Abstract
The critical clinical question in prostate cancer research is: How do we develop means of distinguishing aggressive disease from indolent disease? Using a combination of proteomic and expression array data, we identified a set of 36 genes with concordant dysregulation of protein products that could be evaluated in situ by quantitative immunohistochemistry. Another five prostate cancer biomarkers were included using linear discriminant analysis, we determined that the optimal model used to predict prostate cancer progression consisted of 12 proteins. Using a separate patient population, transcriptional levels of the 12 genes encoding for these proteins predicted prostate-specific antigen failure in 79 men following surgery for clinically localized prostate cancer (P = .0015). This study demonstrates that cross-platform models can lead to predictive models with the possible advantage of being more robust through this selection process.
Keywords: Metastasis, cancer, proteomics, prostate cancer, bioinformatics
Introduction
Expression array technology has led to the development of discrete molecular signatures. These signatures often involve many genes that are able to characterize two or more states, such as a model developed to identify aggressive, diffuse, large B-cell lymphomas [1]. Various computational strategies have been used to reduce these large lists of genes to smaller nonredundant gene lists. A recent example is a 70-gene prognostic panel predicting women at risk of dying from breast cancer [2]. Another example is a 17-gene signature of cancer metastasis [3]. Interestingly, several solid tumors harboring this 17-gene signature were at a significantly greater risk of disease progression. Other expression array studies examining prostate cancer progression have determined that as few as four to five genes could identify men with a greater chance of prostate cancer progression [4,5]. Thus, compelling evidence exists, suggesting that clinically relevant predictive molecular models of cancer progression are composed of less than 100 and more likely significantly fewer genes after the removal of redundant genes from the initial high-throughput screen.
Strategies to identify critical genes are emerging. Meta-analysis of expression array studies has determined robust candidate genes [6]. This approach identifies genes that are differentially expressed regardless of the platform used for discovery and represents one approach to sort through the mounting expression array data being compiled [7]. This general informatics approach to gene selection was recently validated in the classification of diffuse, large B-cell lymphomas by demonstrating that, using genomewide expression array data, one can boil down the principle components to six genes that can predict clinical outcome [8].
In an attempt to further integrate various sources of molecular data with the goal of determining critical genes, we recently combined data from a focused proteomics study and expression array analysis [9]. In this prior study, we interrogated over 1354 antibodies against distinct proteins or post-translational modifications to interrogate tissue extracts derived from benign prostate, clinically localized prostate cancer (LPCa), and metastatic prostate cancer. More weight was given to genes that were both overexpressed at the transcriptional level and simultaneously overexpressed at the protein level. Using integrative analysis of this compendium of proteomic alterations and transcriptome data derived from eight prostate cancer profiling studies, we observed only a 60% concordance between protein and transcriptional levels. One important result was identifying a subset of genes that predicted clinical outcome based on this concordant overexpression at the mRNA and protein levels. An intriguing observation from this initial study was that a subset of 44 genes was sufficient to develop models of cancer progression that are applicable not only to prostate cancer but to other tumors as well. These models could be tested by using expression array data sets that have associated clinical outcome. One of the future goals of the current approach should lead to the development of a clinical tissue-based test that can be assessed in biopsy samples. The current study therefore builds on this work by refining the method of measuring the in situ expression of these proteins using a prostate cancer progression tissue microarray (TMA). Here, we present a molecular signature, composed of 12 genes, that characterizes prostate cancer progression.
Materials and Methods
Case Selection
As previously described, we have developed a prostate cancer progression TMA to test biomarkers [10]. This TMA is composed of benign prostate tissue (BEN), localized prostate cancer, hormone-naïve metastatic prostate cancer (META), and hormone-refractory metastatic prostate cancer (WAP). These cases came from well-fixed radical prostatectomy, lymph node, and metastatic prostate cancer specimens from the University of Michigan (Ann Arbor, MI), the University Hospital of Ulm (Ulm, Germany), and the rapid autopsy program [University of Michigan Specialized Program of Research Excellence (SPORE)] for prostate cancer [11,12]. The metastatic samples from the rapid autopsy program were all histologically confirmed prostatic tumors involving solid organs (e.g., liver and lung) or distant lymph nodes, as recently described [11]. All samples were collected with prior Institutional Review Board approval at each respective institution.
Selection of Biomarkers for Immunohistochemistry
The majority of biomarkers for this study were derived from a large-scale proteomics study where more than 1354 proteins were screened [9]. Refinement of this list of proteins included coordinate overexpression or underexpression by cDNA expression array analysis [6,7,13,14]. The initial selection process identified 50 dysregulated proteins, of which 36 were optimized for in situ tissue evaluation by immunohistochemistry on archival formalin-fixed, paraffin-embedded materials [9]. Five additional prostate cancer biomarkers [Kruppel-like factor 6 (KLF6), Muc1, p63, Ki67, and zinc alpha-2-glycoprotein (ZAG)] were included in this multiplex model based on their association with cancer progression. Forty-one biomarkers are presented in Table 1. This list includes prostate-specific antigen (PSA) [15,16], alpha-methylacyl CoA racemase (AMACR) [17–21], E-cadherin n[22–26], p27 [27–31], fatty acid synthase (FAS) [32–35], Mib1/Ki67 [36–38], and androgen receptor (AR) [39]. This list also includes genes that have been more recently associated with prostate cancer, such as E2F [40–43], enhancer of Zeste 2 (EZH2) [13,44–46], Jagged 1 [47,48], metastasis-associated gene 1 (MTA1) [45,49], p63 [50–52], ZAG [53,54], MUC1 [53,55], and X-linked inhibitor of apoptosis (XIAP) [56,57], which have also been recently associated with prostate cancer progression; tumor protein D 52 (TPD52), a candidate oncogene identified in 8q21 amplicon, was recently identified to be associated with prostate cancer progression and the development of hormonerefractory prostate cancer [58–64]. The remaining genes, such as ABP280 (Filamin A locus, FLNA), JAM1, BM28, and FAS (a protein that had been known to be overexpressed in prostate cancer and consistently seen to be overexpressed in expression array studies), have not been specifically associated with prostate cancer progression [6,33,35].
Table 1.
