenly distributed throughout each nucleus (Figure S3). For YFP::HTZ-1 dots to be considered co-localized, CFP and YFP foci had to overlap and have the same shape. Dots that overlapped, but appeared to have distinct morphologies or locations within the nucleus, were not considered co-localized. Co-localized spots were counted in each embryo and classified as pharyngeal or non-pharyngeal based on position within the embryo as determined by light DIC image.
Expression timing assay.
htz-1 dsRNA [30] was injected into myo-2::GFP::His2B, myo-2Mut::GFP::His2B, or R07B1.9::GFP::His2B transgenic worms at a concentration of 3.2 μg/μl. Comma-stage embryos were picked and examined at 10-min intervals, and initial expression of myo-2::GFP::His2B, myo-2Mut::GFP::His2B, or R07B1.9::GFP::His2B scored at 22 °C. Only embryos with embryonic defects from htz-1 RNAi were compared to uninjected hermaphrodites. htz-1 dsRNA was injected into 3XPRE::GFP::His2B and pax-1::GFP transgenic worms at a concentration of 3.2 μg/μl. Injected worms were dissected the next day, and two-cell embryos were picked and examined at 10-min intervals at 22 °C.
Immunostaining.
Embryos were stained as described previously [65,66]. Antibodies used were polyclonal mouse-generated αHTZ-1 (Bill Kelly [Emory]; B. Kelly, unpublished data) at a dilution of 1:500 and rabbit anti-TBP [65] at a dilution of 1:20.
Crawl assay.
Bacterial plates for GFP, mys-1, ssl-1, or htz-1 RNAi were made as described in the body of the text. Five worms of the genotype smg-1(cc546ts) unc-54(r293), called unc-54(ts), were picked to two of each GFP, mys-1, ssl-1, or htz-1 RNAi plates and incubated at 20 °C or 24 °C. Three days later, four F1 progeny from each plate were picked to a new plate, allowed to crawl for 10 min, and then photographed. As a control, smg-1(cc546ts); pha-4(zu225), called pha-4(ts), animals were put under the same conditions at the same time. L1 lethality was replicated, as in Figure 1A and Figure 2, at 96% with mys-1, 92% with ssl-1, and 48% with htz-1 RNAi (n = 25/strain).
Supporting Information
Cells co-stained with rabbit anti-TBP [65] and DAPI (n = 5). HTZ-1 antibody was a generous donation from Bill Kelly, Emory University; B. Kelly, unpublished data.
(2.4 MB PDF)
unc-54 activity was monitored by a crawling assay at the intermediate temperature of 20 °C or the permissive temperature of 24 °C. After RNAi against the indicated gene, four unc-54(ts) worms were picked to a new plate and allowed to crawl for 10 min. At 24 °C, unc-54(ts) worms were mobile, and this movement was not inhibited when mys-1, ssl-1, or htz-1 were inactivated. At 20 °C, unc-54(ts) worms were largely immobile, and movement was not improved by mys-1, ssl-1, or htz-1 RNAi.
(4.7 MB PDF)
YFP::HTZ-1 associates with chromatin in cells during metaphase and anaphase (arrows). YFP::HTZ-1 is non-randomly distributed in cells during interphase (asterisk), similar to other organisms [10].
(2.9 MB PDF)
(A) Microinjection of 3.2 μg/μl of htz-1 dsRNA caused a highly penetrant late-embryonic arrest. Four terminal htz-1(RNAi) embryos show the pharynx (dotted red line) and intestine appear differentiated, but misshapen morphologically.
(B) Microinjection of 0.5 μg/μl of ssl-1 dsRNA caused a highly penetrant late-embryonic arrest that is indistinguishable from htz-1(RNAi) under the light microscope. Four terminal ssl-1(RNAi) embryos show that the pharynx (dotted-red line) and intestine appear differentiated, but misshapen morphologically. Embryos from injected worms were picked to a new plate after 1 d and allowed to develop 18 h before imaging.
(8.3 MB PDF)
Two-cell embryos were dissected from uninjected hermaphrodites and hermaphrodites injected 2 d prior with 3.2 μg/μl of double-stranded htz-1 RNA. Only embryos with embryonic defects from htz-1(RNAi) (n = 6) were compared to embryos from uninjected hermaphrodites (n = 70).
(1.2 MB PDF)
(A) Onset of pax-1::GFP expression after the two-cell stage. pax-1::GFP expression was not influenced by microinjection of htz-1 dsRNA (p = 0.8622, Student t-test. No RNAi: n = 23 embryos; htz-1 RNAi: n = 12 affected embryos).
