Abstract
A yeast mutant was isolated encoding a single amino acid substitution [serine-53 → proline (S53P)] in transcription factor TFIIB that impairs activation of the PHO5 gene in response to phosphate starvation. This effect is activation-specific because S53P did not affect the uninduced level of PHO5 expression, yet is not specific to PHO5 because Adr1-mediated activation of the ADH2 gene also was impaired by S53P. Pho4, the principal activator of PHO5, directly interacted with TFIIB in vitro, and this interaction was impaired by the S53P replacement. Furthermore, Pho4 induced a conformational change in TFIIB, detected by enhanced sensitivity to V8 protease. The S53P replacement also impaired activation of a lexA(op)-lacZ reporter by a LexA fusion protein to the activation domain of Adr1, thereby indicating that the transcriptional effect on ADH2 expression is specific to the activation function of Adr1. These results define an activation-specific role for TFIIB in vivo and suggest that certain activators induce a conformational change in TFIIB as part of their mechanism of transcriptional stimulation.
Keywords: SUA7, Pho4, PHO5, transcriptional control
Transcriptional activators stimulate gene expression by binding enhancer elements and contacting, either directly or indirectly, components of the RNA polymerase II (RNAPII) transcriptional machinery. There is considerable evidence that activation can occur by a recruitment mechanism (reviewed in ref. 1). Accordingly, activators bind general transcription factors (GTFs) to facilitate formation of the preinitiation complex (PIC). This idea is supported by direct contact in vitro between activators and GTFs, including TATA box-binding protein (TBP) (2), TFIIB (3), TFIIF (4), TFIIH (5), as well as the Rpb5 subunit of RNAPII (6), and TBP-associated factors (7, 8). Furthermore, artificial connections between DNA binding domains and either GTFs or components of the RNAPII holoenzyme complex stimulate transcription (reviewed in ref. 1), yet tethering of an activation domain to GTFs or RNAPII components fails to stimulate transcription (9). Thus, artificial recruitment of the RNAPII machinery to promoter DNA bypasses the activator requirement.
Although a compelling argument can be made for activation by recruitment, it does not account for activation of all genes. Other potential activation mechanisms include activator-induced conformational changes to either the promoter or to components of the PIC, stabilization of the PIC, covalent modification of the PIC, and counteractive effects on the repressive function of chromatin. An especially informative case is the Escherichia coli glnA gene, which is activated by ATP-dependent, NtrC-mediated isomerization of a σ54-holoenzyme complex from a closed to an open promoter complex (reviewed in ref. 1). In another case, the Epstein–Barr virus ZEBRA protein stimulates transcription by isomerization of the TATA-TBP-TFIIA ternary complex (10). Recently, a study of PIC formation and transcriptional activity demonstrated that PIC assembly occurs by at least two steps and that the TATA box and TFIIB also can affect transcription subsequent to PIC assembly (11). Thus, processes other than factor recruitment are potential activator targets.
TFIIB is recruited to the PIC by the acidic activator VP16 and the proline-rich activator CTF1 (3, 12). VP16 directly targets TFIIB (3) and this interaction is required for activation (13, 14). Interestingly, VP16 induces a conformational change in TFIIB that disrupts the intramolecular interaction between the N- and C-terminal domains in solution (15). However, it is not known whether the VP16 effect on TFIIB conformation affects transcription or if this effect is physiologically significant. Another study questioned the functional significance of the VP16-TFIIB interaction (16). Furthermore, the idea that TFIIB recruitment is, in general, a rate-limiting step for transcriptional activation has been challenged by TBP and TFIIB mutations that are defective for TBP-TFIIB-DNA complex formation in vitro, yet respond normally to activators in vivo (17, 18).
In the yeast Saccharomyces cerevisiae, TFIIB is encoded by the essential SUA7 gene (19). Yeast TFIIB is a monomeric protein of 345 aa that is structurally similar to human TFIIB, including a zinc binding motif near the N terminus and two imperfect repeats encompassing the C-terminal two-thirds of the molecule. The C-terminal domain forms a protease-resistant core (cIIB) (20, 21) that interacts with the N-terminal domain (nIIB) (15). In addition to the zinc binding domain, nIIB includes a phylogenetically conserved sequence that links the zinc binding domain to cIIB (22). A solution structure for human cIIB (23), and a cocrystal structure for a TATA-TBP-cIIB ternary complex (24), revealed that the two repeat domains are arranged in pseudo-dyad symmetry, each composed of five α-helices. The structure of free cIIB is comparable to the cocrystal structure, except that free cIIB is more compact and the relative orientation of the two repeats is different, suggesting that TFIIB undergoes a conformational change on assembly into the TATA-TBP-TFIIB complex (25). A tertiary structure for full-length TFIIB is not available, although an NMR structure for the N-terminal region of Pyrococcus TFIIB revealed that the metal binding motif forms a zinc ribbon similar to that of the elongation factor TFIIS (26).
