Abstract
Centrosomes nucleate microtubules and duplicate once per cell cycle. This duplication and subsequent segregation in mitosis results in maintenance of the one centrosome/cell ratio. Centrosome duplication occurs during the G1/S transition in somatic cells and must be coupled to the events of the nuclear cell cycle; failure to coordinate duplication and mitosis results in abnormal numbers of centrosomes and aberrant mitoses. Using both in vivo and in vitro assays, we show that centrosome duplication in Xenopus laevis embryos requires cyclin/cdk2 kinase activity. Injection of the cdk (cyclin-dependent kinase) inhibitor p21 into one blastomere of a dividing embryo blocks centrosome duplication in that blastomere; the related cdk inhibitor p27 has a similar effect. An in vitro system using Xenopus extracts carries out separation of the paired centrioles within the centrosome. This centriole separation activity is dependent on cyclin/cdk2 activity; depletion of either cdk2 or of the two activating cyclins, cyclin A and cyclin E, eliminates centriole separation activity. In addition, centriole separation is inhibited by the mitotic state, suggesting a mechanism of linking the cell cycle to periodic duplication of the centrosome.
The centrosome nucleates the polymerization of microtubules, organizes the ends of those microtubules into functional arrays, and duplicates once per cell cycle (1). In both animal cells and fungi (where the centrosome equivalent is termed the spindle pole body), duplication of the single centrosome is initiated at the G1/S transition and completed before mitosis, where the duplicated centrosomes play a role in organizing the poles of the mitotic spindle. The centrosomes are segregated at mitosis such that each of the two cells resulting from division receives only one. The precise duplication and segregation of the centrosome is required for normal cell cycle progression and accurate segregation of the chromosomes at mitosis. Because defects in the fidelity of chromosome segregation are a common characteristic of cancer cells and are likely to be important in the progression to a cancerous phenotype, an understanding of the mechanism of centrosome duplication is essential.
Although much progress has been made in understanding the composition and function of the centrosome, little is known of how duplication of the centrosome is regulated or of how the organelle is assembled each cell cycle. Much of what is known comes from morphological analysis of duplication of the animal cell centrosome and genetic analysis of the duplication of the fungal spindle pole body. The centrosome consists of a pair of centrioles, typically in a perpendicular orientation, surrounded by pericentriolar material, from which the microtubules grow. Duplication of the centrosome is semiconservative: the paired centrioles split and a new centriole forms in association with each, creating two centrosomes (2). The two centrosomes remain in close contact until prophase of mitosis, when they migrate to opposite sides of the nucleus, ultimately forming the bipolar mitotic spindle (3). The spindle pole body is a laminar plaque in the nuclear envelope with microtubules growing from both the cytoplasmic and nuclear faces. Duplication of the spindle pole body in budding yeast requires the functions of several genes (reviewed in ref. 4), including the Saccharomyces cerevisiae cdc2 homolog CDC28; the centrin homolog CDC31; KAR1, a protein kinase MPS1; and PCS1, a proteasome cap subunit (5).
The major oscillations of the cell cycle are driven by the periodic activation of specific cyclin/cdk (cyclin-dependent kinase) kinases. We have examined centrosome duplication in the simple embryonic cell cycle of Xenopus laevis and in extracts made from Xenopus eggs, in the hope of understanding how the cell cycle controls this process. Fertilized Xenopus embryos divide rapidly and synchronously for approximately 12 divisions with a cell cycle time of 30 min. These divisions require the periodic accumulation and destruction of the mitotic cyclin A and cyclin B proteins, which associate with the cdc2 protein kinase. Treatment of embryos with the protein synthesis inhibitor cycloheximide prevents the accumulation of the mitotic cyclins and results in cessation of the nuclear division cycles. In contrast, centrosome duplication continues under these conditions, resulting in cells with more than two centrosomes (6, 7). Thus, centrosome duplication does not require mitotic cyclin/cdc2 activity. The other major cyclin/cdk activity in frog eggs is cyclin E/cdk2, which has been shown to be required for the initiation of DNA synthesis (8, 9), an event that occurs at approximately the same time in the cell cycle as centrosome duplication. In somatic mammalian cells, both cyclin E levels and cyclin E/cdk2 kinase activity peak at the G1/S transition (10, 11), which is similar to the timing of centrosome duplication and the start of S phase. The frog embryonic cell cycle lacks a G1 phase, but cyclin E and cdk2 are present at constant levels throughout the early divisions (12). Balczon et al. (13) found that in some somatic cell lines, centrosome duplication continues under conditions of S phase arrest, and Hinchcliffe et al. (14) have shown that in sea urchin embryos the potential for multiple rounds of centrosome duplication is unique to S phase. These results led us to test the hypothesis that the cyclin E/cdk2 kinase is driving centrosome duplication.
In this study we show that centrosome duplication in vivo depends on cyclin E/cdk2 activity. In addition, we use an in vitro assay based on extracts of frog eggs to demonstrate that cyclin E/cdk2 drives the separation of centrioles, an early step in centrosome duplication. In this in vitro assay, the presence of high levels of the mitotic cyclin B/cdc2 kinase inhibited centriole separation. These results provide an explanation for the lack of a protein synthesis requirement for centrosome duplication in the embryonic cell cycle and also suggest a mechanism for coordination of centrosome duplication with mitosis.