(B) Percentage of embryos containing one or more co-localized LacI::CFP and YFP::HTZ-1 dots in the pharynx (black) or outside of the pharynx (grey). No significant difference in pharyngeal HTZ-1 association was found when comparing the no-target lines (Figure 4) to the pax-1 line (p > 0.1704, Fisher exact test).A total of 60 embryos were scored for the pax-1 line. Images were taken at the bean, 1.5-, 2-, and 3-fold stages of embryogenesis. pax-1 is activated at the comma stage by GFP reporter (J. Stevenson, A. Chisholm, S. E. Mango, unpublished data).
(211 KB PDF)
Accession Numbers
The National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov) accession numbers for the genes discussed in the paper are as follows: ama-1 (GeneID 177190), hsf-1 (GeneID 173078), htz-1(RNAi) (GeneID 177212), lin-26 (GeneID 3565051), myo-2 (GeneID 181404), mys-1 (GeneID 179096), pax-1 (GeneID 187104), pha-4 (GeneID 180357), R07B1.9 (GeneID 181201), smg-1(cc546ts) (GeneID 172418), ssl-1 (GeneID 190954), tbx-2 (GeneID 175698), and unc-54, (GeneID 259839).
Acknowledgments
We thank M. Maduro and J. Rand for reagents, B. Cairns and A. Schier for critical reading of the manuscript, and B. Kelly (Emory University) for generous donation of αHTZ-1 antibody. Some of the strains used in this study were provided by the Caenorhabditis Genetics Center.
Abbreviations
- dsRNA
double-stranded RNA
- L1
first larval stage
Footnotes
Competing interests. The authors have declared that no competing interests exist.
A previous version of this article appeared as an Early Online Release on August 11, 2006 (DOI: 10.1371/journal.pgen.0020161.eor).
Author contributions. DLU and SEM conceived and designed the experiments. DLU performed the experiments. DLU and SEM analyzed the data. DLU and SEM wrote the paper.
Funding. This work was funded by the National Institutes of Health 5T32 HD07491 to DLU and R01 GM056264 to SEM. Core facilities at the University of Utah were supported by the NIH 2P30CA42014. SEM is an associate investigator of the Huntsman Cancer Institute and an associate professor of the Department of Oncological Sciences at the University of Utah.
References
- Kamakaka RT, Biggins S. Histone variants: Deviants? Genes Dev. 2005;19:295–310. doi: 10.1101/gad.1272805. [DOI] [PubMed] [Google Scholar]
- Stargell LA, Bowen J, Dadd CA, Dedon PC, Davis M, et al. Temporal and spatial association of histone H2A variant hv1 with transcriptionally competent chromatin during nuclear development in Tetrahymena thermophila . Genes Dev. 1993;7:2641–2651. doi: 10.1101/gad.7.12b.2641. [DOI] [PubMed] [Google Scholar]
- Allis CD, Richman R, Gorovsky MA, Ziegler YS, Touchstone B, et al. hv1 is an evolutionarily conserved H2A variant that is preferentially associated with active genes. J Biol Chem. 1986;261:1941–1948. [PubMed] [Google Scholar]
- Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, et al. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. DOI: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, et al. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci U S A. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, et al. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan J, Baxter EM, Corces VG. The role of histone H2Av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes Dev. 2005;19:65–76. doi: 10.1101/gad.1259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach TJ, Mazzeo M, Chotkowski HL, Madigan JP, Wotring MG, et al. Histone H2A.Z is widely but nonrandomly distributed in chromosomes of Drosophila melanogaster . J Biol Chem. 2000;275:23267–23272. doi: 10.1074/jbc.M910206199. [DOI] [PubMed] [Google Scholar]
- Rangasamy D, Berven L, Ridgway P, Tremethick DJ. Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. EMBO J. 2003;22:1599–1607. doi: 10.1093/emboj/cdg160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce K, Myers FA, Mantouvalou E, Lefevre P, Greaves I, et al. The replacement histone H2A.Z in a hyperacetylated form is a feature of active genes in the chicken. Nucleic Acids Res. 2005;33:5633–5639. doi: 10.1093/nar/gki874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Madhani HD. Patterning chromatin: Form and function for H2A.Z variant nucleosomes. Curr Opin Genet Dev. 2006;16:119–124. doi: 10.1016/j.gde.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Ceol CJ, Horvitz HR. A new class of C. elegans synMuv genes implicates a Tip60/NuA4-like HAT complex as a negative regulator of Ras signaling. Dev Cell. 2004;6:563–576. doi: 10.1016/s1534-5807(04)00065-6. [DOI] [PubMed] [Google Scholar]