In an effort to elucidate the role of TFIIB in transcriptional activation, we have generated and characterized a collection of yeast sua7 mutants that encode altered forms of TFIIB. Here we describe one of those mutants, which encodes a serine-53 to proline (S53P) replacement adjacent to the TFIIB zinc ribbon domain. This replacement impairs activation of specific genes in vivo and adversely affects TFIIB interaction with a cognate activator in vitro. Moreover, the transcriptional defect is specific to the activation function of the trans-activator. These results define an activation-specific role for TFIIB in vivo.
MATERIALS AND METHODS
Yeast Strains.
The two isogenic strains used in this study were generated as follows. The SUA7 gene, carried on a CEN HIS3 vector, was mutagenized by PCR under error-prone conditions (27) and introduced into yeast strain YIP91–13B [MATa CYC1 his3-Δ1 leu2–3, 112 trp1–289 ura3–52 ade1–100 sua7∷LEU2 (CEN-SUA7-URA3)] (28). His+ transformants were cured of the CEN-SUA7-URA3 plasmid, selecting for strains resistant to 5-fluoroorotic acid (FOAr). FOAr strains were scored for conditional growth phenotypes (28) and subsequently scored for phenotypes that often are associated with transcriptional defects (29). Strain YMH300 harbors plasmid pM507 (sua7–36 HIS3 CEN), which encodes the S53P derivative of TFIIB. Strain YMH130 carries plasmid pM299 (SUA7+ HIS3 CEN) and is otherwise identical to YMH300. Strains YMH506 [MATa CYC1 his3Δ1 leu2–3, 112 trp1–289 ura3–52 ade1–100 sua7∷LEU2 (SUA7 TRP1 CEN)] and YMH507 (sua7–36 TRP1 CEN) are isogenic strain pairs comparable to YMH130 and YMH300, but include different markers.
Growth Media.
YPD medium consists of 1% yeast extract, 2% peptone, and 2% dextrose. Media depleted of inorganic phosphate (−Pi) were prepared as described (30). The heat- and cold-sensitive phenotypes denote impaired growth of strain YMH300 (sua7–36) relative to strain YMH130 (SUA7+) on YPD medium at 37°C and 11°C, respectively.
DNA Sequence Analysis.
Plasmid DNA was recovered from strain YMH300 and introduced into E. coli strain XL1-Blue by electroporation. Single-stranded DNA was isolated by using the VCS M13 helper bacteriophage in the presence of kanamycin, as described (28). The sua7–36 coding region was sequenced in its entirety by the dideoxy-terminator method using three SUA7-specific primers (19).
Northern Blot Analysis.
Strains YMH130 (SUA7+) and YMH300 (sua7–36) were grown in YPD medium at 30°C to OD600 = 1.0 and diluted to OD600 = 0.05 in either YPD medium or phosphate-depleted YPD medium, grown to OD600 = 0.9, and harvested. Total RNA was extracted and analyzed by Northern blot analysis as described (19). PHO5 and ACT1 probes were α-32P-UTP-labeled RNA run-off transcripts derived from either the 625-bp SalI–BamHI PHO5 DNA fragment (31) in the vector pRS426, or the 1.4-kb BamHI–HindIII ACT1 DNA fragment in the vector pGEM3.
β-Galactosidase Assays.
Strains YMH130 and YMH300 were transformed with the reporter plasmids pMH313 (PHO5UAS-PHO5TATA-lacZ), ADCY4 (ADH2UAS-CYC1TATA-lacZ), or JK103 [lexA(op)-GAL1/10(TATA)-lacZ], which contains four LexA binding sites. Strains harboring JK103 also were transformed with plasmids expressing either LexA alone or LexA fused to the trans-activation region I (amino acids 1–220) of Adr1 (32). Independent transformants were grown at 30°C to OD600 = 0.6, harvested by centrifugation, and resuspended in 500 μl of breaking buffer (100 mM Tris⋅HCl, pH 8/1 mM DTT/20% glycerol/2 mM PMSF). Cell extracts were prepared by vortexing with 0.5-mm glass beads six times in 15-sec bursts. β-galactosidase assays were done as described (33). Activities are reported as (1.7 ml × OD420)/(0.0045 × cell extract [ml] × reaction time [min] × protein [mg/ml]).
Protein–Protein Interaction Assays.
DNA fragments were amplified by PCR using sequence-specific primers and either SUA7, sua7–36 (S53P), or PHO4 DNA as templates and cloned into pGEX-KG for glutathione S-transferase (GST)-fusion proteins, or pET21a for His6-fusion proteins. Constructs were expressed in E. coli strain BL-21. After 2.5-hr induction in the presence of 0.5 mM isopropyl β-d-thiogalactoside, cells extracts were prepared and incubated with glutathione Sepharose-4B beads (Amersham Pharmacia) in buffer A (20 mM Tris⋅HCl, pH 7.6/0.2 mM EDTA/1 mM DTT/1 mM PMSF/1 M NaCl/0.1% NP-40), followed by three washes with buffer A and three washes with buffer C (20 mM Tris⋅HCl, pH 7.6/0.2 mM EDTA/1 mM DTT/1 mM PMSF/100 mM NaCl/20% glycerol). Fusion proteins were eluted in three times 500-μl elution buffer (10 mM reduced glutathione in 50 mM Tris⋅HCl, pH 8.0). His6-tagged proteins were purified as described by Qiagen.
