Abstract
Aberrant methylation in gene promoter regions leads to transcriptional inactivation of cancer-related genes and plays an integral role in tumorigenesis. This alteration has been investigated in lung tumors primarily from smokers, whereas only a few studies involved never-smokers. Here, we applied methylation-specific polymerase chain reaction to compare the frequencies of the methylated promoter of p16 and O6-methylguanine-DNA methyltransferase (MGMT) genes in lung tumors from 122 patients with non-small cell lung cancer, including 81 smokers and 41 never-smokers. Overall, promoter methylation was detected in 52.5% (64 of 122) and 30.3% (37 of 122) of the p16 and MGMT genes, respectively. Furthermore, the frequency of promoter methylation was significantly higher among smokers, compared with never-smokers, for both the p16 [odds ratio (OR) = 3.28; 95% confidence interval (CI) = 1.28-8.39; P = .013] and MGMT (OR = 3.93; 95% CI = 1.27-12.21; P = .018) genes. The trend for a higher promoter methylation frequency of these genes was also observed among female smokers compared with female never-smokers. Our results suggest an association between tobacco smoking and an increased incidence of aberrant promoter methylation of the p16 and MGMT genes in non-small cell lung cancer.
Keywords: Lung tumors, p16, MGMT, promoter methylation, never-smokers
Introduction
Lung cancer is a very common cause of cancer deaths in the United States and in western European countries [1,2]. This disease is associated primarily with tobacco smoking because about 85% of lung cancer patients are smokers, whereas the remaining 10% to 15% have no history of tobacco smoking and consist of mostly women [1]. Other causes of lung cancer include exposure to coal-burning smoke [3] and environmental tobacco smoke [4]. In spite of extensive studies, pathogenic pathways to lung cancer in both smokers and never-smokers remain poorly understood.
Aberrant methylation of 5′ cytosine residues of a guanine residue in CpG islands in the promoter regions of tumorsuppressor genes is an important mechanism of gene transcriptional inactivation and has been associated with tumorigenesis [5–7]. The p16INK4A tumor-suppressor gene plays a key role in cell cycle regulation. This gene codes for a protein that binds to and inhibits cyclin D kinases (CdK4 and CdK6), which normally phosphorylate serine and threonine residues of the retinoblastoma (Rb) protein [8,9]. Therefore, the p16 protein inhibits cell cycle progression through G1 to S phase by maintaining the Rb protein in the unphosphorylated state. Inactivation of the p16 gene expression by aberrant methylation of its promoter region is believed to be a pathway to tumorigenesis [10,11]. Previous studies show that methylation of the p16 gene promoter was detected in a high proportion of primary lung tumors [12,13].
Aberrant methylation in the promoter region of the O6-methylguanine-DNA methyltransferase (MGMT) gene has also been found frequently in many tumor types, including lung tumors, and is implicated as a mechanism of gene silencing leading to tumorigenesis [14,15]. Unlike the p16 protein, however, the MGMT protein is a DNA repair enzyme, which specifically transfers the methyl group and other small alkyl groups at the O6 position of guanine to a cysteine acceptor site of the protein itself in an autoinactivating reaction [16]. Therefore, the repair activity of the MGMT protein helps decrease the probability that the damaged guanine becomes a mutagenic/carcinogenic site. Methylated MGMT gene promoter has been associated with loss or decrease of MGMT expression in tumor tissues of various organs, including lung tumors [14,17–19].
Taken together, both p16 and MGMT genes have been considered as potentially useful candidate biomarkers for lung cancer treatment and early detection. The purpose of this study was to understand the prevalence of aberrant promoter methylation for these two genes in lung tumors from smokers and never-smokers. We have analyzed methylation in the promoter region of the p16 and MGMT genes in lung tumors obtained from lung cancer patients from the Western Pennsylvania region. We compared the frequencies of methylated promoters for these two genes according to smoking status and gender, and we discussed our results in relation to those of previous studies.