- Poulin G, Dong Y, Fraser AG, Hopper NA, Ahringer J. Chromatin regulation and sumoylation in the inhibition of Ras-induced vulval development in Caenorhabditis elegans . Embo J. 2005;24:2613–2623. doi: 10.1038/sj.emboj.7600726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann E, Knochel W. Five years on the wings of fork head. Mech Dev. 1996;57:3–20. doi: 10.1016/0925-4773(96)00539-4. [DOI] [PubMed] [Google Scholar]
- Zaret KS. Regulatory phases of early liver development: Paradigms of organogenesis. Nat Rev Genet. 2002;3:499–512. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- Gaudet J, Muttumu S, Horner M, Mango SE. Whole-genome analysis of temporal gene expression during foregut development. PLoS Biol. 2004;2:e352. doi: 10.1371/journal.pbio.0020352. DOI: 10.1371/journal.pbio.0020352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao W, Gaudet J, Kent WJ, Muttumu S, Mango SE. Environmentally induced foregut remodeling by PHA-4/FoxA and DAF-12/NHR. Science. 2004;305:1743–1746. doi: 10.1126/science.1102216. [DOI] [PubMed] [Google Scholar]
- Kalb JM, Lau KK, Goszczynski B, Fukushige T, Moons D, et al. pha-4 is Ce-fkh-1, a fork head/HNF-3alpha,beta,gamma homolog that functions in organogenesis of the C. elegans pharynx. Development. 1998;125:2171–2180. doi: 10.1242/dev.125.12.2171. [DOI] [PubMed] [Google Scholar]
- Vilimas T, Abraham A, Okkema PG. An early pharyngeal muscle enhancer from the Caenorhabditis elegans ceh-22 gene is targeted by the Forkhead factor PHA-4. Dev Biol. 2004;266:388–398. doi: 10.1016/j.ydbio.2003.10.015. [DOI] [PubMed] [Google Scholar]
- Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, et al. Hepatic specification of the gut endoderm in vitro: Cell signaling and transcriptional control. Genes Dev. 1996;10:1670–1682. doi: 10.1101/gad.10.13.1670. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Kaltenbach LS, Updike DL, Mango SE. Contribution of the amino and carboxyl termini for PHA-4/FoxA function in Caenorhabditis elegans . Dev Dyn. 2005;234:346–354. doi: 10.1002/dvdy.20550. [DOI] [PubMed] [Google Scholar]
- Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- Mango SE. Stop making nonSense: The C. elegans smg genes. Trends Genet. 2001;17:646–653. doi: 10.1016/s0168-9525(01)02479-9. [DOI] [PubMed] [Google Scholar]
- Grimson A, O'Connor S, Newman CL, Anderson P. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA decay in Caenorhabditis elegans . Mol Cell Biol. 2004;24:7483–7490. doi: 10.1128/MCB.24.17.7483-7490.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans . Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Piano F, Schetter AJ, Mangone M, Stein L, Kemphues KJ. RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans . Curr Biol. 2000;10:1619–1622. doi: 10.1016/s0960-9822(00)00869-1. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- Parrish S, Fleenor J, Xu S, Mello C, Fire A. Functional anatomy of a dsRNA trigger: Differential requirement for the two trigger strands in RNA interference. Mol Cell. 2000;6:1077–1087. doi: 10.1016/s1097-2765(00)00106-4. [DOI] [PubMed] [Google Scholar]
- Mango SE, Lambie EJ, Kimble J. The pha-4 gene is required to generate the pharyngeal primordium of Caenorhabditis elegans . Development. 1994;120:3019–3031. doi: 10.1242/dev.120.10.3019. [DOI] [PubMed] [Google Scholar]
- Hajdu-Cronin YM, Chen WJ, Sternberg PW. The L-type cyclin CYL-1 and the heat-shock-factor HSF-1 are required for heat-shock-induced protein expression in Caenorhabditis elegans . Genetics. 2004;168:1937–1949. doi: 10.1534/genetics.104.028423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labouesse M, Sookhareea S, Horvitz HR. The Caenorhabditis elegans gene lin-26 is required to specify the fates of hypodermal cells and encodes a presumptive zinc-finger transcription factor. Development. 1994;120:2359–2368. doi: 10.1242/dev.120.9.2359. [DOI] [PubMed] [Google Scholar]