MATERIALS AND METHODS
Frog Embryo Injections and Imaging.
Animal pole cells from frog embryos at the 16–64 cell stage were injected with the indicated protein and a fluorescein isothiocyanate-conjugated dextran (Molecular Probes) to allow for detection of injected cells. After injection, embryos were placed in 5% MMR + 1 mg/ml cycloheximide for 4 h. They then were fixed in methanol and processed for immunofluorescence according to Gard et al. (6), with the following changes: after bleaching, embryos were rehydrated in PBS (three times for 5 min) and the vegetal pole was removed. The embryos were incubated in primary antibody [anti-α-tubulin DM1α, 1:100 in PBSBT (PBS plus 3% BSA/0.1% Triton/0.02% sodium azide), or anti-γ-tubulin–GTU-88 (Sigma), 1:200 in PBSBT], washed in PBS, and incubated with secondary antibody [rhodamine-conjugated donkey anti-mouse (Jackson ImmunoResearch), 1:100 in PBSBT]. The embryos were dehydrated in 100% methanol (three times for 10 min), cleared in benzyl alcohol/benzyl benzoate (1:2), and mounted on regular slides with a square coverslip. Embryos were examined on a Zeiss Photoscope using a ×16/0.4 numerical aperture objective. Images were obtained with an MRC-1024 confocal system (Bio-Rad).
Protein and Centrosome Purification.
Cdk inhibitor proteins (p21, p21N, p21C, p27N, p27C) were made as glutathione S-transferase fusions according to the method described (8). Human cyclin B, cyclin E, and cdk2 baculoviruses were generously provided by D. O. Morgan (University of California at San Francisco). Sf9 insect cells were coinfected with cyclin E-His6 and cdk2 baculoviruses as described (15), and the cyclin E/cdk2 complex was purified by using nickel affinity chromatography under the conditions described (16). Centrosomes were isolated from XTC cells, a Xenopus epithelial cell line, essentially by a method described for mammalian cells (17). These centrosomes consisted almost exclusively of single centrosomes, each with a pair of centrioles.
In Vitro Centriole Separation Assay.
Frog embryo extracts were made from fertilized eggs that were placed in 1 mg/ml cycloheximide after the first cleavage and incubated for 45 min. Eggs were washed twice with S-lysis buffer (250 mM sucrose/2.5 mM MgCl2/50 mM KCl/10 mM Hepes/KOH, pH 7.5), then placed in S-lysis plus 1 mM DTT, 100 μg/ml cycloheximide, and protease inhibitors, and processed as described (18). The resulting extract was diluted 1:3 into Acetate Buffer Complete (19) and spun at 100,000 × g for 30 min. One hundred microliters of the resulting supernatant was incubated with 10 μl purified XTC centrosomes at 25°C. When appropriate, extract was incubated with indicated proteins or solutions for 30 min at 25°C before the addition of centrosomes. Reactions were stopped by the addition of 300 μl of 10 mM Tris, pH 8.0/nocodazole to 75 μg/ml and placed on ice for 30 min. Next, the sample was spun onto coverslips and processed for immunofluorescence as described (20). The anti-α-tubulin antibody DM1α was used at 1:100 in PBSBT, and the anti-γ-tubulin antibody XGC-1–4 was used at 1:500. Fluorescein isothiocyanate-conjugated donkey anti-mouse and Texas Red-conjugated goat anti-rabbit secondary antibodies (Jackson ImmunoResearch) were used at 1:100. The images were taken on an Applied Precision DeltaVision deconvolution system using an Olympus IX-70 inverted microscope and deconvolved with the Agard and Sedat inverse matrix algorithm.
Extract Manipulations.
Preparation of Suc1p and BSA (for mock depletions) beads and their use in depletion of extracts was as described (8). Double depletions were for approximately 90 min each. The cyclin E, cyclin A, and cdk2 C-peptide antisera and their use in immunodepletions were as described (8). H1 kinase assays (8) were performed to evaluate the depleted extracts. Interphase extract was driven into mitosis by the addition of human cyclin B for 1 h, and cell cycle state was confirmed by morphology of added sperm and kinase activity.
RESULTS
CDK Inhibitors Block Centrosome Duplication in Vivo.
