Abstract
CDC7 is an essential gene required for DNA replication in Saccharomyces cerevisiae. Cdc7p homologs have recently been identified in vertebrates, but their role in DNA replication has not yet been addressed. Here we show that antibodies to the Xenopus laevis homolog, xCdc7, interfere with DNA replication in vivo in developing embryos and in vitro in cycling egg extracts. We also demonstrate cell cycle-dependent association of xCdc7 with the Mcm complex, which binds to replication origins and also is required for DNA synthesis. Taken together, these data indicate that the function of xCdc7 is conserved from fungi to vertebrates. xCdc7 protein accumulates after stimulation of resting oocytes with progesterone, suggesting a molecular explanation for previous observations that the development of the capacity for DNA replication requires protein synthesis late in meiosis I.
Studies over the past several years have provided an increasingly detailed understanding of the proteins governing control of DNA replication in the budding yeast Saccharomyces cerevisiae (1–3). It has been shown that in this organism a group of proteins referred to collectively as the origin recognition complex is associated with conserved sequences at replication origins throughout the cell cycle (4, 5). An additional complex composed of the Mcm (minichromosome maintenance) proteins becomes associated with origins early in G1 (6, 7) through a process depending on the presence of the origin recognition complex and the activity of Cdc6 (8). At this stage, origins are said to be “licensed” for one round of DNA replication. The transition from G1 into S phase is triggered by the proteolysis of the Sic1p cyclin-dependent kinase inhibitor (9–12), relieving inhibition of the Cdc28p cyclin-dependent kinase (associated with either Clb5 or Clb6). Finally, Cdc28/Clb5/Clb6 (13), along with the Cdc7p protein kinase (associated with its regulatory subunit, Dbf4), promote progression through S phase. The precise role of the cyclin-dependent kinase is not fully understood, but the Cdc7/Dbf4 component is required for origin firing throughout S phase (14, 15), probably through phosphorylation of Mcm2 (16), which is released from the origin along with other Mcm proteins before DNA polymerase begins to synthesize a new strand of DNA (6).
Homologs of many components of this pathway have been identified in metazoans, suggesting that mechanisms of replication control are evolutionarily conserved in eukaryotes. Indeed, origin recognition (17) and Mcm (18) complexes are associated with DNA in vertebrates, and Xenopus Mcm (7), origin recognition complex-, and Cdc6-related proteins (19) have been shown to be required for DNA replication in vitro in Xenopus egg extracts. However, it is becoming clear that there are important differences between yeast and metazoan replication control. For example, the mechanisms defining replication origins are not as stringent in vertebrates as they are in yeast. Although some data suggest that metazoan origins are spaced at roughly equal intervals throughout the genome by an unidentified mechanism (20), other data has shown that certain grossly defined sequences can act as replication origins even when transferred to new chromosomal locations (21). Animal cells also prevent premature passage into S phase by employing the anti-mitogenic Rb protein (22), which is not found in yeast. Although a cyclin-dependent kinase/cyclin component is required during S phase, either Cdk2/cyclin E or Cdc2/cyclin A (23) can act in this role, and it has not been established whether the vertebrate proteins have the same function in the pathway as Cdc28/Clb5/6 do in yeast. Moreover, vertebrate embryos employ some mechanisms of S phase control that are distinct from those involved in replication in somatic cell cycles. Cdk4/cyclin D, for example, is clearly required to phosphorylate Rb to promote initiation of S phase in the cells of adult animals, but Rb is not involved in early embryonic cycles, which lack transcription (24).
Vertebrate protein kinases that are ≈26% identical to the yeast Cdc7p have recently been described (25–27), and the human homolog is overexpressed in some tumors and transformed cell lines (27). In addition, the human Cdc7 is capable of phosphorylating Mcm2 and Mcm3 in vitro (25), and its kinase activity (measured by in vitro phosphorylation of histone H1) varies over the cell cycle in a pattern similar to that of Cdk2 (26). However, whether these homologs are involved in DNA replication control has yet to be demonstrated. Here we show functional homology between the yeast and Xenopus S phase kinases by using antibodies to selectively interfere with xCdc7 activity. We also show by coimmunoprecipation experiments that xCdc7 is physically associated with xMcm3 in interphase but not in metaphase. Finally, we show that there is a pronounced increase in xCdc7 protein levels after stimulation of resting oocytes with progesterone, which may explain a well documented requirement for protein synthesis during oocyte maturation (28–31) to allow synthesis of DNA after fertilization.
MATERIALS AND METHODS
Cloning of a Xenopus CDC7 Homolog.