For GST pull-down experiments, 3 μg of fusion protein was incubated with 20-μl bed volume of glutathione-Sepharose beads in 500 μl of buffer C containing 0.05% NP-40. Input protein (10×) was added and incubated at 4°C for at least 4 hr. Beads were collected by centrifugation, washed three times with buffer C containing 500 mM NaCl, and boiled in 20 μl of loading buffer. Proteins were resolved by SDS/PAGE, transferred to nitrocellulose, and probed by Western blot using rabbit polyclonal antiserum directed against yeast TFIIB (28). Antigen-antibody complex was detected by chemiluminescence using goat anti-rabbit IgG conjugated to horseradish peroxidase.
Limited Protease Digestion of TFIIB.
Limited V8 protease digestion of wild-type and S53P TFIIB was done as described (15) with minor modifications. In each reaction, 50 ng of His6-TFIIB or His6-TFIIB(S53P) was incubated with 0, 5, or 25 ng of V8 protease. Reaction mixtures were incubated for 15 min at 30°C in digestion buffer (40 mM Hepes, pH 7.5/120 mM KCl/5 mM MgCl2/0.5 mM EDTA/1 mM DTT/0.05% NP-40). Reactions were stopped by adding 2× volume of sample loading buffer. Proteolytic fragments were separated by SDS/PAGE and analyzed by Western blotting as described for Fig. 5.
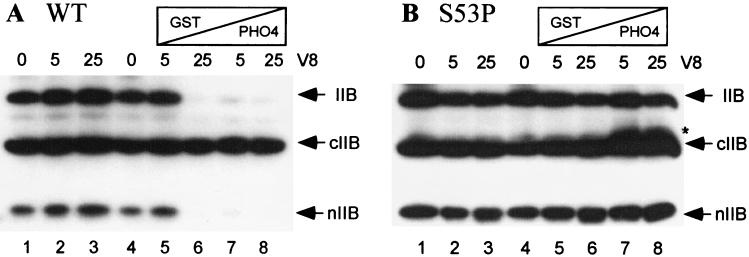
Figure 5.
Pho4-mediated TFIIB conformational change. Susceptibility to V8 protease was used as a probe for TFIIB conformational change. The V8 protease digestion pattern of the wild-type (A) and S53P form (B) of TFIIB is shown. Either 0, 5, or 25 ng of V8 protease was added to each sample, either in the absence (lanes 1–3) or presence (lanes 5–8) of Pho4. Purified Pho4 was added at either 1 (lanes 5 and 7) or 4 μg (lanes 6 and 8); GST was added to bring the total protein concentration to 4 μg. ∗ in B denotes a Pho4-dependent proteolytic fragment unique to the S53P form of TFIIB.
RESULTS
Isolation of the TFIIB S53P Derivative.
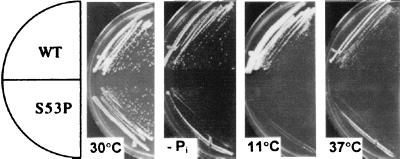
A random collection of sua7-encoded TFIIB mutants was generated by error-prone PCRs (see Materials and Methods). Strains initially were scored for temperature-sensitive growth defects and subsequently scored for pleiotropic phenotypes. One mutant, designated YMH300 (sua7–36), was both cold- and heat-sensitive and exhibited markedly impaired growth on phosphate-depleted medium (Pho− phenotype) (Fig. 1). These phenotypes can be attributed specifically to the sua7–36 allele because all growth defects were rescued by plasmid-borne SUA7+ (data not shown).
Figure 1.
Phenotypes associate with the S53P form of TFIIB. The sua7–36-encoded S53P derivative confers Pho−, cold-sensitive, and heat-sensitive growth phenotypes. An isogenic strain pair expressing either normal TFIIB (WT) or the S53P derivative (S53P) was incubated on rich (YPD) medium at either 11°C, 30°C, or 37°C, or on phosphate-depleted medium (−Pi) at 30°C. Plates were photographed after 2 days of incubation, except for the 11°C plate, which was photographed after 14 days of incubation.
DNA sequence analysis of the entire sua7–36 ORF identified a single base pair substitution encoding replacement of the normal serine at position 53 of TFIIB by proline (S53P). A cold-sensitive S53P replacement also was identified in an independent study, but was not characterized (34). S53 is located near the C terminus of the zinc-ribbon motif (26) and immediately precedes a phylogenetically conserved region involved in transcription start site selection (28). Overexpression of sua7–36 from a high-copy vector does not rescue the sua7–36 phenotypes, nor does the sua7–36 mutation affect the steady-state level of TFIIB, as defined by Western blot analysis (data not shown). Thus, S53P affects function, rather than synthesis or stability, of TFIIB.
TFIIB S53P Blocks PHO5 and ADH2 Activation.
