Abstract
Wild-type and an N-terminal 6-histidine-tagged actin have each been expressed by using a yeast strain that contains the actin gene on a plasmid and not on the chromosome. Yeast strains have also been constructed that use two plasmids, one expressing the wild-type protein and the other the 6-histidine-tagged protein. Yeast cells can be grown with either plasmid alone or with both plasmids together and appear to be normal in that the growth rates of all the yeast strains are quite similar, as is the morphology of the yeast cells. The polymerization properties of the 6-histidine-tagged actin appear almost identical to wild-type actin expressed from the chromosome. When the wild-type and 6-histidine-tagged actin are coexpressed, they can be purified by standard techniques and then separated using nickel-nitrilotriacetate chromatography. The method can be used to prepare actin mutants including those that are nonfunctional or might not support yeast growth for other reasons.
Actin is one of the most important components of the cytoskeleton, and its polymerization and depolymerization control myriad functions of eukaryotic cells. Extensive kinetic studies have been performed on the polymerization characteristics of muscle actin, but there have only been a few studies in which the role of specific amino acid residues on the in vitro polymerization process have been investigated (1–5). Although site-directed mutagenesis of actin expressed in Escherichia coli would be useful, the expressed protein forms inclusion bodies (ref. 6 and unpublished data) and has been difficult to renature (ref. 7 and unpublished data). In the expression of Dictyostelium discoideum actin, for example, the yield of protein was low and the majority of the actin was truncated because of internal initiation of transcription and/or translation (8, 9). Consequently, very little functional actin was obtained after solubilizing the inclusion bodies with detergents (8, 9). Although investigators apparently have not been able to refold denatured actin to a form that binds to ATP and Ca2+, Cowan and coworkers (10–12) have presented evidence for the requirement of chaperones for actin folding, but again the yield of protein was low. Because of these problems, purification of actin from a eukaryotic system is highly desirable. Yeast actin, expressed by the essential gene ACT1 in Saccharomyces cerevisiae (13, 14), is an excellent model for studying actin functions and polymerization properties in vitro and in vivo (15, 16). The yeast actin gene can be easily manipulated, and the actin sequence is highly conserved among eukaryotes. Although overexpression of actin in yeast is lethal (17, 18) and produces insoluble protein (unpublished data), sufficient amounts can be obtained under normal expression conditions to study its polymerization properties by growing large amounts of cells. Functional actin mutants can be purified from yeast, but the preparation of a nonfunctional actin depends on the ability to separate the mutant form from wild-type actin. In this paper, we show that it is possible to histidine tag the N-terminal end of wild-type yeast actin and obtain normal yeast growth in the absence of any other functional ACT1 gene. We also show that it is possible to express the N-terminal 6-histidine-tagged actin in the presence of wild-type untagged actin and then separate the two actin proteins by using nickel-nitrilotriacetate (Ni-NTA) chromatography. The method described should be valuable for obtaining and studying the properties of nonfunctional actin mutants.
MATERIALS AND METHODS
Materials.
The Sculptor in vitro mutagenesis system and enhanced chemiluminescence (ECL) reagents for Western blot analysis were obtained from Amersham Pharmacia Biotech (Piscataway, NJ). Primers were custom-made by Integrated DNA Technologies (Coralville, IA). Plasmid pRSWT and yeast strain PR1 were gifts from Peter Rubenstein (University of Iowa, Iowa City). DNase I was obtained from Boehringer Mannheim and linked to Affi-Gel 10 (Bio-Rad,) according to the procedure by Cook et al. (1). Both the Ni-NTA resin and the tetra-his antibody were products of Qiagen (Chatsworth, CA). The Bradford protein assay kit was obtained from Bio-Rad (Hercules, CA). Double or single amino acid dropout mixes, CSM-TRP-URA and CSM-TRP, used in synthetic minimum dropout media, were obtained from Bio 101 (Vista, CA). 5-Fluoroorotic acid and casein were obtained from Sigma. All other chemicals were reagent grade.
Mutagenesis: N-Terminal 6-Histidine-Tagged Wild-Type Yeast Actin.