The S. cerevisiae Cdc7p amino acid sequence was compared with the sequence of a human homolog (R. Hollingsworth and R. Sclafani, personal communication). An internal cDNA fragment was cloned by PCR amplification using degenerate oligonucleotides specific to the highly conserved sequences in kinase subdomains I and VII (32). The 5′ primer AARATHGGIGARGGIACITTY, corresponding to the amino acid sequence KIGEGTF, was paired in separate reactions with each of four 3′ primers, GCIAGICCRAARTCIACIAG (designated II), GCIAGICCRAARTCIACYAA (designated IY), GCYAAICCRAARTCIACIAG (designated YI), or GCYAAICCRAARTCIACYAA (designated YY) corresponding to the amino acid sequence LVDFGLA. The code for the degenerate nucleotides is H = A, T, or C; r = A or G; and Y = C or T. A total Xenopus oocyte cDNA library was used as a template for reactions containing 50 ng of library DNA, 10 μM each primer, dNTPs, and KlenTaq cDNA polymerase mix (CLONTECH). Samples were subjected to 32 cycles at 94°C for 30 seconds, 47°C for 30 seconds, and 72°C for 15 seconds. A fragment of the expected size (≈410 bp) encoding amino acids with a high degree of homology to the region between subdomains I and VII of the yeast and human Cdc7 proteins was amplified only in reactions containing both the 5′ primer and the YY 3′ primer. The fragment was radiolabeled and used as a probe of the same oocyte cDNA library to obtain a full-length clone. The sequence of the cDNA was determined on both strands and was longer and showed numerous nucleotide differences from the subsequently published sequence (25) in the 5′ and 3′ untranslated regions, but only three nucleotide differences within the coding region. These three nucleotide substitutions encode two amino acid differences between the cloned and the published sequences.
Production and Purification of Rabbit Polyclonal Antibodies Against xCdc7.
The xCdc7 coding sequence was amplified by PCR using the 5′ primer CGTGGGATCCTGATGAGTTCGGGCGATAATTCAGG and the 3′ primer ACTGGGGAATTCCTACCGCATGTTTTTAAACAGAGG, which add BamHI and EcoRI sites, respectively. The resulting fragment was cloned into pGex-3x (Pharmacia), and the resulting plasmid was transformed into BL21(DE3) for expression. The resulting fusion protein was almost completely insoluble, and inclusion bodies were purified by using standard methods. The fusion protein was further purified by using PAGE and was used to immunize two rabbits. Antibodies were affinity purified by using standard methods (33) from the antigen described above that had been eluted from the gel, or, where noted, from a xCdc7 fusion protein purified from Sf9 cells (see below) conjugated to CH-Sepharose 4B (Amersham Pharmacia).
Expression and Purification of Recombinant xCdc7 in Sf9 Cells.
The xCdc7 coding sequence was amplified by PCR using Pfu (Stratagene) with GACGACGACAAGATGAGTTCGGGCGATAATTCAGG as the 5′ primer and GAGGAGAAGCCCGGTCCGCATGTTTTTAAACAGAGGATG as the 3′ primer and inserted by ligation-independent cloning (34) into the baculovirus transfer vector pBAC7 (Novagen). For production of soluble cellulose binding domain-xCdc7 fusion protein (CBD-xCdc7), Sf9 cells expressing the construct were harvested and lysed in EBC (50 mM Tris⋅HCl, pH 8/120 mM NaCl/0.5% Nonidet P-40/1 mM phenylmethylsulfonyl fluoride). The soluble fraction was loaded onto a CBinD300 push column (Novagen), washed once with EBC, once with 20 mM Tris⋅HCl (pH 7.5)/0.8 M NaCl, once with 20 mM Tris⋅HCl (pH 7.5), and eluted with 100% ethylene glycol. The eluate was then dialyzed against injection buffer (20 mM Hepes, pH 7.5/88 mM NaCl/7.5 mM MgCl2/10 mM 2-mercaptoethanol/0.05% Brij-35). For preparation of insoluble CBD-xCdc7 for antibody affinity purification, cells were lysed in Buffer 1 [10 mM Hepes, pH 7.5/150 mM NaCl/0.2% Brij-35/0.1% Nonidet P-40/1 mM EGTA/1× Complete Protease Inhibitors (Boehringer Mannheim)]. Crude lysates were centrifuged at 10,000 × g. The insoluble fraction was washed in Buffer 1, resuspended in 10 mM Tris⋅HCl (pH 8)/1% lauryl sarcosine, and further purified by PAGE.