Data Description: Mean Values and 95% CIs for 41 Markers and 5 Groups.
| BEN | LPCa | META | WAP | SM_CL | ||||||
| Mean | 95% CI | Mean | 95% CI | Mean | 95% CI | Mean | 95% CI | Mean | 95% CI | |
| ABP280 | 0.65 | 0.69 | 0.42 | 1.02 | -1.61 | 0.71 | -0.63 | 1.46 | -0.57 | 0.02 |
| AMACR | -0.87 | 0.22 | 0.76 | 1.05 | -0.20 | 2.05 | 0.64 | 2.82 | -0.36 | 0.32 |
| AR | 0.33 | 1.77 | 0.37 | 1.57 | -0.40 | 1.97 | -0.44 | 2.14 | -1.73 | 1.09 |
| BM28 | -0.63 | 0.44 | 0.11 | 1.41 | 0.07 | 2.98 | 0.54 | 1.90 | 2.27 | 1.64 |
| BUB3 | 0.01 | 0.73 | 0.42 | 1.10 | -0.13 | 2.83 | -0.82 | 3.35 | 0.27 | 1.73 |
| CaMKK | 0.03 | 1.45 | 0.18 | 0.86 | 0.18 | 2.99 | -0.05 | 3.08 | -0.55 | 0.95 |
| CASPASE3 | 1.00 | 0.89 | -0.30 | 1.15 | -0.17 | 1.94 | -1.35 | 0.84 | -1.03 | 0.68 |
| CDK7 | 0.18 | 1.09 | 0.33 | 1.47 | -0.58 | 2.39 | 0.19 | 3.00 | -1.84 | 1.30 |
| DYNAMIN | 0.10 | 1.32 | 0.73 | 1.05 | -0.74 | 1.80 | -0.83 | 2.25 | 0.25 | 2.74 |
| E2F-1 | -0.15 | 1.16 | 0.07 | 1.22 | 0.46 | 2.70 | -0.56 | 2.02 | -0.12 | 5.72 |
| E-cadherin | -0.03 | 1.33 | 0.31 | 1.48 | -0.66 | 2.00 | 0.64 | 2.13 | -2.42 | 1.93 |
| EXPORTIN | 0.02 | 1.25 | 0.24 | 1.44 | -0.18 | 2.41 | -0.21 | 3.96 | -0.40 | 0.95 |
| EZH2 | -0.46 | 0.83 | 0.08 | 1.31 | 0.00 | 2.24 | 0.45 | 1.76 | 2.77 | 3.89 |
| FAS | -0.28 | 2.00 | 0.08 | 1.58 | 0.22 | 2.54 | 0.47 | 2.40 | -0.49 | 1.15 |
| GAS7 | 0.20 | 1.54 | -0.09 | 1.49 | 0.05 | 2.27 | 0.24 | 2.83 | 0.12 | 3.13 |
| GS28 | 0.18 | 0.84 | 0.28 | 0.91 | 0.34 | 0.53 | -1.00 | 4.36 | -0.53 | 2.19 |
| ICBP90 | -0.42 | 0.98 | 0.58 | 1.86 | -0.47 | 2.30 | -0.46 | 1.23 | 1.95 | 2.58 |
| ITGA5 | 0.15 | 1.14 | 0.18 | 1.25 | -1.34 | 1.31 | 0.31 | 1.42 | -0.40 | 0.43 |
| Jagged 1 | -0.77 | 0.71 | -0.30 | 0.47 | 1.11 | 0.98 | 1.23 | 2.20 | 1.15 | 0.74 |
| JAM1 | -0.03 | 1.16 | 0.06 | 0.85 | -1.24 | 2.43 | 0.75 | 1.37 | 1.56 | 3.48 |
| KANADAPTIN | -0.61 | 1.22 | 0.23 | 1.12 | -0.24 | 1.69 | 1.66 | 1.34 | -1.54 | 1.25 |
| KLF6 | -0.09 | 1.41 | -0.05 | 1.24 | -0.62 | 2.66 | 1.11 | 2.60 | -0.02 | 2.50 |
| KRIP1 | -0.35 | 1.43 | 0.33 | 1.58 | 0.02 | 1.66 | -0.15 | 1.90 | 1.57 | 6.30 |
| LAP2 | -0.10 | 0.90 | -0.02 | 1.14 | -0.71 | 2.75 | 0.92 | 2.39 | 1.18 | 4.18 |
| MCAM | -0.40 | 0.86 | 0.05 | 1.50 | 0.93 | 2.72 | -0.16 | 2.38 | -1.32 | 1.42 |
| MIB1 (MKi67) | -0.49 | 0.11 | -0.17 | 0.52 | -0.36 | 0.44 | 1.21 | 3.31 | 3.05 | 0.79 |
| MTA1 | -0.25 | 1.04 | 0.67 | 0.93 | -0.99 | 2.33 | 0.38 | 2.91 | -0.32 | 2.51 |
| MUC1 | -0.33 | 0.26 | -0.30 | 1.28 | -0.24 | 0.87 | 1.16 | 3.21 | 2.75 | 0.71 |
| Myosin VI | -0.48 | 1.04 | 0.24 | 1.73 | 0.58 | 2.10 | 0.49 | 2.66 | -1.40 | 0.68 |
| P27 | 0.62 | 1.42 | 0.20 | 1.78 | -0.26 | 2.05 | -1.30 | 1.06 | 0.04 | 0.11 |
| P63 | 1.30 | 0.48 | -0.76 | 0.26 | -0.77 | 0.24 | -0.27 | 1.84 | -0.34 | 0.48 |
| PAXILLIN | 0.01 | 1.49 | 0.32 | 2.30 | -0.92 | 0.95 | 0.31 | 2.50 | 0.18 | 1.08 |
| PLCLN | -0.51 | 0.73 | 0.63 | 1.21 | 0.05 | 3.16 | -0.29 | 2.27 | 0.04 | 3.39 |
| PSA (KLK3) | 0.65 | 0.18 | 0.36 | 0.54 | -0.27 | 1.84 | -1.48 | 1.99 | -2.57 | 0.07 |
| RAB27 | 0.79 | 0.50 | 0.30 | 1.44 | -0.95 | 1.78 | -1.16 | 0.64 | -1.62 | 0.27 |
| RBBP | 0.33 | 1.37 | 0.16 | 1.44 | 0.01 | 2.21 | -0.75 | 3.52 | -0.60 | 0.52 |
| RIN1 | 1.00 | 0.54 | 0.35 | 1.36 | -1.16 | 0.51 | -1.13 | 0.46 | -1.11 | 0.25 |
| SAPK alpha | -0.05 | 1.18 | 0.05 | 0.99 | -0.09 | 2.78 | 0.22 | 3.14 | 1.36 | 2.75 |
| TPD52 | -0.11 | 0.71 | -0.04 | 1.35 | 1.22 | 2.42 | -1.05 | 2.44 | 0.05 | 1.32 |
| XIAP | -0.43 | 1.28 | 0.74 | 1.02 | -0.90 | 1.58 | 0.58 | 2.35 | -1.63 | 0.04 |
| ZAG | 0.42 | 1.14 | 0.34 | 2.17 | -0.93 | 1.58 | -0.42 | 1.76 | -1.33 | 0.19 |
Key: BEN= benign prostate tissue; LPCA=clinically localized prostate cancer; META= hormone naive metastatic prostate cancer; WAP=hormone refractory metastatic prostate cancer; SM_CL= small cell prostate cancer.