The Sculptor in vitro mutagenesis system was used to insert the 6-histidine tag coding sequence into the wild-type ACT1 gene just after the start codon on pRSWT, a TRP1+ shuttle vector. The primer sequence was 5′-CGA AAA TTT ACT GAA TTA ACA ATG CAC CAT CAC CAC CAT CAT GAT TCT GAG GTT GCT-3′, with the start codon and histidine codons underlined. The order of the histidine codons was chosen to minimize the chance of mismatches and to create a BspHI site at the 3′-joint to use in screening for mutated DNA. The correct insertion was verified by sequencing and this plasmid was named pRSWT6his. Plasmid pRSWT6his was transformed into the yeast strain PR1 that contains an ACT1 gene only on plasmid pCENWT and not on the chromosome. The strain containing both pCENWT and pRSWT6his plasmids was named PR/pCENWT+pRSWT6his. Plasmid pCENWT (URA3+) was then removed by 5-fluoroorotic acid selection (19). The exclusive presence in the selected cells of the act1-6his gene was confirmed by PCR and sequencing, and this strain was named PR/pRSWT6his.
Yeast Growth.
To obtain cells for protein purification, yeast strains PR/pRSWT6his (TRP+) and PR/pCENWT+pRSWT6his (TRP+ and URA+) were each grown to saturation (OD600 ≈ 15–20) in 6 liters of the appropriate selective synthetic dropout medium (20) for 48–72 hr at 30°C. Concentrated medium (100 ml/liter, 10× the normal concentration) was added every 24 h during growth. The cells obtained were pelleted and then washed with cold double-distilled H2O (≈500 ml), and the pellet was stored at −80°C until used for the purification of the actin.
Sensitivity of growth to different temperatures and increased osmotic pressure was analyzed by plating serial 10-fold dilutions of a 15-hr overnight culture on either yeast extract/peptose/dextrose (YPD) (20) or YPD-Osm (450 mM NaCl with YPD) plates. Plates were incubated at room temperature, 30°C and 37°C and observed at 24 and 48 hr.
Fluorescence Microscopy.
Polarization of the actin cytoskeleton was assessed by rhodamine-phalloidin staining of fixed cells. Cultures were grown at 25°C and fixed at 106 cells/ml in 3.7% formaldehyde added directly to the medium from a 37% stock. Cells were stained with rhodamine phalloidin (Molecular Probes) for F-actin using the procedure of Kaiser et al. (21) except that the rhodamine phalloidin concentration was 0.165 μM and staining was done on ice. Cells were imaged on an epifluorescence microscope (Bmax-60F, Olympus) with a 1.35 NA ×100 UPlanApo objective and a U-MNG (rhodamine phalloidin) filter set. Photobleaching was reduced by using a ND25 neutral density filter in the excitation light path. Images were collected with a cooled CCD video camera (RC300, Dage-MIT, Michigan City, IN).
Actin Purification: N-Terminal 6-Histidine-Tagged Wild-Type Yeast Actin.
The purification of 6-histidine-tagged wild-type actin from PR/pRSWT6his was the same as that of untagged wild-type (22) except that the removal of cofilin through high-salt treatment of actin filaments (23) was unnecessary for this particular actin as it had little cofilin associated with it (unpublished data). The N terminus was sequenced to confirm that the 6-histidine tag was present in all the purified actin.
Separation of 6-Histidine-Tagged and Nontagged Actins.
A mixture of 6-histidine-tagged and untagged actins was purified from the two-plasmid strains by using the standard procedure (1). The tagged protein was then removed by batch-binding the mixture of actins to Ni-NTA agarose at 4°C for 1 hr in 10 mM Tris⋅Cl buffer (pH 7.5), containing 0.2 mM ATP, 0.2 mM Ca2+, and 5 mM imidazole. For every 3 μg of actin, 1 μl of the commercial resin suspension (as a 50% slurry) was added. It was found necessary to include imidazole in the binding buffer to prevent the weak binding of untagged wild-type to the resin. The total protein concentration was determined by the Bradford protein assay (24) with muscle actin as the standard. The polymerized actin was treated with KCl (0.7 M) to prevent cofilin from binding to the untagged actin filaments when they were pelleted (23).
Western Blot Analysis.
Western blot analysis with tetra-his antibody (Qiagen) was used to confirm the removal of tagged actin from the mixture of actins because it reacts only with the 6-histidine-tagged protein. This procedure was performed according to the manufacturer’s (Qiagen) instructions. The primary antibody was diluted 1,000-fold and the secondary antibody (horseradish peroxidase-anti-mouse IgG) was diluted 2,000-fold. Enhanced chemiluminescence reagents (Amersham) were used for detection.
Polymerization Assay.
Actin polymerization was followed by decrease of intrinsic fluorescence (excitation at 300 nm, emission at 335 nm) at 20°C at pH 8.0 in G buffer (2 mM Tris⋅Cl/0.2 mM ATP/0.2 mM Ca2+/0.5 mM DTT/0.01% NaN3). The total reaction volume was 200 μl. The reactions were run on a PTI (Alphascan) spectrofluorometer. Actin was preincubated for 5 min with EGTA and a low concentration of Mg2+ (50 μM) to replace the tightly bound Ca2+. In all cases, polymerization was initiated by the addition of 2 mM Mg2+.