Embryo Microinjection and DNA Synthesis Assays.
Embryos were prepared, treated, fixed, and sectioned as described (35). Microinjection volume was 50 nl at an approximate antibody concentration of 50 μg/ml in injection buffer. Cycling-egg extracts were prepared essentially as described (36), except that [α-32P]dCTP [>3,000 Ci/mmol (1 Ci = 37 GBq), Amersham Pharmacia] was added to a final concentration of 5–10 μCi/ml and demembranated sperm nuclei were added to 3,000 per microliter. Where noted, affinity-purified anti-xCdc7 antibodies were added to the noted concentrations in volumes less than or equal to 2% of the total extract, and CBD-xCdc7 (see above) was added to approximately 40 ng/ml. Aliquots were taken at the specified time points, diluted 1:10 in 10 mM Tris (pH 8)/10 mM EDTA/50 μg/ml proteinase K/1% SDS and incubated 1–2 hours at 37°C. The samples were then subjected to agarose gel electrophoresis in the presence of ethidium bromide and analyzed by autoradiography.
Immunoblot and Northern Analyses.
Stage VI oocytes were treated with progesterone at 10 μg/ml to stimulate maturation. Fertilization was simulated (where noted) by transient treatment of fully matured oocytes with 5 μg/ml A23187 (Calbiochem). For analysis of protein content, oocytes or embryos were collected in groups of eight or more per time point and homogenized in 20 mM Tris⋅HCl (pH 7.5)/100 mM NaCl/80 mM glycerol 2-phosphate/5 mM EGTA/1 mM DTT/1 mM sodium orthovanadate/1 μM microcystin/1× Complete Protease Inhibitors (Boehringer Mannheim)/0.5% Nonidet P-40. Crude lysates were centrifuged in a tabletop microcentrifuge at 10,000 × g for 10 min, and the soluble fraction was saved, boiled with 1× sample buffer, subjected to electrophoresis on SDS-12% PAGE gels, and transferred to nitrocellulose by standard methods. Two different polyacrylamide gel formulations were used (37, 38) (as indicated in figures). The Laemmli method resulted in a Mr of approximately 56,000, whereas that of Anderson et al. (37) resulted in a Mr of approximately 50,000. Note that discrepancies such as this have been observed previously with other proteins, for example cyclin B (39). Antibodies to MosXe were from Santa Cruz Biotechnology.
mRNA was analyzed by disrupting eight embryos at the indicated stage in Tri-reagent (Molecular Research Center, Cincinatti) and precipitating the RNA by using standard methods. Approximately 15 μg of total RNA was loaded per lane. After denaturing agarose gel electrophoresis, the RNA was transferred to a nylon membrane and probed with radiolabeled full-length xCdc7 cDNA. The membrane was stripped and reprobed with 18S rRNA as a loading control, indicating that comparable amounts of RNA were loaded in all lanes (data not shown).
xMcm3 and xCdc7 Immunoprecipitations.
Xenopus metaphase-arrested egg extracts were prepared as described (36). Interphase extracts were prepared from these by adding CaCl2 to 0.4 mM and cycloheximide to 100 μg/ml, then incubating at 20°C for 90 minutes. Aliquots of these extracts (10 μl) were diluted into 90 μl of 80 mM glycerol 2-phosphate/15 mM MgCl2 and incubated with 2 μl of affinity-purified xMcm3 or xCdc7 antibodies for 3 hours at 4°C with constant rotation. Protein A Sepharose (20 μl) was added and incubated for 1 hour under the same conditions. Immunoprecipitates were washed eight times with PBS containing 0.2%NP40, resuspended in 1× sample buffer, and electrophoresed on a 10% SDS/PAGE gel. Western blotting was performed with either xCdc7 or xMcm3 antibodies. Horseradish peroxidase-coupled protein A (Zymed) was used for detection by using enhanced chemiluminescence (Amersham).
RESULTS
Inhibition of DNA Synthesis in Embryos by Polyclonal Antibodies to xCdc7.