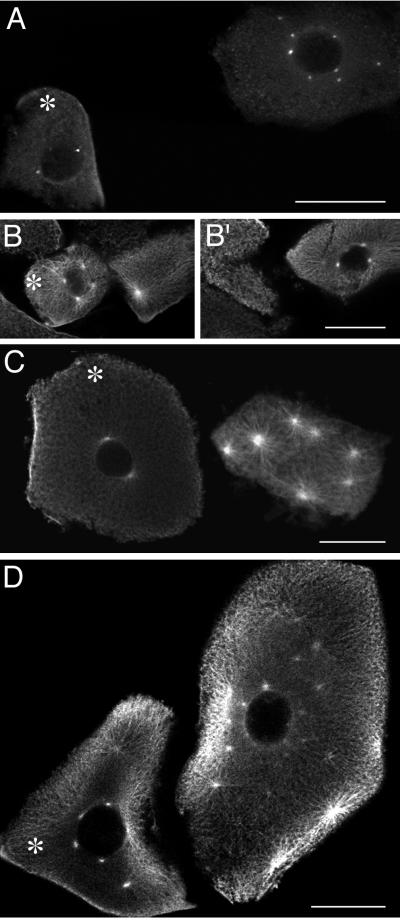
Previous work had shown that cycloheximide treatment of Xenopus embryos blocked cell division and DNA replication, but allowed many rounds of centrosome duplication (6). Although cyclins A and B are present only at low levels under these conditions because of their proteolysis at mitosis, cyclin E and its partner, cdk2, are present at constant levels. To test the hypothesis that cyclin E/cdk2 drives centrosome duplication, single cells from early embryos were injected with specific cdk inhibitor proteins, treated with cycloheximide, and then assayed by confocal microscopy for the extent of centrosome duplication. Fluorescein isothiocyanate-conjugated dextran was coinjected as a visible marker for injected cells. The cdk inhibitor protein p21 binds to and inhibits the activity of cdk2 complexed with either cyclin A or cyclin E (21–24). The cyclin E/cdk2 kinase accounts for the majority of p21-inhibitable cdk activity in the early Xenopus embryo (8, 9). Injection of p21 inhibited the extra rounds of centrosome duplication observed in neighboring uninjected cells. The injected cell shown in Fig. 1A has two centrosomes whereas the neighboring uninjected cell has eight centrosomes. The effect of p21 on centrosome duplication is dose-dependent. Injection of p21 at a final concentration of 3 μM resulted in an average of 1.9 centrosomes (Table 1), whereas p21 at 1 μM resulted in an average of 3.1 centrosomes (significantly different within a 95% confidence level) (Table 1). Cells injected with a control solution had an average of 6.4 centrosomes, similar to the number in uninjected cells (Table 1).
Figure 1.
The cdk inhibitor p21 blocks centrosome duplication in frog embryos. Embryonic cells were injected with various inhibitors and treated with cycloheximide for 4 h to allow for centrosome overduplication. Asterisks mark the injected cells, as visualized by coinjection of a fluorescein-conjugated dextran. (A) p21. (B and B′) p21 and cyclin E (two different sections are shown to view all centrosomes). (C) p21 N terminus. (D) p21 C terminus. (A) γ-Tubulin staining. (B, B′, C, and D) α-Tubulin staining. The difference in cell size is because of the variation in cell stage at the time of injection. (Bar = 100 μm.)
Table 1.
Summary of in vivo injection experiments
| Solution injected | Average number of centrosomes*
|
Ratio, I/U† | |
|---|---|---|---|
| Injected cells | Uninjected cells | ||
| Control‡ | 6.4 ± 0.8 | 7.3 ± 0.6 | 0.88 |
| n = 18 | n = 31 | ||
| p21, 3 μM | 1.9 ± 0.3 | 6.0 ± 0.6 | 0.32§ |
| n = 20 | n = 20 | ||
| p21, 2 μM | 2.1 ± 0.2 | 10.4 ± 0.6 | 0.20§ |
| n = 36 | n = 38 | ||
| p21, 1.5 μM | 2.5 ± 0.2 | 5.5 ± 0.4 | 0.45§ |
| n = 32 | n = 23 | ||
| p21, 1 μM | 3.1 ± 0.3 | 5.6 ± 0.5 | 0.55§ |
| n = 52 | n = 51 | ||
| p21 + cyclin E‖ | 7.7 ± 0.5 | 7.5 ± 0.5 | 1.03 |
| n = 40 | n = 34 | ||
| p21 N terminus, 10 μM | 0.9 ± 0.3 | 5.8 ± 0.4 | 0.16§ |
| n = 11 | n = 15 | ||
| p21 N terminus, 5 μM | 1.2 ± 0.2 | 7.6 ± 0.8 | 0.16§ |
| n = 29 | n = 19 | ||
| p21 C terminus, 8 μM | 5.9 ± 0.3 | 5.4 ± 0.3 | 1.09 |
| n = 33 | n = 36 | ||
| p27 N terminus, 16 μM | 0.9 ± 0.2 | 5.8 ± 0.7 | 0.16§ |
| n = 35 | n = 29 | ||
| p27 N terminus, 8 μM | 2.8 ± 0.2 | 7.5 ± 0.5 | 0.37§ |
| n = 30 | n = 30 | ||
| p27 C terminus, 10 μM | 4.6 ± 0.3 | 5.3 ± 0.4 | 0.87 |
| n = 40 | n = 40 | ||
| p27 C terminus, 5 μM | 6.9 ± 0.5 | 6.9 ± 0.5 | 1.00 |
| n = 29 | n = 30 | ||
n is the number of cells counted for each condition.
The average number of centrosomes is shown ±SEM.
The ratio is centrosome number for injected cells divided by centrosome number for uninjected cells.
The control solution used was PBS.
For these experiments, the average number of centrosomes in injected cells vs. uninjected cells is significantly different within a 99% confidence level.
Cyclin E was added at twice the molarity of p21.