Several of the markers selected for analysis, such as E-cadherin [24,25,65] and XIAP [66], have been associated with cancer outcome. We also included some other biomarkers that have previously been reported to be associated with prostate cancer but were not identified in the screening study, such as KLF6, as this was reported to have a high level of loss of heterozygosity and mutation in prostate cancer [67] and was subsequently found to be one of five genes included in a molecular signature of aggressive prostate cancer [5].
Quantitative Biomarker Analysis
Protein expression was evaluated by immunohistochemistry using a semiautomated quantitative image analysis system ACIS II (Chromavision Medical Systems, Inc., San Juan Capistrano, CA). ACIS II consists of a microscope with a computer-controlled mechanical stage. Proprietary software is used to detect the brown stain intensity of the chromogen used for immunohistochemical analysis and to compare this value to the intensity of the blue counterstain used as background. Intensity levels are recorded as intensity units ranging from 0 to 255. The reproducibility of the ACIS II system was tested and confirmed by the scoring of the same TMA on separate occasions (r2 = 0.997; data not shown). Given the heterogeneity of prostate tissue samples, study pathologists used a computer-based selection tool to highlight areas within each 0.6-mm core for analysis. To account for this heterogeneity, we evaluated four tissue cores for each case. In cases where less than three cores were available, we substituted the data with the median value of the biomarker for histologic subtype. Missing values can arise both from corrupted core sections (i.e., technically inadequate) and from a change of diagnosis. Missing values were present in the data set: 98 times in benign prostate (13.3%), 130 times in localized prostate cancer (17.6%), and 6 times in metastatic prostate cancer samples (0.8%). The change of histologic diagnosis was not a rare event; therefore, it supports the need to review all TMA cores. As a pooling strategy, we adapted the mean TMA core value for each patient.
The diagnosis of the selected area was recorded in the database as either benign prostate, localized prostate cancer, or metastatic prostate cancer. Cores with only stroma or nondiagnostic areas were excluded from further analysis. Hematoxylin and eosin-stained images from this tissue array are available for review at a supplemental web site (http://rubinlab.tch.harvard.edu/supplemental_data/HTMA3_HE/index.jsp).
Statistical Analysis
Clustering Separate hierarchical agglomerative clustering both on samples and genes was carried out using Pearson correlation (as similarity measure) and average linkage method [68]. Clustering was performed using dChip software [69].
Linear discriminant analysis (LDA) LDA was applied on the data set of 41 genes to select genes [70,71] that can be discriminated among diagnostics groups. Discriminant analysis uses both multivariate analysis of variance and discriminant procedure to identify a linear combination of predictor variables that best characterizes differences among the groups. LDA computes so-called canonical variables (or canonical discriminant functions). The first canonical variable is a linear combination of variables that maximizes the differences between the means of groups (one dimension). The second canonical variable represents the maximum dispersion of the means in a direction that is orthogonal to the first canonical variable. The other canonical variables are generated in a similar manner.
By applying a stepwise approach (adding and removing variables on variance evaluation), the most powerful subset of predicting variables can be defined. Stepwise selection begins by identifying the variable for which the means are most different and continues by stepwise addition of the next best variable. Wilks' lambda was used to control the entry or removal of predictor variables from discriminant functions. In discriminant analysis, prior probabilities were computed from group sizes. LDA was performed using R [72] and SPSS (SPSS, Inc., Chicago, IL).
Validation using expression array analysis Expression data from a well-annotated, publicly available data set of 79 localized prostate tumors were obtained from Glinsky et al. [5]. Features representing the 12 genes identified in protein expression analysis were determined for U95Av2 microarrays and mapped to U133A microarrays using the “Best Match” table provided by Affymetrix (Santa Clara, CA). Clustering was performed using the dChip software, as described above [69]. Two major clusters determined using the first branch point of the data set (C0 and C1) were identified, and a chi-square test was performed to determine if the distribution between C0 and C1 was nonrandom with respect to clinical outcome (PSA failure versus nonfailure). These two clusters (C0 vs C1), along with observed class (nonrecurrent versus recurrent) and time to outcome (censorship or recurrence), were imported into GraphPad Prism to generate a Kaplan-Meier plot and to calculate the log rank statistic.
Results
Selection of Genes for Analysis
Using a high-throughput proteomic screen of prostate tissue extracts, we identified a panel of 50 proteins from over 1354 that were differentially expressed [9]. In prior work, this panel was evaluated by Western blot analysis, and the candidate proteins that best distinguished between benign prostate tissue, localized prostate cancer, and metastatic prostate cancer were selected for further analysis. Further selection required that these proteins were also concordantly dysregulated at the transcriptome level, as previously described [9]. We were able to optimize antibodies against 36 of these proteins that also worked on formalin-fixed paraffin-embedded tissue samples. An additional five genes were included based on prior association with prostate cancer progression. Immunohistochemistry was then performed on a prostate cancer progression TMA that has been previously described and consists of BEN, LPCa, META, and WAP [10]. Immunohistochemical staining intensity was scored using an automated image analysis system. The protein expression of these genes, with mean staining intensity scores and 95% confidence intervals (CIs), is presented in Table 1.
Hierarchical Clustering Results
We performed high-level analysis to check the data quality present in the set of 41 selected proteins. In particular, we investigated, through hierarchical clustering, if sufficient protein expression data could distinguish different states of prostate disease. Clustering was separately carried out on the samples and the 41 proteins. The highest levels of the sample tree (Figure 1A) demonstrated a good separation between aggressive prostate cancer states and LPCa. The clustering also reliably distinguished BEN from LPCa. Although metastatic tumors clustered together, no clear subclusters were found for META and WAP, as demonstrated in Figure 1A. Two cases of metastatic small cell prostate cancer (SM_CL) clustered together (Figure 1B, top image). A sample of localized prostate cancer (LPCa_442-GL_7) was naturally grouped with metastatic tumors. Although the overall Gleason score for this case was 7, the sample analyzed for this study (depicted in Figure 1B) demonstrated pure Gleason pattern 4 prostate cancer consistent with a high-grade tumor. When clustering genes based on samples, it is notable that a group of seven genes (p63, ZAG, ABP280, RAB27, RIN1, CASPASE3, and PSA) was overexpressed in benign tissues and underexpressed in aggressive cancer. Extreme overexpression and underexpression of these proteins are present for aggressive cancer types (Figure 1, right side), supporting the hypothesis that the investigated set of markers might distinguish aggressive prostate cancer from indolent prostate cancer. A heat map also suggests that some genes provide redundant or partially redundant information, as confirmed by descriptive statistics presented in Table 1.