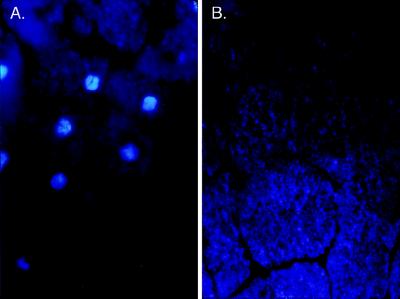
To investigate the role of xCdc7 in the cell cycle, rabbit polyclonal antibodies were generated and affinity-purified on a column containing bacterially expressed recombinant xCdc7. Single-cell embryos were injected with these antibodies, and allowed to divide until controls had reached approximately stage 8 (4,000 cells) before fixing, sectioning, and staining with the fluorescent DNA dye 4′,6-diamidino-2-phenylindole (DAPI). Control embryos injected with buffer showed bright staining nuclei in most sections (Fig. 1A), whereas antibody-injected embryos appeared entirely devoid of nuclear DNA (Fig. 1B) Significantly, cell division was noticeably delayed in antibody-injected embryos compared with controls, as has previously been shown in embryos treated with the DNA replication inhibitor aphidicolin (ref. 40, and data not shown). Because of this effect, cells in the control embryos are smaller than in those treated with antibody (Fig. 1). However, similar results were observed in antibody-injected embryos that were allowed to continue division until their cells were the same size as those of controls.
Figure 1.
xCdc7 is required for DNA replication in vivo. At the single-cell stage, buffer control (A) or anti-xCdc7 antibody (B) was injected and embryos were allowed to develop until controls were stage 8 before fixing, sectioning, and 4′,6-diamidino-2-phenylindole (DAPI) staining.
xCdc7 Is Specifically Required for DNA Synthesis.
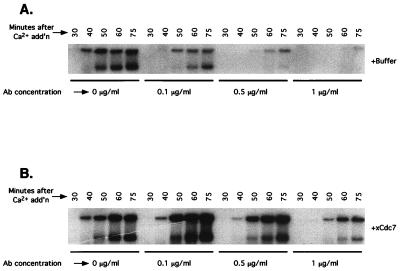
The above data suggest that xCdc7 is required for DNA synthesis in the early embryo. However, the data may instead reflect a failure of karyokinesis in the first mitotic division, which would result in an embryo that lacked nuclei in every cell but one. To distinguish between these possibilities, the antibodies were added to cycling Xenopus egg extracts, and samples were taken at various time points to assess nuclear morphology and the progress of DNA synthesis. As determined by microscopic examination, nuclei formed in the antibody-treated extracts developed normal-appearing nuclear envelopes and were indistinguishable from those formed in control extracts treated with buffer only (data not shown). However, DNA replication assays revealed a dose-dependent inhibition of DNA synthesis in antibody-treated extracts (Fig. 2A). Furthermore, addition of exogenous CBD-xCdc7 from Sf9 cells restored DNA replication (Fig. 2B), demonstrating that the failure of DNA synthesis was specifically caused by loss of xCdc7 function.
Figure 2.
xCdc7 is required for DNA replication in vitro. Xenopus egg extracts arrested in metaphase were supplemented with radiolabeled dCTP and with anti-xCdc7 antibodies at the indicated concentrations. After Ca2+ addition to trigger entry into S phase, aliquots were taken at the indicated time points and subjected to agarose gel electrophoresis and autoradiography. (A) Anti-xCdc7 antibodies; (B) As in A except that recombinant xCdc7 (approximately 40 ng/ml) was added to the extract.
Physical Association of xCdc7 with Components of the Mcm Complex.
Several studies have indicated a link between yeast Cdc7 and the Mcm components of replication “licensing factor,” which must be bound to replication origins by G1 in order for S phase to proceed. In particular, mutations in MCM5 can suppress mutations in CDC7 (41), and Cdc7p interacts with a Mcm2p-affinity matrix (16). If the requirement for xCdc7 in Xenopus is truly analogous to the requirement in yeast, we would expect a similar physical interaction between xCdc7 and components of Xenopus licensing factor. Furthermore, if the model for control of DNA replication in yeast is accurate, we would expect that association to be limited to interphase, around the time of DNA replication.
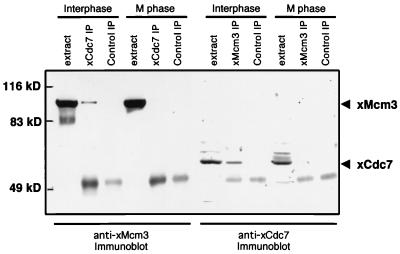
To test these hypotheses, xCdc7 immune complexes were isolated from Xenopus egg extracts and assayed by immunoblot for the presence of xMcm3. Similarly, xMcm3 immune complexes were tested for the presence of xCdc7. The two proteins were found to physically interact when immunoprecipitated from egg extracts arrested in interphase. In contrast, xMcm3 and xCdc7 were not associated in extracts from cells arrested in mitosis (Fig. 3.) It has recently become known that the Mcm proteins form two subcomplexes, one comprised of Mcm3 and Mcm5 and the other comprised of Mcm4, Mcm6, Mcm7, and possibly Mcm2 (42, 43). To test whether xCdc7 is associated with xMcm3 or its subcomplex alone or with the whole Mcm complex, we performed an identical test for cell cycle stage-specific association of xCdc7 and xMcm7. The two proteins were found to be associated in interphase but not in metaphase (data not shown). These data cannot distinguish which member(s) of the Mcm complex xCdc7 interacts with directly, but they do suggest that xCdc7 is associated with the whole complex.