Materials and Methods
Patients and DNA Isolation
Lung tumor tissues analyzed in this study were obtained from 122 patients with non-small cell lung cancer, including 81 smokers and 41 never-smokers, all of whom were whites and lived in the Western Pennsylvania region. They were among the specimens collected between 1988 and 2001 and stored at the University of Pittsburgh Medical Center under an Institutional Review Board-approved protocol. The demographic and clinical profile of these patients is shown in Table 1. All male smokers were current smokers, and packyear data were available for 36 of them. Among female smokers, 38 were current smokers and 1 was a former smoker. Pack-year data were available for 36 of female smokers. One pack-year is defined as an average of one pack of cigarettes smoked per day for 1 year. No significant difference in the number of pack-years was found among male and female smokers. All never-smokers involved in this study had never smoked during their lifetime.
Table 1.
Demographic and Clinical Profile of 122 Lung Cancer Patients.
| Never-Smokers | Smokers | |||
| Female (%) | Male (%) | Female (%) | Male (%) | |
| Total | 31 | 10 | 39 | 42 |
| Age (years) | 66.7 ± 11.0 | 69.2 ± 6.9 | 63.1 ± 11.0 | 64.9 ± 9.1 |
| Smoking (pack-years) | NA | NA | 51.3 ± 20.6 | 59.6 ± 36.3 |
| Tumor types | ||||
| Adenocarcinoma | 19 (61.3) | 4 (40) | 22 (56.4) | 27 (64.2) |
| Squamous cell carcinomas | 3 (9.7) | 3 (30) | 10 (25.6) | 6 (14.3) |
| Bronchoalveolar carcinoma | 6 (19.3) | 5 (12.8) | 4 (9.5) | |
| Adenosquamous carcinoma | 3 (9.7) | 1 (10) | 5 (11.9) | |
| Large cell carcinomas | 2 (5.1) | |||
| Carcinoid | 1 (10) | |||
| Neuroendocrine | 1 (10) | |||
| Tumor stage | ||||
| I | 11 (35.5) | 4 (40) | 22 (56.4) | 17 (40.5) |
| II | 11 (35.5) | 1 (10) | 8 (20.5) | 4 (9.5) |
| III | 3 (9.7) | 2 (20) | 6 (15.4) | 8 (19.0) |
| IV | 5 (16.1) | 1 (10) | 5 (11.9) | |
| Unknown | 1 (3.2) | 2 (20) | 3 (7.7) | 8 (19.0) |
NA, not applicable.
Fresh-frozen tissues were used for DNA extraction using the Proteinase K treatment and phenol/chloroform extraction methods, as described previously [20]. DNA was dissolved in distilled water and stored at -20°C until use.
Methylation-Specific Polymerase Chain Reaction (MSP)
Each genomic DNA sample was treated with sodium bisulfite (Sigma, St. Louis, MO), as described by Herman et al. [21]. Briefly, DNA was denatured by treatment with 0.3 M NaOH and incubation at 37°C for 30 minutes, followed by incubation with 10 mM hydroquinone (Sigma) and 3 M sodium bisulfite (Sigma) at 55°C for 16 to 20 hours. Modified DNA was purified using a Wizard DNA Clean-Up System (Promega Corporation, Madison, WI), recovered by ethanol precipitation, and dissolved in distilled water. Universal methylated human DNA (Chemicon International, Temecula, CA) was treated in the same way and was used as a positive control DNA.