RESULTS
Yeast Growth and Polymerization Properties of Wild-Type and 6-Histidine-Tagged Actin.
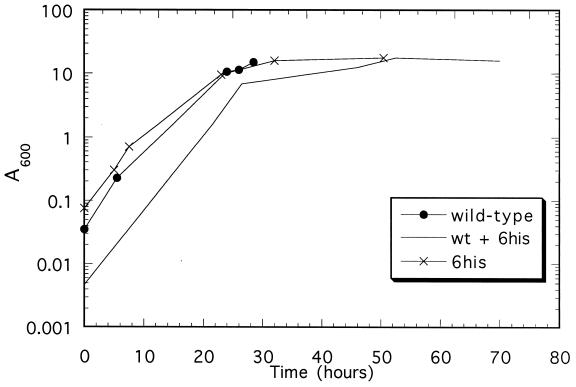
The 6-histidine-tagged ACT1 (act1-6his) gene of yeast was tested for complementation of ACT1 deletion by transforming the plasmid pRSWT6his into strain PR1 and using 5-fluoroorotic acid to exclude pCENWT (carrying the untagged ACT1 gene) from the host. Colonies were readily obtained on 5-fluoroorotic acid plates. Therefore, it is clear that act1-6his is able to support yeast growth. As shown in Fig. 1, the cells containing only the act1-6his gene have a growth rate, using a synthetic dropout media, only slightly slower (2.5 hr doubling time) than cells carrying only the untagged wild-type actin gene under the same conditions (2 hr).
Figure 1.
Growth curves of yeast strains containing only the wild-type actin plasmid (●), the 6-histidine-tagged actin plasmid (×), and both plasmids (no symbol). The cells were grown as described in Materials and Methods. Only a few data points were collected for the strain coexpressing both plasmids.
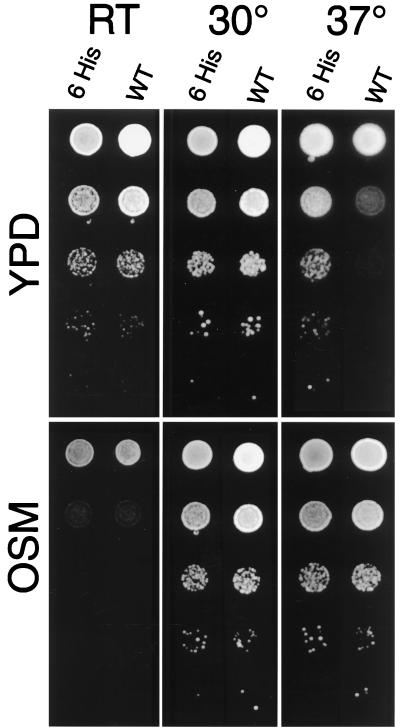
To test whether rescue of the ACT1 deletion is complete, we compared the growth of strains carrying either pRSWT6his or pCENWT plasmids under different conditions. Mutations of the ACT1 gene often lead to temperature sensitivity and osmotic sensitivity. Low gene dosage of ACT1 in ACT1/act1Δ hemizygotes may also lead to these phenotypes (15). Therefore, we tested the temperature sensitivity and osmotic sensitivity of a strain in which the plasmid with the untagged ACT1 gene was replaced by a plasmid carrying 6-histidine-tagged ACT1 gene (Fig. 2). At room temperature and at 30°C, strains with tagged and untagged ACT1 grew similarly. Both strains also displayed similar levels of osmotic sensitivity at room temperature. A slight difference in growth of the two strains was observed only on YPD at 37°C. Therefore, the strain carrying the act1-6his gene is almost indistinguishable in growth characteristics from the wild-type strain.
Figure 2.
Growth of wild-type and strains carrying 6-histidine-tagged ACT1 gene at room temperature (RT), 30°C and 37°C. Labels: 6 His is PR/pRSWT6his strain containing the ACT1 gene expressing 6-histidine-tagged actin. WT is PR/pCENWT strain containing the ACT1 gene expressing wild-type actin. Serial 10-fold dilutions, plated on YPD (20) and OSM (YPD + 450 mM NaCl) media.