To determine whether the effect of p21 injection was a result of cyclin E/cdk2 inhibition, p21 was coinjected with a 2-fold molar excess of cyclin E protein. The injected cells were able to undergo rounds of centrosome duplication to the same extent as neighboring uninjected cells (Fig. 1 B and B′). On average, the cells injected with p21 + cyclin E had 7.7 centrosomes compared with 7.5 centrosomes in uninjected cells (Table 1). Thus, excess cyclin E was able to alleviate p21 inhibition of centrosome duplication. The N terminus of p21 has been shown to specifically inhibit cyclin E/cdk2 activity, whereas the C terminus binds and inhibits PCNA, a DNA-replication factor (25). As an additional test of the specificity of p21 inhibition, cells were injected with N- and C-terminal fragments of p21. The N-terminal fragment of p21 inhibited centrosome duplication to the same extent as the full-length protein (Fig. 1C). Cells injected with the p21 N-terminal fragment had an average of 1.2 centrosomes per cell versus 7.6 in uninjected cells (Table 1). In contrast, the C-terminal fragment of p21 had no effect on centrosome duplication (Fig. 1D), resulting in an average of 5.9 centrosomes versus 5.4 centrosomes in uninjected cells (Table 1).
The cdk inhibitor protein p27 is related to p21 and has similar effects on the activity of cyclin E/cdk2 (26–28); as for p21, the N terminus of p27 is responsible for the cdk inhibition (P.K.J. and A. Dutta, unpublished results). Injection of a p27 N-terminal fragment blocked centrosome duplication, resulting in an average of 0.9 centrosomes per cell vs. 5.8 in uninjected cells (Table 1). The C-terminal fragment of p27, which does not inhibit cdk activity, had no effect on duplication.
An in Vitro Assay for Centriole Separation.
To examine the centrosome duplication process in more detail, we developed an in vitro assay based on that of Tournier et al. (29) that carries out separation of the paired centrioles within the centrosome. Centrosomes purified from Xenopus XTC cells were incubated with a clarified extract made from fertilized Xenopus eggs treated with cycloheximide. After incubation, the extract was treated to depolymerize microtubules, and the centrosomes were fixed and stained for α-tubulin to visualize the centrioles and γ-tubulin to visualize the pericentriolar material. The starting centrosomes consist of paired centrioles (doublets) with sparse pericentriolar material, as revealed by whole-mount electron microscopy (data not shown) and deconvolution immunofluorescence microscopy (see below).
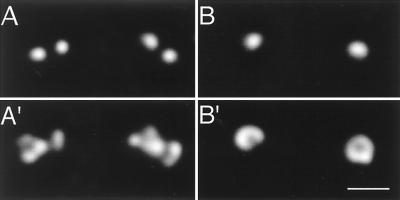
At the zero time point, the majority of centrioles (77%) appear as doublets (Fig. 2 A and A′; Table 2). During the next 30–60 min, intermediate degrees of centriole separation were observed. By 1 h, the majority (77%) of centrioles appear as singlets, with a small percentage of the centrioles appearing as separated doublets (doublets with an increased intercentriolar distance) (Fig. 2 B and B′; Table 2). The total number of individual centrioles at the 0- and 1-h time points of the assay remained approximately the same (average of 14 centrioles/microscope field at 0 h vs. 15.5 centrioles/field at 1 h). Thus, the appearance of singlet centrioles was due to separation of doublets and not coalescence of paired centrioles into a single object. It is convenient to express centriole separation as percent conversion of starting doublet centrioles to nondoublet forms. At the 1-h endpoint, there is 90% conversion. The distinction between centriole doublets, separated doublets, and single centrioles was clear from the microscopic data. The average distance between centrioles in centriole doublets was 0.55 ± 0.008 μm, whereas the average distance between centrioles in separated doublets was 1.07 ± 0.05 μm. The average distance between any two random centrioles in these experiments was 33 μm; thus, centriole doublets and separated doublets are not simply two single centrioles randomly juxtaposed. Although not apparent in Fig. 2, incubation with extract usually resulted in increased γ-tubulin staining, consistent with the described recruitment of soluble γ-tubulin complexes by centrioles (20, 30).
Figure 2.
An in vitro centriole separation assay. Deconvolution images of centriole doublets and singlets. (A and A′) These doublets represent the 0-h starting point of the assay. (B and B′) At the 1-h endpoint, the majority of centrosomes are singlets, as depicted. (A and B) α-Tubulin staining. (A′ and B′) γ-Tubulin staining. (Bar = 1 μm.)
Table 2.
Centriole separation assay data
| Condition | Percentage*
|
Conversion percentage† | ||
|---|---|---|---|---|
| ●● | ● | ● ● | ||
| 0 h | 77 | 23 | — | —‡ |
| 1 h | 8 | 77 | 15 | 90 |
| p21 | 68 | 26 | 6 | 12 |
| p21N | 73 | 25 | 2 | 5 |
| p21C | 17 | 72 | 11 | 78 |
| Hexokinase | 84 | 16 | — | <0.2 |
| EDTA | 79 | 20 | 1 | <0.2 |
| EDTA + MgCl2 | 26 | 66 | 8 | 66 |
| 6-DMAP | 78 | 22 | — | <0.2 |
p21, p21N, and p21C were used at a final concentration of 15 μM. Hexokinase was added at 20 units/ml with 10 mM glucose. EDTA and MgCl2 were added for a final concentration of 5 mM. 6-Dimethylaminopurine (6-DMAP) was used at 500 μM.