Promoter gene methylation was screened using MSP. Two versions of MSP have been previously reported, including an original one-stage MSP [21] and a modified two-stage MSP, which used two rounds of PCR and had a higher level of sensitivity to detect promoter methylation compared with the original one-stage MSP [22]. In this study, we first compared the sensitivity and validity of both MSP methods to analyze the promoter methylation of the p16 and MGMT genes in DNA from a series of 24 lung tumors. The results showed that promoter methylation was detected in the same tumors and at a similar frequency using either the original or modified MSP for both the p16 and MGMT genes, suggesting that the original one-stage MSP was sufficiently sensitive to detect promoter methylation in lung tumors. However, in our experiments, the two-stage MSP method showed MSP products as a clear single band through gel electrophoresis analysis, whereas the one-stage MSP method provided products that usually appeared with other minor bands (data not shown). For this reason, in this study, we applied the two-stage MSP method for promoter methylation analysis. The nucleotide sequences of the primers used for the two-stage MSP for both the p16 and MGMT genes are summarized in Table 2. During round 1 PCR, the primers recognized the bisulfite-modified template but did not discriminate between the methylated and unmethylated alleles. PCR amplification was carried out in a 25-µl reaction mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 100 µM of each kind of dNTP, and 0.2 µM of each kind of primer. The reaction was heated at 95°C for 10 minutes, then amplified for 40 cycles [95°C/30 seconds, 64°C (for p16) or 52°C (for MGMT)/30 seconds, and 72°C/30 seconds], followed by a final 10-minute extension at 72°C. An aliquot of each round 1 PCR product was diluted 10-fold, and 1 µl was used for round 2 PCR, using the same reagents and conditions as for round 1 PCR, except that the MgCl2 concentration was reduced to 1 mM and each sample was amplified in two reactions, with one reaction containing primers specific for methylated C and the other reaction containing primers specific for unmethylated C. Each reaction was heated at 95°C for 10 minutes, then amplified for 40 cycles, each consisting (for the reaction containing methylated primers) of 95°C/30 seconds, 70°C (for p16) or 64°C (for MGMT)/30 seconds, and 72°C/30 seconds, and (for the reaction containing unmethylated primers) of 95°C/30 seconds, 64°C (for both p16 and MGMT)/30 seconds, and 72°C/30 seconds. An aliquot of each round 2 PCR product was separated on an 8% polyacrylamide gel. The gel was stained with ethidium bromide and photographed under UV illumination. The reproducibility of the results was confirmed by repeating MSP analyses for each DNA sample.
Table 2.
Primers Used for MS-PCR.
| Sense Primers (5′ →3′) | Antisense Primers (5′ →3′) | |
| Primers for stage I PCR | ||
| p16 | GAAGAAAGAGGAGGGGTTGG | CCACCTAAATCGACCTCCGACCG |
| MGMT | GGATATGTTGGGATAGTT | CCAAAAACCCCAAACCC |
| Primers for stage II PCR | ||
| M-p16 | TTATTAGAGGGTGGGGCGGATCGC | GACCCCGAACCGCGACCGTAA |
| U-p16 | TTATTAGAGGGTGGGGTGGATTGT | CAACCCCAAACCACAACCATAA |
| M-MGMT | TTTCGACGTTCGTAGGTTTTCGC | GCACTCTTCCGAAAACGAAACG |
| U-MGMT | TTTGTGTTTTGATGTTTGTAGGTTTTTGT | AACTCCACACTCTTCCAAAAACAAAACA |
M, methylated; U, unmethylated.
Round 2 PCR products were also analyzed by restriction fragment length polymorphism to confirm their “methylated” status. In this method, the round 2 PCR products were treated with a specific restriction enzyme that cut only templates containing methylated cytosines (methylated template), whereas in unmethylated templates, unmethylated cytosines were transformed by bisulfite treatment into uracils, which were converted to thymidine after PCR and were unrecognized by the enzyme. For this purpose, a 2-µl aliquot from each round 2 PCR product was treated in a final 10-µl reaction—with the restriction enzyme Fnu4H1 for the p16 gene, and with TaqI and BstU1 for the MGMT gene—using the reagents and conditions provided by the manufacturer (New England Biolabs, Beverly, MA) [22]. Digestion products were separated on an 8% polyacrylamide gel. The gel was stained with ethidium bromide and photographed under UV visualization (data not shown).