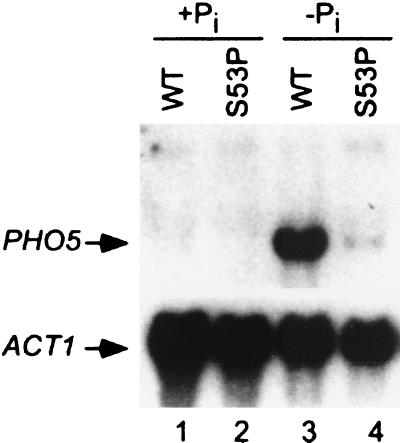
Growth of S. cerevisiae in phosphate-depleted medium depends on the expression of secreted acid phosphatases, encoded by the PHO3, PHO5, PHO10, and PHO11 genes (35). Secreted acid phosphatase is encoded primarily (>90%) by the inducible PHO5 gene, suggesting that PHO5 expression is impaired in the S53P mutant. This hypothesis was confirmed by Northern blot analysis using the isogenic wild-type and S53P strain pairs (Fig. 2). As expected, PHO5 mRNA was barely detectable in either the wild-type strain (lane 1) or the S53P mutant (lane 2) under repressing conditions (+Pi). Under inducing conditions (−Pi), the PHO5 mRNA level was elevated 17-fold in the wild-type strain (lane 3), whereas only 2-fold induction was seen in the S53P mutant (lane 4). Thus, the Pho− phenotype of the S53P mutant correlates with diminished PHO5 transcript levels.
Figure 2.
Northern blot analysis of PHO5 expression. PHO5 transcript levels were determined in isogenic wild-type (WT) and S53P mutant strains grown under repressing (+Pi) or inducing (−Pi) conditions. ACT1 mRNA was used as an internal control. In the presence of inorganic phosphate (+Pi), PHO5 is fully repressed (lanes 1 and 2). In the absence of inorganic phosphate (−Pi), PHO5 is induced in the wild-type strain (lane 3), whereas induction is severely impaired in the S53P mutant (lane 4).
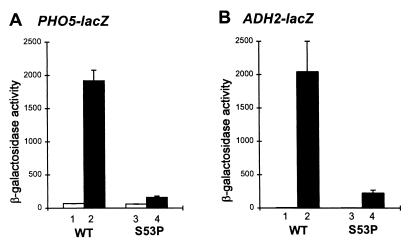
Presumably, the S53P defect impairs PHO5 expression at the transcriptional level. This was demonstrated by assaying β-galactoside expression from a PHO5UAS-PHO5TATA-lacZ reporter plasmid in the wild-type and S53P strains. Results are shown in Fig. 3. Whereas phosphate depletion (−Pi) induced β-galactosidase activity 28-fold in the wild-type strain, identical conditions resulted in only 2-fold stimulation in the S53P mutant. Importantly, both the wild-type and mutant strains expressed comparable levels of β-galactosidase under noninducing (+Pi) conditions, demonstrating that the effect of S53P on PHO5 expression specifically is caused by a defect in the mechanism of PHO5 activation. We conclude that transcriptional activation of the PHO5 gene critically depends on TFIIB and that S53 plays a key role in this process.
Figure 3.
S53P blocks activation of PHO5 and ADH2 reporter plasmids. The PHO5UAS-PHO5TATA-lacZ and ADH2UAS-CYC1TATA-lacZ reporter plasmids were introduced into the isogenic strains YMH130 (SUA7+) and YMH300 (sua7–36) and assayed for β-galactosidase activities under repressing and inducing conditions. (A) PHO5. Repressing (lanes 1 and 3) and inducing (lanes 2 and 4) conditions are described in the legend to Fig. 2. Absolute activities are 68, 1,920, 59 and 160 units of β-galactosidase in columns 1–4, respectively. (B) ADH2. Repressing conditions are 2% glucose (lanes 1 and 3); inducing conditions are 2% ethanol + 2% glycerol (lanes 2 and 4). Absolute activities are 5.2, 2,042, 2.3 and 225 units β-galactosidase in columns 1–4, respectively. All assays were done in duplicate, using three independent transformants. Error bars denote SD and in all cases are less than 25%.
To determine whether the transcriptional effect of the S53P replacement is specific to the PHO5 promoter, we determined whether it also affected expression from an ADH2UAS-CYC1TATA-lacZ reporter. The ADH2 promoter was chosen because the Adr1 activator of ADH2 has been shown to interact directly with TFIIB (32). Comparable to its effect on the PHO5 promoter, S53P impaired expression from this reporter 9-fold under inducing conditions. Again, this effect is activation-specific, because expression under noninduced conditions was reduced only 2-fold. Thus, in addition to its effect on PHO5, S53P impairs activation of the ADH2 gene.
Pho4 Targets the Core Domain of TFIIB.
Transcriptional activation of PHO5 requires Pho4, a basic helix–loop–helix trans-activator that binds two PHO5 UAS elements, UASp1 and UASp2 (36). Pho2, a homeodomain protein, facilitates binding of Pho4 to both UASp1 and UASp2 (37). Pho4 recently was shown to contact TBP, TFIIB, and TFIIEβ, suggesting that Pho4 activates PHO5 transcription, at least in part, by direct interaction with components of the general transcriptional machinery (38). The effect of the TFIIB S53P replacement on PHO5 activation is consistent with this idea and suggests that the Pho4-TFIIB interaction might be critical for PHO5 activation.