Figure 3.
Cell cycle-regulated association of xCdc7 with Mcm complexes. Interphase extracts or metaphase extracts were subjected to immunoprecipitation with xCdc7 antibodies (xCdc7 IP), xMcm3 antibodies (xMcm3 IP) or protein A alone (Control IP) followed by gel electrophoresis according to the method of Laemmli (ref. 38; see Materials and Methods) and immunoblotting with either xMcm3 antibodies or xCdc7 antibodies. The positions of xCdc7 and xMcm3 proteins are indicated by arrowheads on the right. xMcm3 coprecipitated with xCdc7 in interphase but not in metaphase. Control immunoblots show that the xCdc7-IPs from interphase and metaphase precipitated comparable amounts of xCdc7; and the xMcm3-IPs from interphase and metaphase precipitated comparable amounts of xMcm3 (data not shown).
xCdc7 Expression in Oocytes and Embryos.
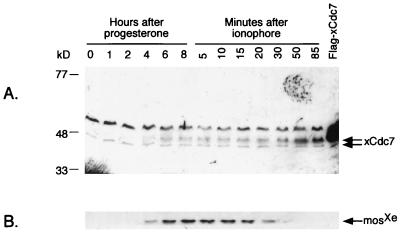
It has long been known that protein synthesis is required late in meiosis I for maturing oocytes to develop the capacity for DNA replication (28–31). To test whether xCdc7 is among the proteins that must be synthesized during this developmental period, its expression was assessed by immunoblot and Northern analysis. Immunoblotting of extracts of maturing oocytes (Fig. 4A) reveals at least two forms of xCdc7. The lower band increases in abundance within an hour after stimulation of these cells with progesterone. The upper band is not present in resting oocytes, (arrested in interphase of meiosis I) but appears after stimulation with progesterone and plateaus by germinal vesicle breakdown. In contrast, MosXe expression continues to increase after germinal vesicle breakdown (ref. 44; Fig. 4B).
Figure 4.
Expression of xCdc7 during oocyte maturation. Resting oocytes (stage VI) were stimulated by progesterone and analyzed by gel electrophoresis according to the method of Anderson et al. (ref. 37; see Materials and Methods) and immunoblotting at the indicated times. Germinal vesicle breakdown occurred at about 4 hours. After 8 hours (approximately meiosis II), the remaining oocytes were treated with calcium ionophore to simulate fertilization. Embryos expressing a Flag-tagged form of xCdc7 (1 kDa greater predicted molecular mass) are indicated by “Flag- xCdc7”. (A) Anti-xCdc7 immunoblot; (B) Same blot as A, reprobed with antibodies to MosXe.
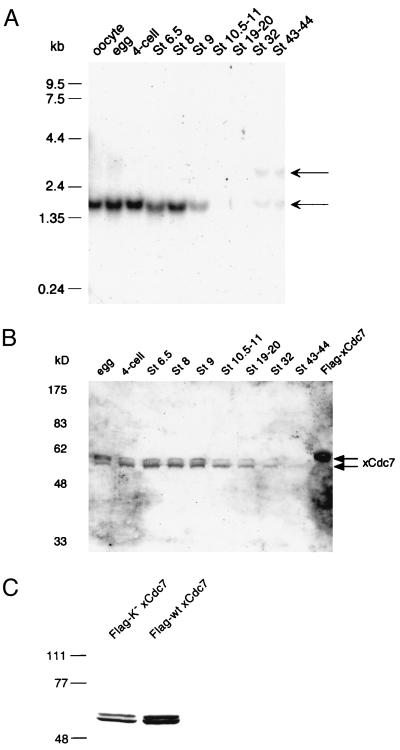
Northern analysis reveals that the xCdc7 transcript is present at relatively constant levels in resting oocytes and through the early embryonic cell cycles (Fig. 5A). The abrupt decline in transcript levels at stage 9 coincides with the midblastula transition, which is characterized by three important developmental phenomena: (i) a pronounced slowing and loss of synchrony in the cell cycle; (ii) the degradation of maternal mRNAs and the beginning of zygotic transcription; and (iii) a change in origins used for replication (45). The reduction in xCdc7 mRNA may be related to the transition from the rapid S phases of pre-midblastula transition divisions to the slower rates of DNA synthesis afterward. However, it should be noted that the level of xCdc7 protein detectable by immunoblotting declined more gradually than the transcript (Fig. 5B), and in some experiments (results not shown), the xCdc7 protein level did not drop until at least stage 20.