Statistical Analysis
Wilcoxon rank sum test and chi-square test (or Fisher's exact test) were used for continuous and categorical variables in univariate analysis, respectively. Logistic regression models were used to assess the effect of multiple variables on methylation status.
Results
Table 1 shows demographic and clinical information for the 122 lung cancer patients, including 81 smokers and 41 never-smokers. Age did not differ significantly between smokers and never-smokers, or between male and female smokers. Adenocarcinomas accounted for 60.5% (49 of 81) of tumors from smokers and for 56.1% (23 of 41) of those from never-smokers.
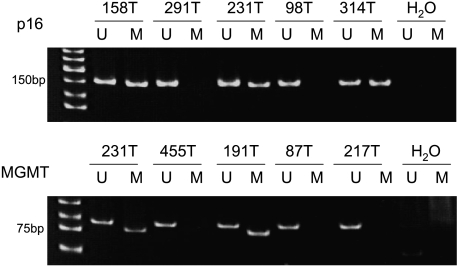
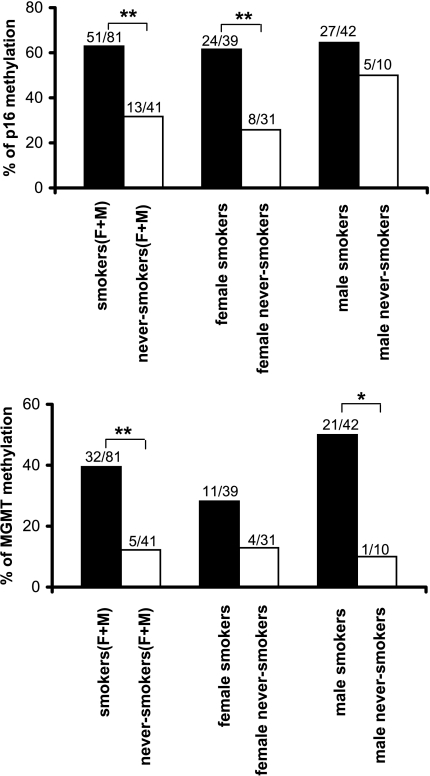
Figure 1 shows a representative example of MSP analysis of lung tumor DNA. Figure 2 summarizes the results on the frequency of promoter methylation for the p16 and MGMT genes. Overall, 52.5% (64 of 122) and 30.3% (37 of 122) of all patients had a methylated promoter for the p16 and MGMT genes, respectively. When grouped according to smoking status, methylated p16 gene promoter was found in 62.9% (51 of 81) of lung tumors from smokers, which is significantly higher than the 31.7% (13 of 41) frequency found in those from never-smokers (P = .001). When grouped according to gender, a significantly higher frequency of methylated p16 gene promoter was also observed among female smokers compared with female never-smokers [61.5% (24 of 39) vs 25.8% (8 of 31), P = .002], but not among male smokers compared with their never-smoking counterparts [64.3% (27 of 42) vs 50% (5 of 10), P = .385], which was probably due to the smaller number of male never-smokers. There was no statistically significant difference in the frequencies of methylated p16 gene promoter in lung tumors by gender [64.3% (27 of 42) in male smokers vs 61.5% (24 of 39) in female smokers, P = .808] and by histology [55.6% (40 of 72) in adenocarcinomas vs 54.5% (12 of 22) in squamous cell carcinomas, P = .922].
Figure 1.
Detection of p16 and MGMT promoter methylation by MSP. M indicates the presence of methylated p16 or MGMT. U indicates the presence of unmethylated p16 or MGMT. All tests were performed twice, and representative data are shown.
Figure 2.
Summary of p16 and MGMT gene hypermethylation in smoking and never-smoking patients. (**P < .01; *P < .05). Chi-square test or Fisher's exact test was used. F, female; M, male.