Approximately 500 centrosomes were counted, categorized, and expressed as a percentage. The three categories depict doublets (D), singlets (S), and separated doublets (SD), respectively.
This represents the percentage of doublets that was converted to singlets or separated doublets, using the formula: [(S + SD) − (S0)]/D0; where S0 = 23 and D0 = 77. If the calculated conversion percentage was ≤0, a minimum value (<1/500 = <0.2) was used.
The conversion value at time 0 is set to zero.
Requirements for Centriole Separation Activity.
We next manipulated the extracts to determine the requirements for centriole separation, first examining the requirement for energy. Depletion of nucleotide triphosphates by addition of hexokinase and glucose inhibited separation, resulting in <0.2% conversion to single centrioles (Table 2). Addition of the divalent cation chelator EDTA also inhibited centriole separation (<0.2% conversion). EDTA chelates Mg2+, which is a necessary cofactor of ATP- and GTP-hydrolyzing enzymes; excess Mg2+ was able to rescue the EDTA-treated extracts, resulting in 66% conversion (Table 2). 6-Dimethyl aminopurine, an inhibitor of cyclin-dependent kinases (31, 32), also blocked centriole separation, resulting in <0.2% conversion. This is consistent with the results implicating the cdk2 kinase in the in vivo reaction.
To test the requirement for cyclin E/cdk2 activity in the centriole separation assay, centrosomes were added to extract that had been incubated with p21 for 30 min. Full-length p21 blocked centriole separation, resulting in only 12% conversion (Table 2). Consistent with the in vivo experiments, the cdk-inhibiting N terminus of p21 blocked centriole separation (5% conversion), whereas the C terminus of p21 allowed separation (78% conversion) (Table 2).
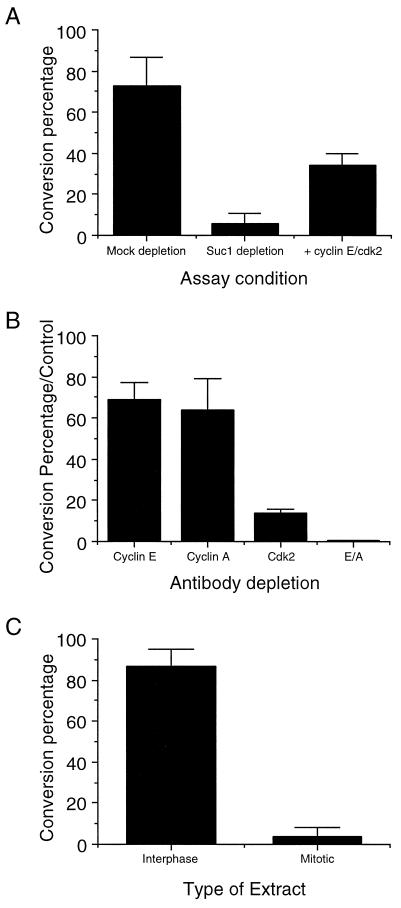
We wished to confirm the results implicating cdk2 and associated cyclins in centrosome duplication by direct means. Two approaches were taken. First, extracts were depleted of cdks and associated proteins by incubation with p13suc1 beads; successful depletion was confirmed by H1 kinase assays. Cdk-depleted extract supported only 6% conversion of centriole pairs to singlets, compared with 73% conversion found with mock-depleted extract (Fig. 3A). Importantly, cyclin E/cdk2 complex, produced by baculovirus expression, was able to restore significant activity to the depleted extract, resulting in 34% conversion (Fig. 3A). Second, extracts were immunodepleted of cdk2, resulting in only 14% of the conversion found with control-depleted extracts (Fig. 3B).
Figure 3.
The dependence of centriole separation on cyclin-dependent kinases. The graphs express centriole separation activity as conversion percentages, as described in Table 2. (A) Depletion and rescue of centriole separation activity with Suc1p and cyclin E/cdk2. (B) Depletion of both cyclin E- and cyclin A-dependent kinases inhibits centriole separation. The conversion percentage has been normalized to the control depletion (uncoupled protein A-Sepharose). (C) The mitotic state inhibits centriole separation.
Cdk2 is known to associate with both cyclin E and cyclin A, although it is predominantly found associated with cyclin E in Xenopus early development (33). Surprisingly, immunodepletion of cyclin E had only a small effect on centriole separation activity; similarly, immunodepletion of cyclin A had only a small effect (Fig. 3B). However, immunodepletion of both cyclin E and cyclin A resulted in a complete loss of separation activity (Fig. 3B).
Thus far, we have shown that inhibition or depletion of cyclin/cdk2, the major interphase cell cycle kinase, prevents centrosome duplication. Because centrosome duplication is not subject to a once-and-only-once control in embryonic systems, there must be a mechanism for coordinating it with other events of the mitotic cycle. One possible mechanism for such coordination would be inhibition of centrosome duplication by the mitotic state. To determine the effect of cell cycle state on centriole separation, we drove interphase extract into mitosis with cyclin B protein. Incubation of centrosomes with this mitotic extract resulted in only 4% conversion of doublet centrioles to single centrioles compared with 87% conversion with an untreated interphase extract (Fig. 3C). We obtained similar results with extracts arrested in mitosis by cytostatic factor (CSF); the inhibition could be relieved by the addition of calcium, which results in the destruction of CSF and entry into interphase (data not shown).