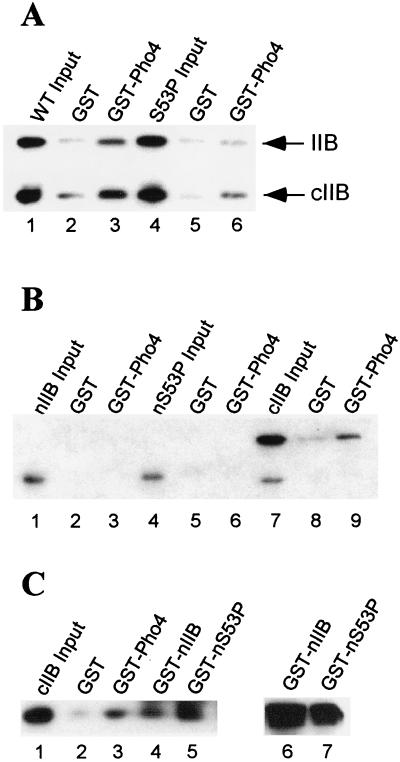
We investigated the potential effect of S53P on Pho4-TFIIB interaction by GST pull-down experiments followed by Western blot analysis using antibodies specific for TFIIB. Input proteins in these experiments were His6-tagged derivatives of the wild-type and S53P form of TFIIB purified from E. coli. GST-Pho4 bound specifically to TFIIB (Fig. 4A, lanes 1–3), resulting in recovery of 51% of the wild-type TFIIB input sample. In contrast, the same amount of GST-Pho4 recovered only 3% of TFIIB S53P input sample (Fig. 4A, lanes 4–6). It is also noteworthy that GST-Pho4 bound the cIIB form of TFIIB, which was generated spontaneously from full-length TFIIB, yielding cIIB from both the wild-type and S53P samples (Fig. 4A, lanes 3 and 6). This result was confirmed by repeating the pull-down experiments using nIIB and cIIB as input proteins. In this case GST-Pho4 failed to interact with nIIB of either the wild-type (Fig. 4B, lanes 1–3) or S53P form of TFIIB (lanes 4–6), but instead bound to cIIB (lanes 7–9). Thus, Pho4 binds cIIB but not nIIB, which includes the S53P replacement.
Figure 4.
Protein–protein interactions. (A) GST-Pho4 pull-down of normal (WT input) and the S53P form (S53P input) of TFIIB. Equal amounts of either GST alone or GST-Pho4 were incubated with either the normal (WT) or the S53P form of TFIIB. Immobilized samples were washed (500 mM NaCl), eluted, and analyzed by Western blot analysis using polyclonal yeast TFIIB antibody as described in Materials and Methods. (B) GST-Pho4 pull-down of the nIIB input and the S53P form (nS53P input) of TFIIB, and cIIB of normal TFIIB. Equal amounts of GST-Pho4 protein were incubated with the indicated input proteins and analyzed by Western blot as described in A. (C) Intramolecular TFIIB interaction. GST-Pho4, GST-nIIB (WT), and GST-nIIB (S53P) were incubated with cIIB and analyzed as described in A. Western blot analysis (TFIIB antibody) was used to confirm that equal amounts of GST-nIIB (lane 6) and GST-nS53P (lane 7) were used in this experiment.
One interpretation of these results is that the S53P effect on Pho4-TFIIB interaction is manifest through an intramolecular interaction between nIIB and cIIB. There is precedent for this possibility from the work of Roberts and Green on human TFIIB (15). The results presented in Fig. 4C establish that this is indeed the case. Like GST-Pho4 (lane 3), GST-nIIB bound input cIIB (lane 4). The S53P form of nIIB also bound cIIB (lane 5), perhaps with slightly greater affinity than normal nIIB. These results demonstrate that yeast TFIIB, like its human counterpart, forms an intramolecular interaction between the N- and C-terminal domains.
Pho4 Promotes a TFIIB Conformational Change.
The preceding results suggest that TFIIB undergoes a Pho4-mediated conformational change as part of the mechanism of PHO5 activation. Again, there is precedent for this possibility from the effect of VP16 on the conformation of human TFIIB (15). We monitored TFIIB conformation by assaying the susceptibility of TFIIB to limited V8 protease digestion in the absence and presence of Pho4. The results clearly demonstrated that wild-type TFIIB undergoes Pho4-dependent V8 protease degradation (Fig. 5A). Under the same conditions the S53P form of TFIIB is resistant to degradation (Fig. 5B), presumably because Pho4 interaction with TFIIB is impaired by the S53P replacement (Fig. 4A).
The TFIIB S53P Defect Is Specific to the Transcriptional Activation Function of Adr1.
The data in this paper clearly demonstrate that the S53P form of TFIIB impairs transcriptional activation of PHO5 in vivo and that the Pho4 activator of PHO5 interacts with TFIIB in vitro, an effect that is impaired by the S53P replacement. Similarly, activation of ADH2 is impaired by S53P in vivo (Fig. 3B) and Adr1 interacts with TFIIB in vitro (32). However, are the PHO5 and ADH2 transcriptional defects specific to the activator functions of Pho4 and Adr1? Given the general role of TFIIB in transcription, it is conceivable that these effects are indirect. As an initial experiment to address this issue, we asked whether S53P affected PHO4 expression. Western blot analysis using antibodies directed against Pho4 revealed that the steady-state levels of Pho4 were identical in the wild-type and S53P strains (data not shown). Thus, the effect of S53P on PHO5 activation is not the result of an indirect effect on the level of the Pho4 activator.