Methylated MGMT gene promoter was found in 39.5% (32 of 81) of lung tumors from smokers—significantly higher than the 12.2% (5 of 41) frequency found in those from never-smokers (P = .002). This trend was also observed among female smokers versus female never-smokers [28.2% (11 of 39) vs 12.9% (4 of 31), P = .127], and male smokers versus male never-smokers [50.0% (21 of 42) vs 10.0% (1 of 10), P = .023]. Among smokers, the methylated MGMT gene promoter occurred more frequently in male [50.0% (21 of 42)] than in female [28.2% (11 of 39)] patients (P = .045). The frequencies of MGMT gene promoter methylation did not differ between adenocarcinomas (30.6%, 22 of 72) and squamous cell carcinomas (27.3%, 6 of 22) (P = .749).
Multivariate logistic regression models were employed to control for potential confounding effects of variables, such as age, gender, histology, and tumor stage. As shown in Table 3, smokers had a significantly increased risk for promoter methylation, compared with never-smokers, for both the p16 [odds ratio (OR) = 3.28; 95% confidence interval (CI) = 1.28-8.39; P = .013] and MGMT (OR = 3.93; 95% CI = 1.27-12.21; P = .018) genes.
Table 3.
Logistic Regression Models of p16 and MGMT Promoter Methylation (n = 122).
| p16 | MGMT | |||
| OR (95% CI) | P | OR (95% CI) | P | |
| Age | 1.02 (0.97–1.06) | .502 | 1.02 (0.97–1.07) | .455 |
| Sex | 0.69 (0.29–1.66) | .404 | 0.61 (0.25–1.52) | .287 |
| Smoking | 3.28 (1.28–8.39) | .013 | 3.93 (1.27–12.2) | .018 |
| Histology | 0.66 (0.45–0.96) | .032 | 0.98 (0.66–1.46) | .927 |
| Stage | 0.76 (0.50–1.16) | .206 | 1.26 (0.82–1.95) | .291 |
Discussion
In the present study, we compared the frequencies of aberrant promoter methylation for the p16 and MGMT genes with the smoking status and gender of the patients. Our results show that the frequency of promoter methylation was significantly higher among smokers, compared with never-smokers, for both the p16[62.9% (51 of 81) vs 31.7% (13 of 41), P = .001] and MGMT [39.5% (32 of 81) vs 12.2% (5 of 41), P = .002] genes (Figure 2). Furthermore, the trend for a higher frequency of promoter methylation was also observed among the group of female smokers compared with the group of female never-smokers. The difference in the frequencies of promoter methylation was not associated with the presence of a different proportion of lung tumor types between the two patient groups analyzed. For instance, in this study, adenocarcinoma accounted for 60.5% (49 of 81) among smokers and for 56.1% (23 of 41) among never-smokers (Table 1), whereas the frequency of promoter methylation in this tumor type was higher among smokers, compared with never-smokers, for both the p16 [63.3% (31 of 49) vs 39.1% (9 of 23), P = .054] and MGMT [36.7% (18 of 49) vs 17.4% (4 of 23), P = .094] genes. Logistic regression analysis showed that tobacco smoking was significantly related to promoter methylation of both the p16 (OR = 3.28; 95% CI = 1.28-8.39; P = .013) and MGMT (OR = 3.93; 95% CI = 1.27-12.21; P = .018) genes. Taken together, these results suggest that aberrant methylation of the promoter region of the p16 and MGMT genes may be influenced by tobacco smoking status.
So far, there have been only a few studies involving both smoking and never-smoking lung cancer patients, and the results showed some disagreements among studies. For instance, a study [23] of lung cancer patients from multiple centers, including the United States, Australia, Japan, and Taiwan, showed a significantly higher rate of methylated p16gene promoter in ever-smokers compared with never-smokers (P = .007). Kim et al. observed the same trend in their study of 172 smoking and 13 nonsmoking lung cancer patients from the Massachusetts General Hospital (Boston, MA) (P = .05). Furthermore, the authors showed that the rate of methylated p16 gene promoter was significantly associated with pack-years smoked (P = .007) [24]. However, Sanchez-Cespedes et al. reported a frequency of methylated p16 gene promoter that was not significantly different between smoking (21.2%, 7 of 33) and nonsmoking (36%, 5 of 14) lung cancer patients from the Johns Hopkins Hospital (Baltimore, MD), the Johns Hopkins Bayview Medical Center (Baltimore, MD), and the Medical College of Wisconsin (Milwaukee, WI) (P = .33) [25].