DISCUSSION
We have demonstrated here that inhibition of cyclin/cdk2 blocks centrosome duplication in vivo and have used an in vitro assay to show that, of the cdk complexes, cyclin E/cdk2, in particular, is sufficient for this activity. In this in vitro assay, purified centrosomes were incubated with egg extract. The centriole pairs within the centrosomes separated into single centrioles; this centriole separation activity was heat- and dilution-sensitive, ATP-dependent, and was specific to interphase extracts. We believe that the centriole separation that occurs in this assay is likely to correspond to the centriole “disorientation” step described in the centrosome duplication cycle of somatic mammalian cells (3), followed by physical separation of the centrioles in our preparation of them for microscopy. Although single centriole centrosomes are not usually observed in vivo, we suppose that the ability to separate the centrioles in vitro reflects the loss of “pairing” that must be an early step in centrosome duplication. We note that our assay was designed to carry out a simplified version of centrosome duplication, in which only the centriole separation step occurs; other in vitro systems have been shown to carry out the later step of new centriole formation (29, 34). Our results clearly show that cdk2 is required for centriole separation and suggest that cyclin E is the cdk2-binding partner involved.
Both cyclin E and cyclin A have been shown to bind to cdk2 in vitro; however, in the early divisions of Xenopus, only cyclin E is found associated with cdk2 (33). Why then was depletion of both cyclin E and cyclin A required to inhibit centriole separation in our experiments? Although cyclin E is the preferred binding partner of cdk2, we suppose that in the absence of cyclin E, the cyclin A present in interphase extracts as prepared here would bind to cdk2 (P.K.J., unpublished results). We consider it most likely that cyclin E is the relevant cyclin for centrosome duplication in vivo. The dependence on cyclin E/cdk2 activity would explain the observed lack of a protein synthesis requirement for centrosome duplication; cyclin E protein levels do not decrease until after the midblastula transition (≈12th division) (12). Thus, inhibiting protein synthesis in early embryos would have no effect on the amount of cyclin E or, by extension, the amount of the cyclin E/cdk2 kinase.
It is useful to compare centrosome duplication with DNA replication, the only other known discrete duplication event in cells. In embryonic systems with rapid cell division cycles, S phase, during which both DNA replication and centrosome duplication occur, alternates with M phase. Like centrosome duplication, DNA replication depends on cyclin E/cdk2 activity (8, 35–37) and is inhibited in M phase (38). DNA replication makes use of the antiparallel, complementary nature of double-stranded DNA to provide a template for precise duplication. Centrioles are the only known discrete structure in the centrosome, yet, it is not clear how they could act as a template for the growth of another centriole, given that new centrioles typically grow near, but not directly attached to, old centrioles. Also, fungi lack centrioles, yet duplication of the fungal spindle pole body occurs with timing and fidelity similar to that in animal cells. A striking difference is that DNA replication occurs only once after inhibition of protein synthesis in embryos, whereas centrosome duplication continues for many rounds under the same conditions. One explanation for this difference is that DNA replication is known to have a second level of control, known as licensing, which ensures that replication does not occur without an intervening mitosis (39, 40). It appears that there is no similar once-and-only-once control for centrosome duplication in the cycloheximide-treated frog embryos that we have examined here.
If cyclin E/cdk2 is responsible for the initiation of centrosome duplication and is present throughout early divisions, how, then, is duplication coordinated with the other events of the cell cycle? Our results demonstrate that the active mitotic state inhibits the duplication process, similar to results in sea urchin embryos (14). If the mechanics of centrosome duplication required a significant portion of S phase in the rapid embryonic cycles, then the mitotic inhibition normally would prevent the initiation of a second round and ensure an oscillation of duplication and division. In somatic cells there are likely to be additional controls on centrosome duplication. First, activity of cyclin E/cdk2 is restricted to the G1/S transition (10, 11). Second, somatic cells do not have the large stockpiles of centrosomal components found in eggs and must rely on new synthesis; in yeast, transcription of genes for the spindle pole body components γ-tubulin and Spc110p is limited to the G1/S period in which duplication takes place. It is interesting to note that Chinese hamster ovary cells, in which extra rounds of centrosome duplication have been observed to occur under conditions of S phase arrest (13), continue to synthesize protein and grow while arrested, whereas HeLa cells do not (41). A third difference is that somatic cells have cell cycle checkpoint controls that are absent in the Xenopus egg. Particularly interesting is the observation that p53−/− mouse cells display a high frequency of excess centrosomes (42). One of the roles of p53 is to induce p21 and to enforce a cell cycle checkpoint after DNA damage (43). Although in our experiments we have used p21 simply as a reagent to inhibit cyclin E/cdk2 activity, it is possible that in somatic cells, the p53/p21 checkpoint helps to coordinate centrosome duplication with the other events of the cell cycle.