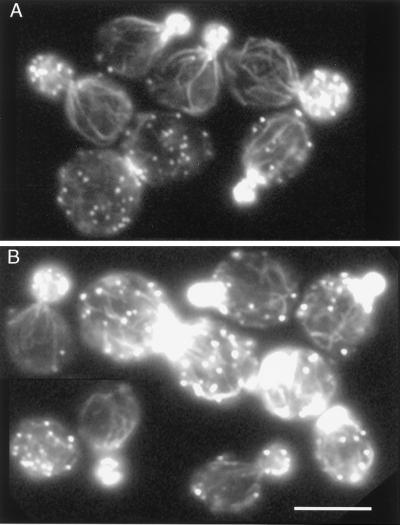
The morphology of the actin cytoskeleton was also compared in the same strains. In growing cells of wild-type strains the F-actin-containing structures—cables and patches—are distributed in a polar fashion. The actin patches are clustered in the growing bud, and the actin cables in the mother cell are oriented toward the patch cluster (25). Temperature-sensitive mutations in the ACT1 gene lead to disruption of polarization of the actin cytoskeleton (26). The presence of the 6-histidine-tagged actin does not disrupt polarization of the actin cytoskeleton (Fig. 3). Some of the cells of both strains were abnormally large and contained randomly distributed patches. This phenotype was expected because some of the cells of the strain containing autonomously replicating plasmid may lose this plasmid. Cells that lose the plasmid with the ACT1 gene are inviable and are expected to have abnormal polarization of the actin cytoskeleton. There was no difference in frequency of cells that were large or had randomly distributed patches between strains carrying either the tagged or untagged ACT1 gene. Therefore, 6-histidine-tagged actin almost completely complements loss of wild-type actin.
Figure 3.
Normal polarization of the actin cytoskeleton in a strain carrying 6-histidine-tagged ACT1 gene. (A) Control PR/pCENWT strain, carrying the ACT1 gene for wild-type actin. (B) Strain PR/pRSWT6his, carrying the ACT1 gene expressing 6-histidine-tagged actin. Rhodamine-phalloidin staining. (Bar = 5 μm.)
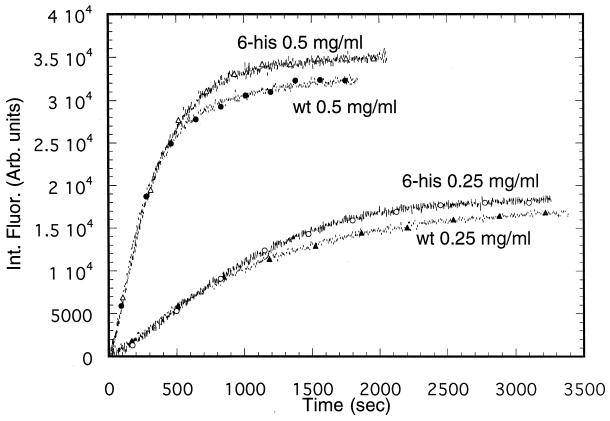
The 6-histidine-tagged actin was purified from yeast containing only the act1-6his gene by using exactly the same procedure that was used for wild-type actin including binding to a DNase I column, indicating that the 6-histidine-tagged actin has binding properties similar to those of wild-type actin. To further compare the proteins, the 6-histidine-tagged and wild-type actin were examined for their ability to polymerize at two different protein concentrations and under a given set of conditions in G buffer with polymerization induced by the addition of 2 mM Mg2+. As shown in Fig. 4, the rate of polymerization (using intrinsic fluorescence) of 6-histidine-tagged actin under these conditions is essentially identical to that of wild-type actin.
Figure 4.
Polymerization of wild-type and 6-histidine-tagged yeast actin in G buffer at pH 8.0 and 20°C at two different actin concentrations. The reactions were initiated by the addition of 2 mM Mg2+ at pH 8.0 after preincubation with 50 μM Mg2+ and 1 mM EGTA to replace Ca2+ with Mg2+ as described in Materials and Methods. Polymerization was measured using changes in the intrinsic fluorescence. The decrease of intrinsic fluorescence upon polymerization has been redrawn to appear as an increase.
Separation of 6-Histidine-Tagged and Nontagged Actin with Ni-NTA Resin.