There have been even fewer studies of the MGMT gene aberrant promoter methylation in lung tumors from never-smokers. Pulling et al. analyzed the MGMT gene promoter methylation in DNA extracted from paraffin-embedded lung adenocarcinoma obtained from 157 smokers and 46 never-smokers—consisting mostly of non-Hispanic whites (about 80%) and also of Hispanics and African Americans—from various centers, including the New Mexico Tumor Registry (Albuquerque, NM), the Saint Mary's Hospital Tumor Registry (Grand Junction, CO), and the Metropolitan Detroit Cancer Surveillance System (Detroit, MI). The authors reported a significantly increased incidence of methylated promoter for the MGMT gene among never-smokers compared with smokers (66% vs 47%, respectively, P = .02) [19]. This result is, however, in disagreement with reports by Toyooka et al. [23] that the frequency of promoter methylation for the MGMT gene was higher than—but not significantly different from—that observed in nonsmokers. Our data on the frequencies of MGMT promoter gene methylation are consistent with those of Toyooka et al. but are in disagreement with those of Pulling et al.
The reason for the disagreement on the frequencies of aberrant promoter methylation for the p16 and MGMT genes between smokers and never-smokers among different studies remains unclear. One possible contributing factor to these inconsistencies may be the smaller number of never-smokers involved in these studies, due to the smaller incidence of lung cancer occurrence among never-smokers compared with smokers. Furthermore, a never-smoker or a nonsmoker has been defined in some studies as a patient who has smoked less than 100 cigarettes [19,23,25,26], whereas in our study, it refers to a patient who smoked no cigarettes during his lifetime. Geographical and/or ethnic differences of lung cancer patients among various studies may also be a factor in the disagreement. For instance, in the study by Toyooka et al. [23], the majority of nonsmokers were from Japan and Taiwan, whereas only 20 of them were from the United States and Australia. Furthermore, it remains unclear whether the use of DNA from paraffin-embedded tumors, such as those used in the study of Pulling et al. [19], as opposed to DNA from fresh-frozen tumors may affect the results on the frequencies of gene promoter methylation.
Our data suggest that tobacco smoking correlated with an increased frequency of promoter methylation for the p16 and MGMT genes in lung tumors. The reason for this observation is unknown. Some previous studies showed that tobacco smoking was associated with alterations of only some genes in lung cancer. For instance, in lung adenocarcinomas, K-ras mutations are identified primarily in smokers [27], whereas mutations in the epidermal growth factor receptor (EGFR) gene are associated with mostly nonsmokers [28,29]. Lung cancer patients whose lung tumors had EGFR mutations responded better than those without such mutations to gefitinib therapy [30]. The precise mechanism(s) of the association between tobacco smoking and increased frequencies of p16 and MGMT gene promoter methylation remains to be determined. Tobacco smoke contains many carcinogens, some of which have been shown to affect gene promoter methylation [12,31]. For instance, a study of rats treated with tobacco-specific 4-methynitrosamino-1-(3-pyridy)-butanone (NNK) showed that hypermethylated p16 gene promoter was detected not only in lung adenocarcinomas but also in adenomas and hyperplastic lesions, which represented precursor lesions to the tumors, indicating a link between exposure to NNK and p16 gene promoter aberrant methylation in lung tumors and implicating this epigenetic alteration as an early event in lung carcinogenesis [12].
It remains unclear how tobacco smoke carcinogens may affect the methylation of the MGMT gene promoter. Grafstrom et al. [32] showed that human epithelial cells treated with acetaldehyde (another carcinogen present in tobacco smoke) had a significantly decreased activity of the MGMT gene. However, so far, there have been no reports linking acetaldehyde or other tobacco smoke carcinogens to aberrant methylation of the MGMT gene promoter.