A common theme emerging in the cell cycle is that important transitions are made irreversible by protein destruction. For example, destruction of the yeast cdk inhibitor p40 is essential for progression into S phase (44), destruction of mitotic cyclins is necessary for exit from M phase (45), and destruction of Pds1p is required for sister chromatid separation in yeast mitosis (46). In addition, a component of the destruction machinery, a cap subunit of the proteasome, is required for spindle pole body duplication in yeast (5). Because an early step in centrosome duplication is the separation of the centrioles, it is easy to imagine that destruction of a proteinaceous link between the centrioles could be the initiating event. If so, cyclin E/cdk2 might be required to phosphorylate and, thus, mark for destruction, a particular protein, which is similar to the role of the yeast G1/S cyclin/cdk in the destruction of the cell cycle inhibitor p40 (47). We note that the nature of the link between centrioles is not known, but that a fibrous network has been observed between centrioles in electron microscopy preparations of purified centrosomes (48). Fry et al. (49) have identified a coiled-coil protein, C-Nap1, that is located at the proximal ends of centrioles and can be phosphorylated by Nek2, a kinase that also is located at the centrosome. Remarkably, overexpression of Nek2 results in “centrosome splitting” (50), which might be analogous to the centriole separation that we have observed in our assay. It is not known whether there is any link between cyclin E/cdk2 and these centrosomal proteins; determination of the substrates of cyclin E/cdk2 relevant to centrosome duplication ultimately will be necessary to establish the mechanism of cell cycle control of centrosome duplication.
Acknowledgments
We thank David Morgan for providing the human cyclin and cdk baculoviruses and Sigrid Reinsch for helpful advice with extract preparations. We thank Janet Carminati for critical reading of the manuscript and Steve Murphy and Lenore Urbani for technical advice on baculovirus experiments and centrosome preparations. This work was supported by grants to T.S. from the American Cancer Society and the Searle Foundation. K.R.L. was supported by a National Institutes of Health predoctoral training grant.
ABBREVIATION
- cdk
cyclin-dependent kinase
Note Added in Proof
Hinchcliffe et al. (51) have recently reported similar results concerning the role of cyclin E/cdk2 in centrosome duplication.
References
- 1.Kellogg D R, Moritz M, Alberts B M. Annu Rev Biochem. 1994;63:639–674. doi: 10.1146/annurev.bi.63.070194.003231. [DOI] [PubMed] [Google Scholar]
- 2.Kochanski R S, Borisy G G. J Cell Biol. 1990;110:1599–1605. doi: 10.1083/jcb.110.5.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuriyama R, Borisy G G. J Cell Biol. 1981;91:814–821. doi: 10.1083/jcb.91.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winey M, Byers B. Trends Genet. 1993;9:300–304. doi: 10.1016/0168-9525(93)90247-f. [DOI] [PubMed] [Google Scholar]
- 5.McDonald H B, Byers B. J Cell Biol. 1997;137:539–553. doi: 10.1083/jcb.137.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gard D L, Hafezi S, Zhang T, Doxsey S J. J Cell Biol. 1990;110:2033–2042. doi: 10.1083/jcb.110.6.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sluder G, Miller F J, Cole R, Rieder C L. J Cell Biol. 1990;110:2025–2032. doi: 10.1083/jcb.110.6.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson P K, Chevalier S, Philippe M, Kirschner M W. J Cell Biol. 1995;130:755–769. doi: 10.1083/jcb.130.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strausfeld U P, Howell M, Rempel R, Maller J L, Hunt T, Blow J J. Curr Biol. 1994;4:876–883. doi: 10.1016/s0960-9822(00)00196-2. [DOI] [PubMed] [Google Scholar]
- 10.Koff A, Giordano A, Desai D, Yamashita K, Harper J W, Elledge S, Nishimoto T, Morgan D O, Franza B R J, Roberts J M. Science. 1992;257:1689–1694. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- 11.Dulic V, Lees E, Reed S I. Science. 1992;257:1958–1961. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- 12.Hartley R S, Rempel R E, Maller J L. Dev Biol. 1996;173:408–419. doi: 10.1006/dbio.1996.0036. [DOI] [PubMed] [Google Scholar]
- 13.Balczon R, Bao L, Zimmer W E, Brown K, Zinkowski R P, Brinkley B R. J Cell Biol. 1995;130:105–115. doi: 10.1083/jcb.130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinchcliffe E H, Cassels G O, Rieder C L, Sluder G. J Cell Biol. 1998;140:1417–1426. doi: 10.1083/jcb.140.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgan D O, Kaplan J M, Bishop J M, Varmus H E. Methods Enzymol. 1991;200:645–660. doi: 10.1016/0076-6879(91)00177-x. [DOI] [PubMed] [Google Scholar]