Figure 1.
Protein expression of 41 genes selected for differential expression in prostate cancer progression. (A) A “heat map” showing the relative protein expression of 41 genes selected as highly likely to demonstrate differential expression in prostate tissue samples along the spectrum of cancer progression. Protein expression was determined using antibodies directed against gene products and measured by immunohistochemistry and using a semiautomated image analysis system (ACIS II; Chromavision Medical Systems, Inc.), which measures staining intensity along a continuous scale from 0 to 255. Low and high expressions are depicted using light green and bright red, respectively. Hierarchical clustering of the samples demonstrates a good—but not perfect—ability of this 41-gene panel to distinguish between classes. (B) Although some subtypes of metastatic prostate cancer (small cell cancer) (upper figure; original magnification, x 200) had discrete profiles, the clustering did not accurately distinguish between all of the hormone-naïve and hormone-refractory prostate tumor samples. Interestingly, a high-grade LPCa (Gleason pattern 4 prostate cancer) (lower figure; original magnification, x 200) was found to cluster more closely to the metastatic samples using this 41-gene profile.
LDA
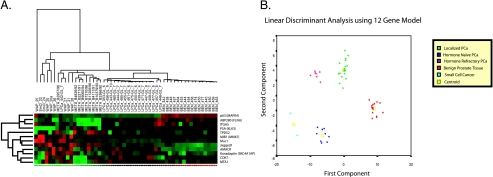
To verify the discriminative power of genes in terms of cancer progression and to identify gene profiles specific for localized prostate cancer and advanced prostate cancer, we developed a predictive model based on protein expression. LDA was applied to identify a linear combination of predictor variables that best characterizes differences among the groups. A clear separation of the groups was found, as depicted in Figure 2. The first and the second canonical variables cumulatively account for 91.7% of the variance (68.1 % and 23.6%, respectively). This result suggests that different groups (benign, localized cancer, hormone-naive metastases, hormone-refractory metastases, and small cell cancer) are linearly separable in gene space.
Figure 2.
Stepwise LDA identified a group of 12 genes that best predicts prostate cancer progression (Table 2). (A) The heat map for these 12 genes. (B) Cases along the first and the second canonical components of the LDA, which account for a cumulative variance of 87.9%. The discriminative power of the 12-gene model was not decreased with respect to the 41-gene model, confirming also the redundancy of information provided by some genes.
Stepwise LDA identified a set of 12 genes from the original set of 41 studied genes that best predicted tumor progression (Table 2) using 52 cases. Figure 2B represents the cases along the first and the second canonical components, which account for a cumulative variance of 87.9%. The discriminative power of the 12-gene model was not decreased with respect to the 41-gene model, confirming also the redundancy of information provided by some genes; alternative subsets of genes from the 41-gene set could be selected. Even though the model accuracy evaluated by crossvalidation (both using training and test sets 2/3 to 1/3 and leave-one-out) was very good, reliable performances need to be assessed on a different larger data set.
Table 2.
Stepwise LDA Data: Fisher Linear Discriminant Function Coefficients (Classification Model).
| Groups | |||||
| BEN | LPCa | META | WAP | SM_CL | |
| ABP280(FLNA) | 12.05 | 1.74 | -16.81 | -10.88 | -8.60 |
| AMACR | 3.18 | 3.80 | -10.10 | -1.78 | -14.03 |
| CDK7 | 3.39 | 1.51 | -2.94 | -3.56 | -19.28 |
| ITGA5 | -7.43 | 3.77 | -2.73 | 9.44 | 0.27 |
| Jagged 1 | -10.04 | -1.60 | 11.47 | 11.76 | 13.77 |
| KANADAPTIN | -2.57 | 0.49 | 0.00 | 6.20 | -3.41 |
| MIB1 (MKi67) | -6.06 | -4.08 | 5.75 | 8.76 | 32.84 |
| MTA1 | 2.18 | 2.32 | -8.73 | 0.02 | -3.26 |
| MUC1 | -12.92 | 3.43 | 4.59 | 12.31 | 19.73 |
| p63 | 41.51 | -10.06 | -24.94 | -31.61 | -49.72 |
| PSA(KLK3) | 11.82 | 2.26 | -7.45 | -14.78 | -38.07 |
| TPD52 | 2.07 | -4.91 | 6.93 | -3.88 | 12.23 |
| Constant | -42.68 | -9.22 | -42.86 | -47.72 | -170.25 |
Expression Array Validation
The genes identified from the original proteomics screen were selected because they demonstrated either overexpression or underexpression at both the protein and transcriptional levels [9]. In the current study, we have refined this discriminatory panel to 12 genes. To determine if RNA expression of these 12 genes could discriminate between local prostate cancers that progress following radical prostatectomy from those that do not, a previously published data set of 79 tumors was analyzed [5]. In an approach described by Ramaswamy et al. [3], we used the dChip software to develop a hierarchical cluster of all samples using features representing the 12 genes included in our model or 15 features on U133A (Figure 3A). When the 79 cases were clustered using the dChip software, recurrent and nonrecurrent samples were nonrandomly distributed between the two major clusters (P < .01) (Figure 3A). When these major clusters (C0 and C1) were used as a categorical variable to divide the set for Kaplan-Meier analysis (Figure 3B), a significant separation was observed between samples within the two clusters with respect to PSA failure following surgery (P = .0021). Thus, RNA expression of this group of genes also appears to distinguish between localized tumors that are likely to be aggressive from those that are cured by surgery.
Figure 3.
Expression array clustering of 79 clinical cases and Kaplan-Meier analysis. To validate if the expression of these 12 genes distinguishes men with more aggressive prostate cancer, we employed an independent data set from a previously published study of 79 tumors analyzed at the Memorial Sloan Kettering Cancer Center (New York, NY). (A) The first hierarchical clustering of the 79 samples, using the features representing the 12 genes included in our model (15 features on U133A), was performed. When the 79 cases were clustered using the dChip software, recurrent and nonrecurrent samples were nonrandomly distributed between the two major clusters (P < .01). (B) When these major clusters (C0 and C1) were used as categorical variables to divide the set for Kaplan-Meier analysis, a significant separation was observed between samples within the two clusters with respect to PSA failure following surgery (P = .0021). Thus, RNA expression of this group of genes also appears to distinguish between localized tumors that are likely to be aggressive from those that are cured by surgery.