- Roy Chowdhuri S, Crum T, Woollard A, Aslam S, Okkema PG. The T-box factor TBX-2 and the SUMO conjugating enzyme UBC-9 are required for ABa-derived pharyngeal muscle in C. elegans . Dev Biol. 2006;295:664–677. doi: 10.1016/j.ydbio.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Miyahara K, Suzuki N, Ishihara T, Tsuchiya E, Katsura I. TBX2/TBX3 transcriptional factor homologue controls olfactory adaptation in Caenorhabditis elegans . J Neurobiol. 2004;58:392–402. doi: 10.1002/neu.10299. [DOI] [PubMed] [Google Scholar]
- Sanford T, Golomb M, Riddle DL. RNA polymerase II from wild type and alpha-amanitin-resistant strains of Caenorhabditis elegans . J Biol Chem. 1983;258:12804–12809. [PubMed] [Google Scholar]
- Carmi I, Kopczynski JB, Meyer BJ. The nuclear hormone receptor SEX-1 is an X-chromosome signal that determines nematode sex. Nature. 1998;396:168–173. doi: 10.1038/24164. [DOI] [PubMed] [Google Scholar]
- Fukushige T, Hendzel MJ, Bazett-Jones DP, McGhee JD. Direct visualization of the elt-2 gut-specific GATA factor binding to a target promoter inside the living Caenorhabditis elegans embryo. Proc Natl Acad Sci U S A. 1999;96:11883–11888. doi: 10.1073/pnas.96.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro MF, Lin R, Rothman JH. Dynamics of a developmental switch: Recursive intracellular and intranuclear redistribution of Caenorhabditis elegans POP-1 parallels Wnt-inhibited transcriptional repression. Dev Biol. 2002;248:128–142. doi: 10.1006/dbio.2002.0721. [DOI] [PubMed] [Google Scholar]
- Santisteban MS, Kalashnikova T, Smith MM. Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000;103:411–422. doi: 10.1016/s0092-8674(00)00133-1. [DOI] [PubMed] [Google Scholar]
- Jones D, Dixon DK, Graham RW, Candido EP. Differential regulation of closely related members of the hsp16 gene family in Caenorhabditis elegans . DNA. 1989;8:481–490. doi: 10.1089/dna.1.1989.8.481. [DOI] [PubMed] [Google Scholar]
- Miller DM, Stockdale FE, Karn J. Immunological identification of the genes encoding the four myosin heavy chain isoforms of Caenorhabditis elegans . Proc Natl Acad Sci U S A. 1986;83:2305–2309. doi: 10.1073/pnas.83.8.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans . Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Russnak RH, Candido EP. Locus encoding a family of small heat shock genes in Caenorhabditis elegans: Two genes duplicated to form a 3.8-kilobase inverted repeat. Mol Cell Biol. 1985;5:1268–1278. doi: 10.1128/mcb.5.6.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara H, Motohashi T, Kohara Y. A multi-well version of in situ hybridization on whole mount embryos of Caenorhabditis elegans . Nucleic Acids Res. 1996;24:2119–2124. doi: 10.1093/nar/24.11.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway P, Brown KD, Rangasamy D, Svensson U, Tremethick DJ. Unique residues on the H2A.Z containing nucleosome surface are important for Xenopus laevis development. J Biol Chem. 2004;279:43815–43820. doi: 10.1074/jbc.M408409200. [DOI] [PubMed] [Google Scholar]
- Ang SL, Rossant J. HNF-3 beta is essential for node and notochord formation in mouse development. Cell. 1994;78:561–574. doi: 10.1016/0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- Weinstein DC, Ruiz i Altaba A, Chen WS, Hoodless P, Prezioso VR, et al. The winged-helix transcription factor HNF-3 beta is required for notochord development in the mouse embryo. Cell. 1994;78:575–588. doi: 10.1016/0092-8674(94)90523-1. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Hogan BL. HNF-3 beta as a regulator of floor plate development. Cell. 1994;76:103–115. doi: 10.1016/0092-8674(94)90176-7. [DOI] [PubMed] [Google Scholar]
- Babiarz JE, Halley JE, Rine J. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae . Genes Dev. 2006;20:700–710. doi: 10.1101/gad.1386306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh M-C, Mennella TA, Sawa C, Berthelet S, Krogan NJ, et al. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar CB, Xu F, Zhang K, Grunstein M. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 2006;20:711–722. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WH, Alami S, Luk E, Wu CH, Sen S, et al. Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat Struct Mol Biol. 2005;12:1064–1071. doi: 10.1038/nsmb1023. [DOI] [PubMed] [Google Scholar]