Figure 5.
Developmental expression of xCdc7. (A) Developmental Northern analysis of xCdc7. To confirm that equal amounts of RNA were loaded in all lanes, the blot was stripped and reprobed with 18S rRNA (data not shown). (B) Developmental immunoblot of xCdc7 from the same set of embryos as in A. The gel was run according to the method of Laemmli (ref. 38; see Materials and Methods). (C) Expression of recombinant xCdc 7. mRNA encoding Flag-tagged wild-type (wt) or kinase-dead (K−) was injected into embryos, and the expressed protein is a doublet when analyzed by immunoblotting with anti-Flag antibodies.
DISCUSSION
To explore the mechanisms of S phase control in metazoans, we cloned a homolog of the CDC7 gene from a Xenopus oocyte cDNA library. We obtained a sequence encoding a predicted 53.5-kDa protein (xCdc7) that is 26.4% identical to the S. cerevisiae Cdc7p and almost identical to a Xenopus homolog that was reported during the course of this work (ref. 25; see Materials and Methods). Although these proteins are the closest relatives of Cdc7p yet obtained from animal cells, their similarity is limited and is most evident only in the highly conserved protein kinase subdomains (32). Indeed, the putative vertebrate homologs do not possess the unusually large gap between subdomains I and II observed in Cdc7p and have a substantially larger gap between subdomains VII and VIII than is seen in the yeast protein. Therefore, further testing was required to establish whether these metazoan kinases are functional homologs of Cdc7 or merely similar proteins. Accordingly, we generated antibodies to xCdc7 and found they blocked DNA replication in embryos and egg extracts, providing both in vivo and in vitro evidence that a requirement for Cdc7 is conserved from yeast to animals.
When yeast cells carrying a temperature-sensitive cdc7 mutation are grown at a semipermissive temperature, S phase initiation is not significantly delayed, but S phase duration is significantly increased; furthermore, Cdc7p function is still required after release of the cells from a block with hydroxyurea, which interrupts DNA replication early in S phase (14). When cdc7 mutants are arrested at restrictive temperature then returned briefly to permissive temperature before being returned to the restrictive condition, S phase is similarly prolonged, and DNA synthesis is shown to proceed exclusively from early firing origins (15). These data demonstrate that at least in S. cerevisiae, Cdc7p is required for the firing of replication origins rather than the transition from G1 into S phase of the cell cycle. Our data do not provide specific evidence of xCdc7 involvement in origin firing but are consistent with that possibility. Indeed, our data show cell cycle-specific physical association of xCdc7 with the Mcm complex, as predicted by the yeast model. Furthermore, the increase in duration of DNA synthesis over the course of embryonic development is known to be a direct result of the decrease in the number of origins that fire (46). In Xenopus, this reduction coincides with a decline in xCdc7 mRNA and protein levels (Fig. 5) as well as the appearance of a distinct xCdc7-hybridizing transcript. At present it is not clear whether there is a causal connection between these two developmental processes. However, our data could indicate a difference between the mode of xCdc7 regulation in early embryonic versus later somatic cell cycles.
The human homolog of CDC7 (25–27) differs from the Xenopus homolog in that it has a very large 3′-untranslated region. Although it encodes a protein that is 55.7% identical to the Xenopus kinase, it also possesses 31 additional amino acids at the N terminus and 55 more amino acids between subdomains VII and VIII (32) than are seen in the yeast and Xenopus homologs. These characteristics combine to explain the difference in transcript length (3.2 kb for human and 1.7 kb for Xenopus). Notably, the human Cdc7 was cloned from adult tissues or cultured cell lines, whereas the Xenopus Cdc7 was cloned from oocytes. The Northern data could indicate a switch to a zygotically transcribed CDC7 homolog distinct from the gene characterized here; indeed, transcripts migrating at about 3 kb and 1.7-kb transcript are evident by stage 32 (Fig. 5). This could indicate either the existence of such a zygotic homolog or an uncharacterized splice isoform of the cloned maternal gene. Thus, it seems likely that vertebrate embryos employ distinct Cdc7 homologs in earlier and later stages of development. Interestingly, the only other example of this sort of regulation is for another component of the DNA replication pathway in Xenopus, the Mcm6 protein (47).