Discussion
Prostate cancer progression is a complex process involving many genes and pathways [73,74]. Although some alterations may be critical during early development (e.g., 8p loss), more advanced metastatic prostate cancers demonstrate numerous molecular alterations that may not be causative but may instead be seen later during progression as a consequence of genetic instability (e.g., PTEN mutations). It is also evident from multiple expression array studies that the genes identified to distinguish different disease states (such as cancer versus benign) may differ. This may be due to different molecular platforms, samples used for investigation, treatment effects, or analytic approaches used. One approach used to develop a robust model has been the determination of which genes are consistently differentially regulated from experiment to experiment using a metaanalysis of expression array data [6,7]. Using this approach, we have focused on genes that were consistently dysregulated both at the protein and expression array levels [9] (Figure 4). This has now led to a focused model, including 12 genes from a starting point of over 1354 genes used in the initial proteomics screening [9]. It is intriguing that this 12-gene model of prostate cancer progression, which was initially developed using a TMA, was able to distinguish men with LPCa who were at highest risk of developing PSA failure following surgery. This testing and validation study crossed platforms but was still able to predict outcomes on an entirely independent clinical cohort.
Figure 4.
The stages of biomarker development are presented schematically in this figure. The first critical step is the identification of high-quality samples used to perform high-throughput analysis, including proteomics and expression array analysis. In this study, a wide range of prostate cancer samples was used from several sources. Meta-analysis was performed to ensure that the genes identified in one study are genes that also have been determined to be dysregulated in other studies. The data were then tested using TMAs, with the goal of developing a robust model. In the current study, a panel of 12 genes was determined. This panel of genes was then validated on an independent data set to discriminate aggressive from indolent forms of prostate cancer. Throughout the entire process, bioinformatics is required to prioritize and refine the molecular models.
Larger numbers of genes also might predict outcome. However, as seen in the analysis of microarray studies, there are many redundant genes. The reason for associated gene expression patterns may be explained by the activation of similar molecular pathways or general processes such as proliferation or apoptosis. This observation supports the view that one should, in fact, be surprised if all studies came up with the same sets of genes. For example, in the current study, after removing the 12-gene model from the original 41 genes, we can also identify good models from the remaining genes. Therefore, the approach used in this study should be considered as a paradigm to identify predictive gene sets, but should not be viewed as the only possible solution. This study further supports the view that most of the large molecular profiles have redundancy built into them. Methods such as integrative proteomic and genomic analysis demonstrate how critical it is to develop strategies to refine large gene sets [9]. Other methods such as gene set enrichment analysis [75–77] can also help define pathways of dysregulated genes, allowing us to focus on sets of genes or gene interactions as opposed to lists of nonannotated genes.
Recent work from Glinsky et al. [78], which used a stem cell-like expression profile selected by comparing two cell lines, led to the identification of an 11-gene model. Unlike the current study that used human tumor samples to identify and refine genes for progression profiling, Glinsky et al. identified their focused profile by comparing BMI-1-overexpressing cells to a sister cell line that expresses BMI-1 at lower levels. Previous work had demonstrated that BMI-1 plays a critical role in the ability of stem cells to maintain self-renewal [79,80]. Their novel signature was associated with poor clinical outcome in 11 different types of cancer, including prostate, breast, lung, ovarian, and bladder cancers. It is intriguing that the initial study of Varambally et al. [9] and Glinsky et al. [78] using expression array profiles determined a small set associated with poor clinical outcome. Our current study focused on developing a tissue-based protein profile, which also identified a small set of genes required to best define prostate cancer progression.
In summary, we have developed a model of prostate cancer progression that was developed using a multistage approach. The first selection process sorted through over 1354 genes by evaluating a combination of cDNA expression array analysis and a high-throughput proteomic screening, which led to the identification of 50 differentially expressed genes. The second stage described in the current study tested a prostate cancer progression model using a quantitative analysis of protein expression using immunohistochemistry on a TMA. This led to a 12-gene model that was validated on a separate patient cohort using PSA failure following surgery with LPCa as endpoint. This study demonstrates that cross-platform models can lead to predictive models. More importantly, this smaller model can be feasibly used in a clinical setting. Future work will test this model in a prospective manner.
Acknowledgements
The authors would like to thank Martina Storz-Schweizer and Lela Schumacher for excellent technical support.
Abbreviations
- PSA
prostate-specific antigen
- AMACR
alpha-methylacyl CoA racemase
- FAS
fatty acid synthase
- EZH2
enhancer of Zeste 2
- ZAG
zinc alpha-2-glycoprotein
- XIAP
X-linked inhibitor of apoptosis
- TPD52
tumor protein D 52
- KLF6
Kruppel-like factor 6
- MTA1
metastasis-associated gene 1
Footnotes
This work was supported by SPORE National Cancer Institute (NCI) grants P50CA90381 (M.A.R.), P50CA69568 (M.A.R. and A.M.C.), CA 97063 (A.M.C. and M.A.R.), and R01AG21404 (M.A.R.), American Cancer Society grant RSG-02-179-MGO (A.M.C. and M.A.R.), Department of Defense (PC051081 to A.M.C. and S.V.), Early Detection Research Network (U01 CA111275-01 to A.M.C. and M.A.R.), NIH Prostate Specialized Program of Research Excellence (SPORE) (P50CA69568 to A.M.C.), and the UM Cancer Center Support Grant (5P30 CA46592). A.M.C. is a Pew Biomedical Scholar.
Drs. Chinnaiyan and Rubin share cosenior authorship.