The limitations of this study may be the small number of lung tumors from never-smokers, due to the low incidence of lung cancer among never-smoking individuals. Furthermore, because the DNA samples analyzed were obtained from surgically resected tumors, the incidence of promoter methylation observed may not indicate the incidence in the whole non-small cell lung cancer population.
In conclusion, our study showed a higher frequency of promoter methylation for the p16 and MGMT genes in lung tumors from smokers compared with never-smokers, indicating an association between tobacco use and the increased incidence of promoter methylation of these genes in lung cancer. In spite of the relatively small number of never-smokers available for our study, the strength of our data was the homogeneity of the lung cancer patient population, all of whom were white and from the Western Pennsylvania region. These results may be useful for the future study of smoking-related epigenetic changes in lung carcinogenesis. It will be of interest to further investigate methylation differences between smokers and never-smokers in other genes that have been found to be frequently hypermethylated in lung tumors and in a larger number of patients.
Footnotes
This work was supported by grants from the American Cancer Society (grants RPG-99-16101-CNE and RSG-99-161-04-CNE).
References
- 1.American Cancer Society Cancer facts and figures 2004. 2004 http://www.cancer.org/
- 2.Boyle P, Ferlay J. Cancer incidence and mortality in Europe, 2004. Ann Oncol. 2005;16:481–488. doi: 10.1093/annonc/mdi098. [DOI] [PubMed] [Google Scholar]
- 3.Mumford JL, He XZ, Chapman RS, Cao SR, Harris DB, Li XM, Xian YL, Jiang WZ, Xu CW, Chuang JC, et al. Lung cancer and indoor air pollution in Xuan Wei, China. Science. 1987;235:217–220. doi: 10.1126/science.3798109. [DOI] [PubMed] [Google Scholar]
- 4.Fontham ET, Correa P, WuWilliams A, Reynolds P, Greenberg RS, Buffler PA, Chen VW, Boyd P, Alterman T, Austin DF, et al. Lung cancer in nonsmoking women: a multicenter case-control study. Cancer Epidemiol Biomark Prev. 1991;1:35–43. [PubMed] [Google Scholar]
- 5.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 6.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 7.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 8.Sherr CJ. G1 phase progression: cycling on cue. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 9.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 10.Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 11.Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, Davidson NE, Sidransky D, Baylin SB. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 12.Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, Baylin SB, Herman JG. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci USA. 1998;95:11891–11896. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bearzatto A, Conte D, Frattini M, Zaffaroni N, Andriani F, Balestra D, Tavecchio L, Daidone MG, Sozzi G. p16(INK4A) hypermethylation detected by fluorescent methylation-specific PCR in plasmas from non-small cell lung cancer. Clin Cancer Res. 2002;8:3782–3787. [PubMed] [Google Scholar]
- 14.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyl-transferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 15.Esteller M, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, Watkins DN, Issa JP, Sidransky D, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyl-transferase by promoter hypermethylation is associated with G to A mutations in K-ras in colorectal tumorigenesis. Cancer Res. 2000;60:2368–2371. [PubMed] [Google Scholar]
- 16.Pegg AE. Repair of O(6)-alkylguanine by alkyltransferases. Mutat Res. 2000;462:83–100. doi: 10.1016/s1383-5742(00)00017-x. [DOI] [PubMed] [Google Scholar]