In normal oocyte maturation, genome duplication does not occur following meiosis I. However, data from several laboratories have indicated that resting frog oocytes are not competent for later DNA replication until a protein or proteins have been synthesized before germinal vesicle breakdown (28–31). The timing of xCdc7 protein synthesis is sufficient to explain these data, although it is possible that another protein (such as a homolog of the yeast Dbf4 protein, for example) must also be synthesized during the same developmental period.
Acknowledgments
We thank Andrea Lewellyn for help with embryo injections and cytology, Jan Kyes for help with antibody purification and characterization, and Eleanor Erikson and Yue-Wei Qian for providing a sample of the oocyte cDNA library. We thank Robert Hollingsworth (Upjohn-Pharmacia) for communicating unpublished data and Robert Sclafani (University of Colorado Health Sciences Center Department of Biochemistry) for helpful discussion. DNA samples were sequenced by the University of Colorado Cancer Center DNA Sequencing and Analysis Core Facility, which is supported by the National Institutes of Health/National Cancer Institute Cancer Core Support Grant (CA46934). This work was supported by grants from the National Institutes of Health to J.L.M. (GM26743 and DK28353). J.L.M. is an Investigator and B.T.R. is an Associate of the Howard Hughes Medical Institute. J.G. is an Irma T. Hirsch Scholar and is sponsored by a Career Development Award from the U.S. Army (DAMD 17-97-1-7071).
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF080068).
References
- 1.Marx J. Science. 1995;270:1585–1587. doi: 10.1126/science.270.5242.1585. [DOI] [PubMed] [Google Scholar]
- 2.Nasmyth K. Trends Genet. 1996;12:405–412. doi: 10.1016/0168-9525(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 3.Toone W M, Aerne B L, Morgan B A, Johnston L H. Annu Rev Microbiol. 1997;51:125–149. doi: 10.1146/annurev.micro.51.1.125. [DOI] [PubMed] [Google Scholar]
- 4.Micklem G, Rowley A, Harwood J, Nasmyth K, Diffley J F. Nature (London) 1993;366:87–89. doi: 10.1038/366087a0. [DOI] [PubMed] [Google Scholar]
- 5.Foss M, McNally F J, Laurenson P, Rine J. Science. 1993;262:1838–1844. doi: 10.1126/science.8266071. [DOI] [PubMed] [Google Scholar]
- 6.Aparicio O M, Weinstein D M, Bell S P. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 7.Madine M, Khoo C-Y, Mills A, Laskey R. Nature (London) 1995;375:421–424. doi: 10.1038/375421a0. [DOI] [PubMed] [Google Scholar]
- 8.Cocker J H, Piatti S, Santocanale C, Nasmyth K, Diffley J F. Nature (London) 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- 9.Schwob E, Bhàm T, Mendenhall M D, Nasmyth K. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 10.Nugroho T T, Mendenhall M D. Mol Cell Biol. 1994;14:3320–3328. doi: 10.1128/mcb.14.5.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendenhall M D. Science. 1993;259:216–219. doi: 10.1126/science.8421781. [DOI] [PubMed] [Google Scholar]
- 12.Schneider B L, Yang Q H, Futcher A B. Science. 1996;272:560–562. doi: 10.1126/science.272.5261.560. [DOI] [PubMed] [Google Scholar]
- 13.Epstein C B, Cross F R. Genes Dev. 1992;6:1695–1706. doi: 10.1101/gad.6.9.1695. [DOI] [PubMed] [Google Scholar]
- 14.Bousset K, Diffley J. Genes Dev. 1998;12:480–490. doi: 10.1101/gad.12.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donaldson A, Fangman W, Brewer B. Genes Dev. 1998;12:491–501. doi: 10.1101/gad.12.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lei M, Kawasaki Y, Young M, Kihara M, Sugino A, Tye B. Genes Dev. 1997;11:3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carpenter P B, Mueller P R, Dunphy W G. Nature (London) 1996;379:357–360. doi: 10.1038/379357a0. [DOI] [PubMed] [Google Scholar]
- 18.Romanowski P, Madine M A, Rowles A, Blow J J, Laskey R A. Curr Biol. 1996;6:1416–1425. doi: 10.1016/s0960-9822(96)00746-4. [DOI] [PubMed] [Google Scholar]