References
- 1.Shipp M-A, Ross K-N, Tamayo P, Weng A-P, Kutok J-L, Aguiar R-CT, Gaasenbeek M, Angelo M, Reich M, Pinkus G-S, et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 2002;8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- 2.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 3.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 4.Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA, D'Amico AV, Richie JP, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 5.Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest. 2004;113:913–923. doi: 10.1172/JCI20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhodes DR, Barrette TR, Rubin MA, Ghosh D, Chinnaiyan AM. Meta-analysis of microarrays: interstudy validation of gene expression profiles reveals pathway dysregulation in prostate cancer. Cancer Res. 2002;62:4427–4433. [PubMed] [Google Scholar]
- 7.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A Cancer Microarray Database and Integrated Data-Mining Platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lossos IS, Czerwinski DK, Alizadeh AA, Wechser MA, Tibshirani R, Botstein D, Levy R. Prediction of survival in diffuse largeB-cell lymphoma based on the expression of six genes. N Engl J Med. 2004;350:1828–1837. doi: 10.1056/NEJMoa032520. [DOI] [PubMed] [Google Scholar]
- 9.Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Shah RB, Chandran U, Monzon FA, Becich MJ, Wei JT, et al. Integrative proteomic and genomic analysis of prostate cancer progression. Cancer Cell. 2005;8(5):393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Rubin MA, Zerkowski MP, Camp RL, Kuefer R, Hofer MD, Chinnaiyan AM, Rimm DL. Quantitative determination of expression of the prostate cancer protein alpha-methylacyl-CoA racemase using Automated Quantitative Analysis (AQUA): a novel paradigm for automated and continuous biomarker measurements. Am J Pathol. 2004;164:831–840. doi: 10.1016/s0002-9440(10)63171-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah RB, Mehra R, Chinnaiyan AM, Shen R, Ghosh D, Zhou M, Macvicar S, Varambally S, Harwood J, Bismar TA, et al. Androgen-independent prostate cancer is a heterogeneous group of diseases: lessons from a rapid autopsy program. Cancer Res. 2004;64:9209–9216. doi: 10.1158/0008-5472.CAN-04-2442. [DOI] [PubMed] [Google Scholar]
- 12.Rubin MA, Putzi M, Mucci N, Smith DC, Wojno K, Korenchuk S, Pienta KJ. Rapid (“warm”) autopsy study for procurement of metastatic prostate cancer. Clin Cancer Res. 2000;6:1038–1045. [PubMed] [Google Scholar]
- 13.Rhodes DR, Sanda MG, Otte AP, Chinnaiyan AM, Rubin MA. Multiplex biomarker approach for determining risk of prostatespecific antigen-defined recurrence of prostate cancer. J Natl Cancer Inst. 2003;95:661–668. doi: 10.1093/jnci/95.9.661. [DOI] [PubMed] [Google Scholar]
- 14.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Large-scale metaanalysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci USA. 2004;101:9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stein BS, Vangore S, Petersen RO. Immunoperoxidase localization of prostatic antigens. Comparison of primary and metastatic sites. Urology. 1984;24:146–152. doi: 10.1016/0090-4295(84)90416-3. [DOI] [PubMed] [Google Scholar]
- 16.Brawn PN, Speights VO, Kuhl D, Riggs M, Spiekerman AM, McCord RG, Coffield KS, Stewart DT, Lind ML. Prostate-specific antigen levels from completely sectioned, clinically benign, whole prostates. Cancer. 1991;68:1592–1599. doi: 10.1002/1097-0142(19911001)68:7<1592::aid-cncr2820680721>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 17.Xu J, Stolk JA, Zhang X, Silva SJ, Houghton RL, Matsumura M, Vedvick TS, Leslie KB, Badaro R, Reed SG. Identification of differentially expressed genes in human prostate cancer using subtraction and microarray. Cancer Res. 2000;60:1677–1682. [PubMed] [Google Scholar]
- 18.Jiang Z, Woda BA, Rock KL, Xu Y, Savas L, Khan A, Pihan G, Cai F, Babcook JS, Rathanaswami P, et al. P504S: a new molecular marker for the detection of prostate carcinoma. Am J Surg Pathol. 2001;25:1397–1404. doi: 10.1097/00000478-200111000-00007. [DOI] [PubMed] [Google Scholar]
- 19.Rubin MA, Zhou M, Dhanasekaran SM, Varambally S, Barrette TR, Sanda MG, Pienta KJ, Ghosh D, Chinnaiyan AM. alpha-Methylacyl coenzyme A racemase as a tissue biomarker for prostate cancer. JAMA. 2002;287:1662–1670. doi: 10.1001/jama.287.13.1662. [DOI] [PubMed] [Google Scholar]
- 20.Luo J, Zha S, Gage WR, Dunn TA, Hicks JL, Bennett CJ, Ewing CM, Platz EA, Ferdinandusse S, Wanders RJ, et al. alpha-Methylacyl-CoA racemase: a new molecular marker for prostate cancer. Cancer Res. 2002;62:2220–2226. [PubMed] [Google Scholar]
- 21.Kuefer R, Varambally S, Zhou M, Lucas PC, Loeffler M, Wolter H, Mattfeldt T, Hautmann RE, Gschwend JE, Barrette TR, et al. alpha-Methylacyl-CoA racemase: expression levels of this novel cancer biomarker depend on tumor differentiation. Am J Pathol. 2002;161:841–848. doi: 10.1016/s0002-9440(10)64244-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bussemakers MJ, van Moorselaar RJ, Giroldi LA, Ichikawa T, Isaacs JT, Takeichi M, Debruyne FM, Schalken JA. Decreased expression of E-cadherin in the progression of rat prostatic cancer. Cancer Res. 1992;52:2916–2922. [PubMed] [Google Scholar]
- 23.Otto T, Rembrink K, Goepel M, Meyer-Schwickerath M, Rubben H. E-cadherin: a marker for differentiation and invasiveness in prostatic carcinoma. Urol Res. 1993;21:359–362. doi: 10.1007/BF00296837. [DOI] [PubMed] [Google Scholar]
- 24.Umbas R, Schalken JA, Aalders TW, Carter BS, Karthaus HF, Schaafsma HE, Debruyne FM, Isaacs WB. Expression of the cellular adhesion molecule E-cadherin is reduced or absent in high-grade prostate cancer. Cancer Res. 1992;52:5104–5109. [PubMed] [Google Scholar]
- 25.De Marzo AM, Knudsen B, Chan-Tack K, Epstein JI. E-cadherin expression as a marker of tumor aggressiveness in routinely processed radical prostatectomy specimens. Urology. 1999;53:707–713. doi: 10.1016/s0090-4295(98)00577-9. [DOI] [PubMed] [Google Scholar]