- 17.Oue N, Shigeishi H, Kuniyasu H, Yokozaki H, Kuraoka K, Ito R, Yasui W. Promoter hypermethylation of MGMT is associated with protein loss in gastric carcinoma. Int J Cancer. 2001;93:805–809. doi: 10.1002/ijc.1403. [DOI] [PubMed] [Google Scholar]
- 18.Whitehall VL, Walsh MD, Young J, Leggett BA, Jass JR. Methylation of O-6-methylguanine DNA methyltransferase characterizes a subset of colorectal cancer with low-level DNA microsatellite instability. Cancer Res. 2001;61:827–830. [PubMed] [Google Scholar]
- 19.Pulling LC, Divine KK, Klinge DM, Gilliland FD, Kang T, Schwartz AG, Bocklage TJ, Belinsky SA. Promoter hypermethylation of the O6-methylguanine-DNA methyltransferase gene: more common in lung adenocarcinomas from never-smokers than smokers and associated with tumor progression. Cancer Res. 2003;63:4842–4848. [PubMed] [Google Scholar]
- 20.Gealy R, Zhang L, Siegfried JM, Luketich JD, Keohavong P. Comparison of mutations in the p53 and K-ras genes in lung carcinomas from smoking and nonsmoking women. Cancer Epidemiol Biomark Prev. 1999;8:297–302. [PubMed] [Google Scholar]
- 21.Herman JG, Graff JR, Myohane S, Nelkin BFD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmisano WA, Divine KK, Saccomanno G, Gilliland FD, Baylin SB, Herman JG, Belinsky SA. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000;60:5954–5958. [PubMed] [Google Scholar]
- 23.Toyooka S, Maruyama R, Toyooka KO, Mclerran D, Feng Z, Fukuyama Y, Virmani AK, Zochauer-Muller S, Tsukuda K, Sugio K, et al. Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int J Cancer. 2003;103:153–160. doi: 10.1002/ijc.10787. [DOI] [PubMed] [Google Scholar]
- 24.Kim DH, Nelson HH, Wiebcke JK, Zheng S, Christiani DC, Wain JC, Mark EJ, Kelsey KT. p16(INK4a) and histology-specific methylation of CpG islands exposure to tobacco smoke in non-small cell lung cancer. Cancer Res. 2001;61:3419–3424. [PubMed] [Google Scholar]
- 25.Sanchez-Cespedes M, Decker PA, Doffek KM, Esteller M, Westra WH, Alawi EA, Herman JG, Demeure MJ, Sidransky D, Ahrendt SA. Increased loss of chromosome 9p21 but not p16 inactivation in primary non-small cell lung cancer from smokers. Cancer Res. 2001;61:2092–2096. [PubMed] [Google Scholar]
- 26.Divine KK, Pulling LC, Marron-Terada PG, Liechty KC, Kang T, Schwartz AG, Bocklage TJ, Coons TA, Gilliland FD, Belinsky SA. Multiplicity of abnormal promoter methylation in lung adenocarcinomas from smokers and never smokers. Int J Cancer. 2005;114:400–405. doi: 10.1002/ijc.20761. [DOI] [PubMed] [Google Scholar]
- 27.Keohavong P, DeMichele MA, Melacrinos AC, Landreneau RJ, Weyant RJ, Siegfried JM. Detection of K-ras mutations in lung carcinomas: relationship to prognosis. Clin Cancer Res. 1996;2:411–418. [PubMed] [Google Scholar]
- 28.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, Fong KM, Lee H, Toyooka S, Shimizu N, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 29.Sonobe M, Manabe T, Wada H, Tanaka F. Mutations in the epidermal growth factor receptor gene are linked to smoking-independent, lung adenocarcinoma. Br J Cancer. 2005;93:355–363. doi: 10.1038/sj.bjc.6602707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 31.Issa JP, Baylin SB, Belinsky SA. Methylation of the estrogen receptor CpG island in lung tumors is related to the specific type of carcinogen exposure. Cancer Res. 1996;56:3655–3658. [PubMed] [Google Scholar]
- 32.Grafstrom RC, Dypbukt JM, Sundqvist K, Atzori L, Nielsen I, Curren RD, Harris CC. Pathobiological effects of acetaldehyde in cultured human epithelial cells and fibroblasts. Carcinogenesis. 1994;15:985–990. doi: 10.1093/carcin/15.5.985. [DOI] [PubMed] [Google Scholar]