- 19.Coleman T R, Carpenter P B, Dunphy W G. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- 20.Walter J, Newport J W. Science. 1997;275:993–995. doi: 10.1126/science.275.5302.993. [DOI] [PubMed] [Google Scholar]
- 21.Aladjem M I, Rodewald L W, Kolman J L, Wahl G M. Science. 1998;281:1005–1009. doi: 10.1126/science.281.5379.1005. [DOI] [PubMed] [Google Scholar]
- 22.Bartek J, Bartkova J, Lukas J. Exp Cell Res. 1997;237:1–6. doi: 10.1006/excr.1997.3776. [DOI] [PubMed] [Google Scholar]
- 23.Strausfield U, Howell M, Descombes P, Chevalier S, Rempel R, Adamczewski J, Maller J, Hunt T, Blow J. J Cell Sci. 1996;109:1555–1563. doi: 10.1242/jcs.109.6.1555. [DOI] [PubMed] [Google Scholar]
- 24.Philpott A, Friend S H. Mol Cell Biol. 1994;14:5000–5009. doi: 10.1128/mcb.14.7.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato N, Arai K-I, Masai H. EMBO J. 1997;16:4340–4351. doi: 10.1093/emboj/16.14.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang W, Hunter T. Proc Natl Acad Sci USA. 1997;94:14320–14325. doi: 10.1073/pnas.94.26.14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hess G F, Drong R F, Weiland K L, Slightom J L, Sclafani R A, Hollingsworth R E. Gene. 1998;211:133–140. doi: 10.1016/s0378-1119(98)00094-8. [DOI] [PubMed] [Google Scholar]
- 28.Benbow R M, Ford C C. Proc Natl Acad Sci USA. 1975;72:2437–2441. doi: 10.1073/pnas.72.6.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gurdon J B, Speight V A. Exp Cell Res. 1969;55:253–256. doi: 10.1016/0014-4827(69)90488-1. [DOI] [PubMed] [Google Scholar]
- 30.Furuno N, Nishizawa M, Okazaki K, Tanaka H, Iwashita J, Nakajo N, Ogawa Y, Sagata N. EMBO J. 1994;13:2399–2410. doi: 10.1002/j.1460-2075.1994.tb06524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gurdon J B. Proc Natl Acad Sci USA. 1967;58:545–552. doi: 10.1073/pnas.58.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanks S K, Quinn A M. Methods Enzymol. 1991;200:38–62. doi: 10.1016/0076-6879(91)00126-h. [DOI] [PubMed] [Google Scholar]
- 33.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 34.Huan R S, Servanti I M, Moss J. BioTechniques. 1992;13:515–518. [PubMed] [Google Scholar]
- 35.Anderson J, Lewellyn A, Maller J. Mol Biol Cell. 1997;8:1195–1206. doi: 10.1091/mbc.8.7.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray A W. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- 37.Anderson C W, Baum P R, Gesteland R F. J Virol. 1973;12:241–252. doi: 10.1128/jvi.12.2.241-252.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 39.Gautier J, Minshull J, Lohka M, Glotzer M, Hunt T, Maller J L. Cell. 1990;60:487–494. doi: 10.1016/0092-8674(90)90599-a. [DOI] [PubMed] [Google Scholar]
- 40.Hartley R, Sible J, Lewellyn A, Maller J. Dev Biol. 1997;188:312–321. doi: 10.1006/dbio.1997.8647. [DOI] [PubMed] [Google Scholar]
- 41.Hardy C, Dryga O, Seematter S, Pahl P, Sclafani R. Proc Natl Acad Sci USA. 1997;94:3151–3155. doi: 10.1073/pnas.94.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sherman D A, Forsburg S L. Nucleic Acids Res. 1998;26:3955–3960. doi: 10.1093/nar/26.17.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherman D A, Pasion S G, Forsburg S L. Mol Biol Cell. 1998;9:1833–1845. doi: 10.1091/mbc.9.7.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roy L, Haccard O, Izumi T, Lattes B, Lewellyn A, Maller J. Oncogene. 1996;12:2203–2211. [PubMed] [Google Scholar]
- 45.Hyrien O, Maric C, Mechali M. Science. 1995;270:994–997. doi: 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- 46.Blumenthal A B, Kriegstein H J, Hogness D S. Cold Spring Harb Symp Quant Biol. 1974;38:205–223. doi: 10.1101/sqb.1974.038.01.024. [DOI] [PubMed] [Google Scholar]
- 47.Sible J C, Erikson E, Hendrickson M, Maller J L, Gautier J. Curr Biol. 1998;8:347–350. doi: 10.1016/s0960-9822(98)70136-8. [DOI] [PubMed] [Google Scholar]