- 16.Strausfeld U P, Howell M, Descombes P, Chevalier S, Rempel R E, Adamczewski J, Maller J L, Hunt T, Blow J J. J Cell Sci. 1996;109:1555–1563. doi: 10.1242/jcs.109.6.1555. [DOI] [PubMed] [Google Scholar]
- 17.Mitchison T J, Kirschner M W. Methods Enzymol. 1986;134:261–268. doi: 10.1016/0076-6879(86)34094-1. [DOI] [PubMed] [Google Scholar]
- 18.Murray A W. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- 19.Reinsch S, Karsenti E. Curr Biol. 1997;7:211–214. doi: 10.1016/s0960-9822(97)70092-7. [DOI] [PubMed] [Google Scholar]
- 20.Stearns T, Kirschner M. Cell. 1994;76:623–637. doi: 10.1016/0092-8674(94)90503-7. [DOI] [PubMed] [Google Scholar]
- 21.El-Deiry W S, Tokino T, Velculesco V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 22.Gu Y, Turck C W, Morgan D O. Nature (London) 1993;366:707–710. doi: 10.1038/366707a0. [DOI] [PubMed] [Google Scholar]
- 23.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 24.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. Nature (London) 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 25.Chen J, Jackson P K, Kirschner M, Dutta A. Nature (London) 1995;374:386–388. doi: 10.1038/374386a0. [DOI] [PubMed] [Google Scholar]
- 26.Polyak K, Lee M-H, Erdjument-Bromage H, Koff A, Roberts J M, Tempst P, Massagué J. Cell. 1994;78:59–66. doi: 10.1016/0092-8674(94)90572-x. [DOI] [PubMed] [Google Scholar]
- 27.Polyak K, Kato J, Solomon M, Sherr C J, Massagué J, Roberts J M, Koff A. Genes Dev. 1994;8:9–22. doi: 10.1101/gad.8.1.9. [DOI] [PubMed] [Google Scholar]
- 28.Toyoshima H, Hunter T. Cell. 1994;78:67–74. doi: 10.1016/0092-8674(94)90573-8. [DOI] [PubMed] [Google Scholar]
- 29.Tournier F, Cyrklaff M, Karsenti E, Bornens M. Proc Natl Acad Sci USA. 1991;88:9929–9933. doi: 10.1073/pnas.88.22.9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Felix M-A, Antony C, Wright M, Maro B. J Cell Biol. 1994;124:19–31. doi: 10.1083/jcb.124.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meijer L, Pondaven P. Exp Cell Res. 1988;174:116–129. doi: 10.1016/0014-4827(88)90147-4. [DOI] [PubMed] [Google Scholar]
- 32.Neant I, Guerrier P. Exp Cell Res. 1988;176:68–79. doi: 10.1016/0014-4827(88)90121-8. [DOI] [PubMed] [Google Scholar]
- 33.Rempel R E, Sleight S B, Maller J L. J Biol Chem. 1995;270:6843–6855. doi: 10.1074/jbc.270.12.6843. [DOI] [PubMed] [Google Scholar]
- 34.Palazzo R E, Vaisberg E, Cole R W, Rieder C L. Science. 1992;256:219–221. doi: 10.1126/science.1566068. [DOI] [PubMed] [Google Scholar]
- 35.Blow J J, Nurse P. Cell. 1990;62:855–862. doi: 10.1016/0092-8674(90)90261-c. [DOI] [PubMed] [Google Scholar]
- 36.Fang F, Newport J W. Cell. 1991;66:731–742. doi: 10.1016/0092-8674(91)90117-h. [DOI] [PubMed] [Google Scholar]
- 37.Fang F, Newport J W. J Cell Sci. 1993;106:983–994. doi: 10.1242/jcs.106.3.983. [DOI] [PubMed] [Google Scholar]
- 38.Blow J J, Sleeman A M. J Cell Sci. 1990;95:383–391. doi: 10.1242/jcs.95.3.383. [DOI] [PubMed] [Google Scholar]
- 39.Blow J J, Laskey R A. Nature (London) 1988;332:546–548. doi: 10.1038/332546a0. [DOI] [PubMed] [Google Scholar]
- 40.Coverley D, Downes C S, Romanowski P, Laskey R A. J Cell Biol. 1993;122:985–992. doi: 10.1083/jcb.122.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kung A L, Sherwood S W, Schimke R T. J Biol Chem. 1993;268:23072–23080. [PubMed] [Google Scholar]
- 42.Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude G F. Science. 1996;271:1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- 43.Dulic V, Kaufmann W K, Wilson S J, Tlsty T D, Lees E, Harper J W, Elledge S J, Reed S I. Cell. 1994;76:1013–1023. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- 44.Schwob E, Bohm T, Mendenhall M D, Nasmyth K. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 45.Murray A W, Solomon M J, Kirschner M W. Nature (London) 1989;339:280–286. doi: 10.1038/339280a0. [DOI] [PubMed] [Google Scholar]
- 46.Cohen-Fix O, Peters J-M, Kirschner M W, Koshland D. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 47.Feldman R M R, Correll C C, Kaplan K B, Deshaies R J. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 48.Paintrand M, Moudjou M, Delacroix H, Bornens M. J Struct Biol. 1992;108:107–128. doi: 10.1016/1047-8477(92)90011-x. [DOI] [PubMed] [Google Scholar]
- 49.Fry A M, Mayor T, Meraldi P, Stierhof Y D, Tanaka K, Nigg E A. J Cell Biol. 1998;141:1563–1574. doi: 10.1083/jcb.141.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fry A M, Meraldi P, Nigg E A. EMBO J. 1998;17:470–481. doi: 10.1093/emboj/17.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hinchcliffe E H, Li C, Thompson E A, Maller J L, Sluder G. Science. 1999;283:851–854. doi: 10.1126/science.283.5403.851. [DOI] [PubMed] [Google Scholar]