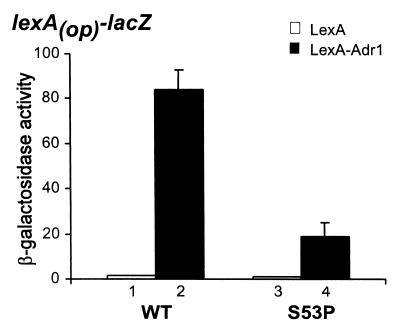
Second, we asked whether S53P would impair expression of a lexA(op)-lacZ reporter using a LexA-Pho4 fusion protein as the activator. Unfortunately, none of the LexA-Pho4 fusion proteins, which included several protein fragments encompassing the Pho4 activation domain (39), stimulated lexA(op)-lacZ expression in the wild-type strains (data not shown). Because the trans-activation domain I (TADI) of Adr1 also interacts with TFIIB (32) and this interaction is impaired by the S53P replacement (data not shown), we performed the analogous experiment by using a LexA-Adr1 (TADI) fusion protein. In this case, the S53P replacement impaired lexA(op)-lacZ expression by 4.4-fold (Fig. 6). This result demonstrates that the effect of S53P on activation of ADH2 in vivo is specific to the activator function of Adr1.
Figure 6.
Effect of S53P on LexA-Adr1 activation of a lexA(op)-lacZ reporter. The lexA(op)-lacZ reporter plasmid was introduced into the isogenic strains YMH506 (SUA7+) and YMH507 (sua7–36) and assayed for β-galactosidase activities. Absolute activities are 1.6, 84, 1.1, and 19 units of β-galactosidase in columns 1–4, respectively. All assays were done in duplicate, using three independent transformants. Error bars denote SD and in all cases are less than 30%.
DISCUSSION
The results presented in this paper define a TFIIB defect that blocks expression of the PHO5 and ADH2 genes in vivo and demonstrate that this effect occurs at the level of transcriptional activation. Biochemical experiments designed to address the mechanism of this effect confirmed that the Pho4 activator of PHO5 interacts directly with TFIIB. Surprisingly, Pho4 interacts specifically with cIIB, rather than nIIB, which includes the S53P replacement. Furthermore, nIIB and cIIB form an intramolecular interaction, and Pho4-TFIIB interaction induces a conformational change that might represent disruption of the nIIB-cIIB interaction. Furthermore, we demonstrate that the transcriptional defect associated with the S53P replacement in vivo is specific to the activation function of the Adr1 activator of ADH2. These results define an activation-specific role for TFIIB in vivo and support a previous model for gene activation involving an activator-induced TFIIB conformational change that facilitates PIC assembly (15).
According to the model of Roberts and Green (15), nIIB and cIIB are engaged in an intramolecular interaction that is targeted by specific activators. This model is also consistent with recent structural studies of human TFIIB showing that interaction of either VP16 or nIIB with cIIB induce distinct changes in the orientation of the two repeat domains of cIIB relative to each other (25). However, the studies of neither Roberts and Green (15) nor Hayashi et al. (25) addressed the potential role of TFIIB conformational changes on transcription. The results presented here substantiate the intramolecular interaction between nIIB and cIIB, as well as the suggestion that this interaction can be targeted by specific activators to alter TFIIB conformation. Indeed, if the S53P replacement affects this interaction, then the results presented in this study argue that the nIIB-cIIB interaction is physiologically relevant for transcriptional activation in vivo.
The region of human TFIIB that interacts with the C-terminal core was mapped to residues 24–65 (15). This sequence corresponds to residues 35–77 of yeast TFIIB and includes S53. S53 is not phylogenetically invariant, but is highly conserved, either serine in yeast TFIIB or glycine in metazoan TFIIB. By comparison with the NMR structure for the zinc binding domain of the Pyrococcus TFIIB homolog, S53 would lie just downstream of the fourth cysteine (C48) of the metal binding motif and within the third β-sheet of the zinc ribbon. This finding suggests that the TFIIB intramolecular interaction might be mediated by the zinc ribbon binding to an undefined region within the cIIB. Furthermore, activators that stimulate transcription by inducing a conformational change in TFIIB might do so by affecting the zinc ribbon-core domain interaction. Although the effect of S53P is presumably a consequence of the proline residue on the structure of the zinc ribbon, further structural studies are necessary to define how this replacement affects nIIB-cIIB and activator-cIIB interactions.