- 26.Rubin MA, Mucci NR, Figurski J, Fecko A, Pienta KJ, Day ML. E-cadherin expression in prostate cancer: a broad survey using high-density tissue microarray technology. Hum Pathol. 2001;32:690–697. doi: 10.1053/hupa.2001.25902. [DOI] [PubMed] [Google Scholar]
- 27.Guo Y, Sklar GN, Borkowski A, Kyprianou N. Loss of the cyclin-dependent kinase inhibitor p27(Kip1) protein in human prostate cancer correlates with tumor grade. Clin Cancer Res. 1997;3:2269–2274. [PubMed] [Google Scholar]
- 28.Cheville JC, Lloyd RV, Sebo TJ, Cheng L, Erickson L, Bostwick DG, Lohse CM, Wollan P. Expression of p27kip1 in prostatic adenocarcinoma. Mod Pathol. 1998;11:324–328. [PubMed] [Google Scholar]
- 29.Cordon-Cardo C, Koff A, Drobnjak M, Capodieci P, Osman I, Millard SS, Gaudin PB, Fazzari M, Zhang ZF, Massague J, et al. Distinct altered patterns of p27KIP1 gene expression in benign prostatic hyperplasia and prostatic carcinoma. J Natl Cancer Inst. 1998;90:1284–1291. doi: 10.1093/jnci/90.17.1284. [DOI] [PubMed] [Google Scholar]
- 30.Tsihlias J, Kapusta LR, DeBoer G, Morava-Protzner I, Zbieranowski I, Bhattacharya N, Catzavelos GC, Klotz LH, Slingerland JM. Loss of cyclin-dependent kinase inhibitor p27Kip1 is a novel prognostic factor in localized human prostate adenocarcinoma. Cancer Res. 1998;58:542–548. [PubMed] [Google Scholar]
- 31.Yang RM, Naitoh J, Murphy M, Wang HJ, Phillipson J, deKernion JB, Loda M, Reiter RE. Low p27 expression predicts poor disease-free survival in patients with prostate cancer. J Urol. 1998;159:941–945. [PubMed] [Google Scholar]
- 32.Epstein JI, Carmichael M, Partin AW. OA-519 (fatty acid synthase) as an independent predictor of pathologic state in adenocarcinoma of the prostate. Urology. 1995;45:81–86. doi: 10.1016/s0090-4295(95)96904-7. [DOI] [PubMed] [Google Scholar]
- 33.Shurbaji MS, Kalbfleisch JH, Thurmond TS. Immunohistochemical detection of a fatty acid synthase (OA-519) as a predictor of progression of prostate cancer. Hum Pathol. 1996;27:917–921. doi: 10.1016/s0046-8177(96)90218-x. [DOI] [PubMed] [Google Scholar]
- 34.Swinnen JV, Roskams T, Joniau S, Van Poppel H, Oyen R, Baert L, Heyns W, Verhoeven G. Overexpression of fatty acid synthase is an early and common event in the development of prostate cancer. Int J Cancer. 2002;98:19–22. doi: 10.1002/ijc.10127. [DOI] [PubMed] [Google Scholar]
- 35.Baron A, Migita T, Tang D, Loda M. Fatty acid synthase: a metabolic oncogene in prostate cancer? J Cell Biochem. 2004;91:47–53. doi: 10.1002/jcb.10708. [DOI] [PubMed] [Google Scholar]
- 36.Bubendorf L, Sauter G, Moch H, Schmid HP, Gasser TC, Jordan P, Mihatsch MJ. Ki67 labelling index: an independent predictor of progression in prostate cancer treated by radical prostatectomy. J Pathol. 1996;178:437–441. doi: 10.1002/(SICI)1096-9896(199604)178:4<437::AID-PATH484>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 37.Botticelli AR, Casali AM, Botticelli L, Zaffe D. Immunohistochemical detection of cell-cycle associated markers on paraffin embedded and formalin fixed needle biopsies of prostate cancer: correlation of p120 protein expression with AgNOR, PCNA/cyclin, Ki-67/MIB1 proliferation-scores and Gleason gradings. Eur J Histochem. 1998;42:41–48. [PubMed] [Google Scholar]
- 38.Bubendorf L, Tapia C, Gasser TC, Casella R, Grunder B, Moch H, Mihatsch MJ, Sauter G. Ki67 labeling index in core needle biopsies independently predicts tumor-specific survival in prostate cancer. Hum Pathol. 1998;29:949–954. doi: 10.1016/s0046-8177(98)90199-x. [DOI] [PubMed] [Google Scholar]
- 39.Gelmann EP. Molecular biology of the androgen receptor. J Clin Oncol. 2002;20:3001–3015. doi: 10.1200/JCO.2002.10.018. [DOI] [PubMed] [Google Scholar]
- 40.Park C, Lee I, Kang WK. Lovastatin-induced E2F-1 modulation and its effect on prostate cancer cell death. Carcinogenesis. 2001;22:1727–1731. doi: 10.1093/carcin/22.10.1727. [DOI] [PubMed] [Google Scholar]
- 41.Farhana L, Dawson M, Rishi AK, Zhang Y, Van Buren E, Trivedi C, Reichert U, Fang G, Kirschner MW, Fontana JA. Cyclin B and E2F-1 expression in prostate carcinoma cells treated with the novel retinoid CD437 are regulated by the ubiquitin-mediated pathway. Cancer Res. 2002;62:3842–3849. [PubMed] [Google Scholar]
- 42.Mu X, Chang C. TR3 orphan nuclear receptor mediates apoptosis through up-regulating E2F1 in human prostate cancer LNCaP cells. J Biol Chem. 2003;278:42840–42845. doi: 10.1074/jbc.M305594200. [DOI] [PubMed] [Google Scholar]
- 43.Foster CS, Falconer A, Dodson AR, Norman AR, Dennis N, Fletcher A, Southgate C, Dowe A, Dearnaley D, Jhavar S, et al. Transcription factor E2F3 overexpressed in prostate cancer independently predicts clinical outcome. Oncogene. 2004 doi: 10.1038/sj.onc.1207800. [DOI] [PubMed] [Google Scholar]
- 44.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 46.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin DB, Gifford DR, Wright ME, Keller A, Yi E, Goodlett DR, Aebersold R, Nelson PS. Quantitative proteomic analysis of proteins released by neoplastic prostate epithelium. Cancer Res. 2004;64:347–355. doi: 10.1158/0008-5472.can-03-2062. [DOI] [PubMed] [Google Scholar]
- 48.Santagata S, Demichelis F, Riva A, Varambally S, Hofer MD, Kutok JL, Kim R, Tang J, Montie JE, Chinnaiyan AM, et al. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res. 2004;64:6854–6857. doi: 10.1158/0008-5472.CAN-04-2500. [DOI] [PubMed] [Google Scholar]
- 49.Hofer MD, Kuefer R, Varambally S, Li H, Ma J, Shapiro GI, Gschwend JE, Hautmann RE, Sanda MG, Giehl K, et al. The role of metastasis-associated protein 1 in prostate cancer progression. Cancer Res. 2004;64:825–829. doi: 10.1158/0008-5472.can-03-2755. [DOI] [PubMed] [Google Scholar]