- Fan JY, Gordon F, Luger K, Hansen JC, Tremethick DJ. The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat Struct Biol. 2002;9:172–176. doi: 10.1038/nsb767. [DOI] [PubMed] [Google Scholar]
- Abbott DW, Ivanova VS, Wang X, Bonner WM, Ausio J. Characterization of the stability and folding of H2A.Z chromatin particles: Implications for transcriptional activation. J Biol Chem. 2001;276:41945–41949. doi: 10.1074/jbc.M108217200. [DOI] [PubMed] [Google Scholar]
- Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans . Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand JB. Genetic analysis of the cha-1-unc-17 gene complex in Caenorhabditis . Genetics. 1989;122:73–80. doi: 10.1093/genetics/122.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbach L, Horner MA, Rothman JH, Mango SE. The TBP-like factor CeTLF is required to activate RNA polymerase II transcription during C. elegans embryogenesis. Mol Cell. 2000;6:705–713. doi: 10.1016/s1097-2765(00)00068-x. [DOI] [PubMed] [Google Scholar]
- Fan JY, Rangasamy D, Luger K, Tremethick DJ. H2A.Z alters the nucleosome surface to promote HP1alpha-mediated chromatin fiber folding. Mol Cell. 2004;16:655–661. doi: 10.1016/j.molcel.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Horner MA, Quintin S, Domeier ME, Kimble J, Labouesse M, et al. pha-4, an HNF-3 homolog, specifies pharyngeal organ identity in Caenorhabditis elegans . Genes Dev. 1998;12:1947–1952. doi: 10.1101/gad.12.13.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cells co-stained with rabbit anti-TBP [65] and DAPI (n = 5). HTZ-1 antibody was a generous donation from Bill Kelly, Emory University; B. Kelly, unpublished data.
(2.4 MB PDF)
unc-54 activity was monitored by a crawling assay at the intermediate temperature of 20 °C or the permissive temperature of 24 °C. After RNAi against the indicated gene, four unc-54(ts) worms were picked to a new plate and allowed to crawl for 10 min. At 24 °C, unc-54(ts) worms were mobile, and this movement was not inhibited when mys-1, ssl-1, or htz-1 were inactivated. At 20 °C, unc-54(ts) worms were largely immobile, and movement was not improved by mys-1, ssl-1, or htz-1 RNAi.
(4.7 MB PDF)
YFP::HTZ-1 associates with chromatin in cells during metaphase and anaphase (arrows). YFP::HTZ-1 is non-randomly distributed in cells during interphase (asterisk), similar to other organisms [10].
(2.9 MB PDF)
(A) Microinjection of 3.2 μg/μl of htz-1 dsRNA caused a highly penetrant late-embryonic arrest. Four terminal htz-1(RNAi) embryos show the pharynx (dotted red line) and intestine appear differentiated, but misshapen morphologically.
(B) Microinjection of 0.5 μg/μl of ssl-1 dsRNA caused a highly penetrant late-embryonic arrest that is indistinguishable from htz-1(RNAi) under the light microscope. Four terminal ssl-1(RNAi) embryos show that the pharynx (dotted-red line) and intestine appear differentiated, but misshapen morphologically. Embryos from injected worms were picked to a new plate after 1 d and allowed to develop 18 h before imaging.
(8.3 MB PDF)
Two-cell embryos were dissected from uninjected hermaphrodites and hermaphrodites injected 2 d prior with 3.2 μg/μl of double-stranded htz-1 RNA. Only embryos with embryonic defects from htz-1(RNAi) (n = 6) were compared to embryos from uninjected hermaphrodites (n = 70).
(1.2 MB PDF)
(A) Onset of pax-1::GFP expression after the two-cell stage. pax-1::GFP expression was not influenced by microinjection of htz-1 dsRNA (p = 0.8622, Student t-test. No RNAi: n = 23 embryos; htz-1 RNAi: n = 12 affected embryos).
(B) Percentage of embryos containing one or more co-localized LacI::CFP and YFP::HTZ-1 dots in the pharynx (black) or outside of the pharynx (grey). No significant difference in pharyngeal HTZ-1 association was found when comparing the no-target lines (Figure 4) to the pax-1 line (p > 0.1704, Fisher exact test).A total of 60 embryos were scored for the pax-1 line. Images were taken at the bean, 1.5-, 2-, and 3-fold stages of embryogenesis. pax-1 is activated at the comma stage by GFP reporter (J. Stevenson, A. Chisholm, S. E. Mango, unpublished data).
(211 KB PDF)