TFIIB has been implicated as the target of a number of transcriptional activators in vitro. A role for yeast TFIIB in activation is supported by a TBP derivative that fails to interact with TFIIB and is defective for activation (40). However, these experiments were done in vitro and do not necessarily reflect activation in vivo. In another study of TBP-TFIIB interactions, a human TBP derivative was rescued by a compensatory mutation in TFIIB, restoring activation in vivo (41). In this case the TBP-TFIIB interaction was important for the response to several activators, but not to Sp1 (41). These results suggest that the TBP-TFIIB interaction is targeted by specific activators, although the possibility cannot be excluded that these mutants affect basal rather than activated expression. Mutational analysis of a species-specific region of yeast TFIIB also implicated TFIIB in the process of activation from specific promoters in vivo (42). In contrast, amino acid replacements in either TBP or TFIIB that adversely affect TATA-TBP-TFIIB ternary complex formation in vitro respond normally to several different transcriptional activators in vivo (17, 18). These results were interpreted to mean that recruitment of TFIIB is not generally rate limiting for transcriptional activation in vivo. However, an alternative explanation is that defective interactions between TBP and TFIIB in vitro are compensated by other interactions within the PIC in vivo. Indeed, this possibility is supported by human TBP mutations that impair activator-TBP interaction in vitro, yet are without effect on activation in vivo (43).
It is clear from both biochemical and structural studies that TFIIB is conformationally pliable and that certain transcriptional activators induce TFIIB conformational changes. However, the activator-induced conformational changes described here (Fig. 5) or previously (15) do not necessarily represent the transition from the structure of free TFIIB to the form found in the ternary complex. Moreover, we do not know that the observed TFIIB conformational change is part of the mechanism of Pho4-mediated transcriptional activation. Therefore, our results do not necessarily imply that Pho4-mediated isomerization is rate limiting for PHO5 activation. Furthermore, we cannot distinguish between isomerization and recruitment as mechanisms to account for PHO5 activation. Nonetheless, the results presented here clearly demonstrate that TFIIB can be a physiologically relevant target of specific activators in vivo.
Preliminary experiments indicate that S53P affects expression from promoters in addition to PHO5 and ADH2 (unpublished results). However, in some cases these effects appear to be manifest at the basal level. The differential effects of S53P on basal and activated expression might be explained by a general requirement for isomerization of TFIIB from the free to the bound form. In some cases, this transition is targeted by activators, whereas in other cases it occurs spontaneously. In the former case, S53P would affect activation, whereas in the latter case it would affect basal expression. Further characterization of the S53P mutant, as well as other activation-defective TFIIB mutants, will provide additional insight into the role of TFIIB in transcriptional activation.
Acknowledgments
M.H. dedicates this paper to Professor G. B. Kohlhaw on the occasion of his retirement. We are especially grateful to K. Struhl for valuable suggestions. We also thank D. Gross, D. Reinberg, and Z.-W. Sun for valuable discussions throughout the course of this work and for comments on the manuscript; Y. Zhang for advice regarding GST pull-down experiments; C. Denis, M. Grunstein, W. Horz, and S. Johnson for plasmids; and E. O’Shea for Pho4 antiserum. This work was supported by National Institutes of Health Grant GM39484 to M.H.
ABBREVIATIONS
- TF
transcription factor
- S53P
serine-53 → proline
- RNAPII
RNA polymerase II
- PIC
preinitiation complex
- TBP
TATA box-binding protein
- cIIB
C-terminal core domain of TFIIB
- nIIB
N-terminal domain of TFIIB
- YPD
yeast extract/peptone/dextrose
- GST
glutathione S-transferase
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Ptashne M, Gann A. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 2.Stringer K F, Ingles C J, Greenblatt J. Nature (London) 1990;345:783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- 3.Lin Y S, Ha I, Maldonado E, Reinberg D, Green M R. Nature (London) 1991;353:569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- 4.Joliet V, Demma M, Prywes R. Nature (London) 1995;373:632–635. doi: 10.1038/373632a0. [DOI] [PubMed] [Google Scholar]
- 5.Xiao H, Pearson A, Coulombe B, Truant R, Zhang S, Regier J L, Triezenberg S J, Reinberg D, Flores O, Ingles C J, Greenblatt J. Mol Cell Biol. 1994;14:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin Y, Nomura T, Cheong J, Dorjsuren D, Iida K, Murakami S. J Biol Chem. 1997;272:7132–7139. doi: 10.1074/jbc.272.11.7132. [DOI] [PubMed] [Google Scholar]