- 50.Signoretti S, Waltregny D, Dilks J, Isaac B, Lin D, Garraway L, Yang A, Montironi R, McKeon F, Loda M. p63 is a prostate basal cell marker and is required for prostate development. Am J Pathol. 2000;157:1769–1775. doi: 10.1016/S0002-9440(10)64814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parsons JK, Gage WR, Nelson WG, De Marzo AM. p63 protein expression is rare in prostate adenocarcinoma: implications for cancer diagnosis and carcinogenesis. Urology. 2001;58:619–624. doi: 10.1016/s0090-4295(01)01311-5. [DOI] [PubMed] [Google Scholar]
- 52.Davis LD, Zhang W, Merseburger A, Young D, Xu L, Rhim JS, Moul JW, Srivastava S, Sesterhenn IA. p63 expression profile in normal and malignant prostate epithelial cells. Anticancer Res. 2002;22:3819–3825. [PubMed] [Google Scholar]
- 53.Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hale LP, Price DT, Sanchez LM, Demark-Wahnefried W, Madden JF. Zinc alpha-2-glycoprotein is expressed by malignant prostatic epithelium and may serve as a potential serum marker for prostate cancer. Clin Cancer Res. 2001;7:846–853. [PubMed] [Google Scholar]
- 55.Kirschenbaum A, Itzkowitz SH, Wang JP, Yao S, Eliashvili M, Levine AC. MUC1 expression in prostate carcinoma: correlation with grade and stage. Mol Urol. 1999;3:163–168. [PubMed] [Google Scholar]
- 56.Nomura T, Mimata H, Takeuchi Y, Yamamoto H, Miyamoto E, Nomura Y. The X-linked inhibitor of apoptosis protein inhibits Taxol-induced apoptosis in LNCaP cells. Urol Res. 2003;31:37–44. doi: 10.1007/s00240-003-0300-y. [DOI] [PubMed] [Google Scholar]
- 57.Ng CP, Bonavida B. X-linked inhibitor of apoptosis (XIAP) blocks Apo2 ligand/tumor necrosis factor-related apoptosis-inducing ligand-mediated apoptosis of prostate cancer cells in the presence of mitochondrial activation: sensitization by overexpression of second mitochondria-derived activator of caspase/direct IAP-binding protein with low pl (Smac/DIABLO) Mol Cancer Ther. 2002;1:1051–1058. [PubMed] [Google Scholar]
- 58.Cher ML, MacGrogan D, Bookstein R, Brown JA, Jenkins RB, Jensen RH. Comparative genomic hybridization, allelic imbalance, and fluorescence in situ hybridization on chromosome 8 in prostate cancer. Genes Chromosomes Cancer. 1994;11:153–162. doi: 10.1002/gcc.2870110304. [DOI] [PubMed] [Google Scholar]
- 59.Macoska JA, Trybus TM, Wojno KJ. 8p22 loss concurrent with 8c gain is associated with poor outcome in prostate cancer. Urology. 2000;55:776–782. doi: 10.1016/s0090-4295(00)00468-4. [DOI] [PubMed] [Google Scholar]
- 60.Macoska JA, Trybus TM, Sakr WA, Wolf MC, Benson PD, Powell IJ, Pontes JE. Fluorescence in situ hybridization analysis of 8p allelic loss and chromosome 8 instability in human prostate cancer. Cancer Res. 1994;54:3824–3830. [PubMed] [Google Scholar]
- 61.Macoska JA, Trybus TM, Benson PD, Sakr WA, Grignon DJ, Wojno KD, Pietruk T, Powell IJ. Evidence for three tumor suppressor gene loci on chromosome 8p in human prostate cancer. Cancer Res. 1995;55:5390–5395. [PubMed] [Google Scholar]
- 62.Macoska JA, Micale MA, Sakr WA, Benson PD, Wolman SR. Extensive genetic alterations in prostate cancer revealed by dual PCR and FISH analysis. Genes Chromosomes Cancer. 1993;8:88–97. doi: 10.1002/gcc.2870080205. [DOI] [PubMed] [Google Scholar]
- 63.Wang R, Xu J, Saramaki O, Visakorpi T, Sutherland WM, Zhou J, Sen B, Lim SD, Mabjeesh N, Amin M, et al. PrLZ, a novel prostatespecific and androgen-responsive gene of the TPD52 family, is amplified in chromosome 8q21.1 and overexpressed in human prostate cancer. Cancer Res. 2004;64:1589–1594. doi: 10.1158/0008-5472.can-03-3331. [DOI] [PubMed] [Google Scholar]
- 64.Rubin MA, Varambally S, Beroukhim R, Tomlins SA, Rhodes DR, Paris PL, Hofer MD, Storz-Schweizer M, Kuefer R, Fletcher JA, et al. Overexpression, amplification, and androgen regulation of TPD52 in prostate cancer. Cancer Res. 2004;64:3814–3822. doi: 10.1158/0008-5472.CAN-03-3881. [DOI] [PubMed] [Google Scholar]
- 65.Umbas R, Isaacs WB, Bringuier PP, Schaafsma HE, Karthaus HF, Oosterhof GO, Debruyne FM, Schalken JA. Decreased E-cadherin expression is associated with poor prognosis in patients with prostate cancer. Cancer Res. 1994;54:3929–3933. [PubMed] [Google Scholar]
- 66.Krajewska Maryla KS, Banares S, Huang X, Turner B, Bubendorf L, Kallioniemi O-P, Shabaik A, Vitiello A, Peehl D, Gao G-J, et al. Elevated expression of inhibitor of apoptosis proteins in prostate cancer. Clin Cancer Res. 2003;9:4914–4925. [PubMed] [Google Scholar]
- 67.Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC, Glucksman MJ, Narla J, Eng FJ, Chan AM, et al. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science. 2001;294:2563–2566. doi: 10.1126/science.1066326. [DOI] [PubMed] [Google Scholar]
- 68.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiang H, Deng Y, Chen HS, Tao L, Sha Q, Chen J, Tsai CJ, Zhang S. Joint analysis of two microarray gene-expression data sets to select lung adenocarcinoma marker genes. BMC Bioinformatics. 2004;5:81. doi: 10.1186/1471-2105-5-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnson RA, Wichern DW. Applied Multivariate Statistical Analysis. 5th ed. Prentice Hall: Upper Saddle River; 2002. p. 767. (xviii) [Google Scholar]
- 72.Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 73.Rubin MA, De Marzo AM. Molecular genetics of human prostate cancer. Mod Pathol. 2004;17:380–388. doi: 10.1038/modpathol.3800051. [DOI] [PubMed] [Google Scholar]
- 74.De Marzo AM, Nelson WG, Isaacs WB, Epstein JI. Pathological and molecular aspects of prostate cancer. Lancet. 2003;361:955–964. doi: 10.1016/S0140-6736(03)12779-1. [DOI] [PubMed] [Google Scholar]
- 75.Monti S, Savage KJ, Kutok JL, Feuerhake F, Kurtin P, Mihm M, Wu B, Pasqualucci L, Neuberg D, Aguiar RC, et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood. 2004 doi: 10.1182/blood-2004-07-2947. [DOI] [PubMed] [Google Scholar]
- 76.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1 alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 77.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. From the cover: gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115:1503–1521. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 80.Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]