- 7.Goodrich J A, Hoey T, Thut C J, Admon A, Tjian R. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- 8.Gill G, Pascal E, Tseng Z H, Tjian R. Proc Natl Acad Sci USA. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keaveney M, Struhl K. Mol Cell. 1998;1:917–924. doi: 10.1016/s1097-2765(00)80091-x. [DOI] [PubMed] [Google Scholar]
- 10.Chi T H, Lieberman P, Ellwood K, Carey M. Nature (London) 1995;377:254–257. doi: 10.1038/377254a0. [DOI] [PubMed] [Google Scholar]
- 11.Ranish J A, Yudkovsky N, Hahn S. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim T K, Roeder R G. Proc Natl Acad Sci USA. 1994;91:4170–4174. doi: 10.1073/pnas.91.10.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts S G E, Ha I, Maldonado E, Reinberg D, Green M R. Nature (London) 1993;363:741–744. doi: 10.1038/363741a0. [DOI] [PubMed] [Google Scholar]
- 14.Roberts S G E, Choy B, Walker S S, Lin Y S, Green M R. Curr Biol. 1995;5:508–516. doi: 10.1016/s0960-9822(95)00103-5. [DOI] [PubMed] [Google Scholar]
- 15.Roberts S G E, Green M R. Nature (London) 1994;371:717–720. doi: 10.1038/371717a0. [DOI] [PubMed] [Google Scholar]
- 16.Gupta R, Emili A, Pan G, Xiao H, Shales M, Greenblatt J, Ingles C J. Nucleic Acids Res. 1996;24:2324–2330. doi: 10.1093/nar/24.12.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou S, Struhl K. Mol Cell Biol. 1997;17:6794–6802. doi: 10.1128/mcb.17.12.6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee M, Struhl K. Mol Cell Biol. 1997;17:1336–1345. doi: 10.1128/mcb.17.3.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinto I, Ware D E, Hampsey M. Cell. 1992;68:977–988. doi: 10.1016/0092-8674(92)90040-j. [DOI] [PubMed] [Google Scholar]
- 20.Barberis A, Muller C W, Harrison S C, Ptashne M. Proc Natl Acad Sci USA. 1993;90:5628–5632. doi: 10.1073/pnas.90.12.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malik S, Lee D K, Roeder R G. Mol Cell Biol. 1993;13:6253–6259. doi: 10.1128/mcb.13.10.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Na J G, Hampsey M. Nucleic Acids Res. 1993;21:3413–3417. doi: 10.1093/nar/21.15.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bagby S, Kim S J, Maldonado E, Tong K I, Reinberg D, Ikura M. Cell. 1995;82:857–867. doi: 10.1016/0092-8674(95)90483-2. [DOI] [PubMed] [Google Scholar]
- 24.Nikolov D B, Chen H, Halay E D, Usheva A A, Hisatake K, Lee D K, Roeder R G, Burley S K. Nature (London) 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 25.Hayashi F, Ishima R, Liu D, Tong K I, Kim S, Reinberg D, Bagby S, Ikura M. Biochemistry. 1998;37:7941–7951. doi: 10.1021/bi9801098. [DOI] [PubMed] [Google Scholar]
- 26.Zhu W L, Zeng Q D, Colangelo C M, Lewis L M, Summers M F, Scott R A. Nat Struct Biol. 1996;3:122–124. doi: 10.1038/nsb0296-122. [DOI] [PubMed] [Google Scholar]
- 27.Muhlrad D, Hunter R, Parker R. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 28.Pinto I, Wu W-H, Na J G, Hampsey M. J Biol Chem. 1994;269:30569–30573. [PubMed] [Google Scholar]
- 29.Hampsey M. Yeast. 1997;13:1099–1133. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1099::AID-YEA177>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 30.Almer A, Rudolph H, Hinnen A, Horz W. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyhack B, Bajwa W, Rudolph H, Hinnen A. EMBO J. 1982;1:675–680. doi: 10.1002/j.1460-2075.1982.tb01229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiang Y C, Komarnitsky P, Chase D, Denis C L. J Biol Chem. 1996;271:32359–32365. doi: 10.1074/jbc.271.50.32359. [DOI] [PubMed] [Google Scholar]
- 33.Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 34.Knaus R, Pollock R, Guarente L. EMBO J. 1996;15:1933–1940. [PMC free article] [PubMed] [Google Scholar]
- 35.Svaren J, Horz W. Trends Biochem Sci. 1997;22:93–97. doi: 10.1016/s0968-0004(97)01001-3. [DOI] [PubMed] [Google Scholar]
- 36.Svaren J, Schmitz J, Horz W. EMBO J. 1994;13:4856–4862. doi: 10.1002/j.1460-2075.1994.tb06812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barbaric S, Munsterkotter M, Svaren J, Horz W. Nucleic Acids Res. 1996;24:4479–4486. doi: 10.1093/nar/24.22.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magbanua J P, Ogawa N, Harashima S, Oshima Y. J Biochem. 1997;121:1182–1189. doi: 10.1093/oxfordjournals.jbchem.a021713. [DOI] [PubMed] [Google Scholar]
- 39.McAndrew P C, Svaren J, Martin S R, Horz W, Goding C R. Mol Cell Biol. 1998;18:5818–5827. doi: 10.1128/mcb.18.10.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim T K, Hashimoto S, Kelleher R R, Flanagan P M, Kornberg R D, Horikoshi M, Roeder R G. Nature (London) 1994;369:252–255. doi: 10.1038/369252a0. [DOI] [PubMed] [Google Scholar]
- 41.Tansey W P, Herr W. Science. 1997;275:829–831. doi: 10.1126/science.275.5301.829. [DOI] [PubMed] [Google Scholar]
- 42.Shaw S P, Carson D J, Dorsey M J, Ma J. Proc Natl Acad Sci USA. 1997;94:2427–2432. doi: 10.1073/pnas.94.6.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tansey W P, Herr W. Proc Natl Acad Sci USA. 1995;92:10550–10554. doi: 10.1073/pnas.92.23.10550. [DOI] [PMC free article] [PubMed] [Google Scholar]