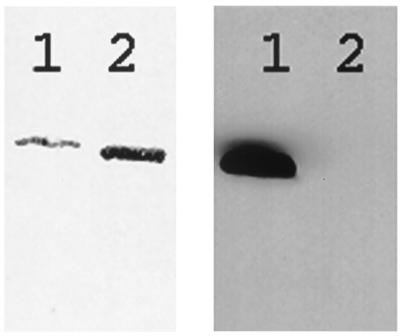
In the above experiments, the wild-type and 6-histidine-tagged wild-type actins were isolated from yeast containing only a single ACT1 gene (tagged or untagged). To study actin mutants incapable of supporting yeast growth (unpublished work), it was necessary to show that the 6-histidine-tagged and nontagged actins could be separated when coexpressed in the same host. To do this, the 6-histidine-tagged and wild-type actins were coexpressed from two different plasmids. In these experiments, about half of the total actin expressed was tagged (data not shown), although the total actin concentration was similar to that in which only one actin gene was expressed. The growth rate of the coexpressing strain (as shown in Fig. 1) was similar to those strains containing only a single plasmid. Because 6-histidine-tagged actin can bind to DNase I as does wild-type actin, the two coexpressed actins were purified together as if the untagged wild-type was being prepared alone. Batch binding to Ni-NTA agarose was then used to remove the 6-histidine-tagged protein as described in Materials and Methods. The results of Coomassie staining and Western blot analysis showed that almost all of the tagged actin was removed in this manner (Fig. 5). The concentration of imidazole used during binding to the resin was chosen to avoid both unnecessary loss of untagged protein and inefficient binding of tagged actin. The amount of resin was also crucial for efficient separation such that the total resin binding capacity was enough to bind the tagged protein but not the untagged actin. In all cases, similar amounts of tagged and untagged actins were present in the mixture before the batch binding.
Figure 5.
Separation of 6-histidine-tagged wild-type actin from untagged wild-type actin using Ni-NTA chromatography. Lane 1, wild-type plus 6-histidine-tagged actin before binding to Ni-NTA resin; lane 2, wild-type plus 6-histidine-tagged actin after binding to Ni-NTA resin. Total protein was detected by Coomassie blue staining (Left). The 6-histidine-tagged protein was detected by Western blot analysis of the same samples (Right) using tetra-his antibody as described in Materials and Methods.
For strains expressing only a single actin the final yield of purified wild-type actin from 6 liters of culture is ≈4 mg and slightly less for the 6-histidine-tagged actin. Somewhat less total material was obtained from cultures containing plasmids for both the wild-type and the 6-histidine-tagged actins. Similar results were seen with other actin mutants (unpublished data).
DISCUSSION
The ability to study actin mutants may help to define the role of this cytoskeleton protein in numerous physiological responses. Indeed the properties of some mutant actins isolated from yeast strains that are phenotypically similar to wild-type strains have been studied (1–5). However, there are problems in obtaining mutant actins that might have very different properties from the wild-type protein. Alanine scanning, for example, has indicated that there are many mutations of actin that result in different phenotypic properties of yeast (15, 16). If the mutation does not support yeast growth, it has not been possible, until now, to examine the properties of the mutant protein. A solution to this problem would be to express the mutant protein in bacteria, and it is possible to express actin in E. coli. The expressed material, however, always appears in inclusion bodies, and the usual procedure of dissolving the inclusion bodies in denaturant and then refolding the protein is problematic because refolding gives rise to a nonviable form that does not bind ATP, Ca2+, or DNase I (ref. 7 and unpublished work). Although it is known that actin can be refolded in the presence of chaperones (10, 11), the yield of material has always been very small. In this work, we have made and purified a 6-histidine-tagged actin that supports yeast growth, thus allowing us to coexpress and to isolate mutant actins that do not support growth. Actin with the 6-histidine tag on the N-terminal end appears to be very similar to wild-type actin in that it supports yeast growth. The growth rate of yeast (Fig. 1) is only slightly affected when histidine-tagged actin is the sole actin source. This is somewhat surprising because there are interactions with various actin binding proteins, including cofilin (27) and myosin (28–31), that are known to bind at or near the N terminus. Indeed, it appears that the tag does affect interaction with at least one protein because contamination with cofilin is less in the histidine-tagged protein compared with wild-type implying a weakened interaction of cofilin to actin. It is of interest that actin tagged at the C-terminus with green fluorescent protein could not function as the sole actin source in yeast (32). None of the actin-GFP fusion proteins was capable of complementing the delection of chromosomal actin. Although some appeared to be incorporated into the cytoskeleton, the actin was localized to patches but not cables (32).
Our purpose in tagging actin was to provide a means for separation of wild type actin and mutants that cannot support growth alone. We have designed a system in which two actins, a tagged actin with nearly wild-type activity and untagged wild type are expressed in yeast cells on separate, low copy number plasmids, at roughly equal concentrations. Following our standard purification procedure using DNase I binding of the two proteins, we can remove the wild-type 6-histidine tagged actin by Ni-NTA chromatography and obtain the untagged mutant actin in pure form.
Acknowledgments
We thank Dr. Peter Rubenstein (University of Iowa) for gifts of the plasmid pRSWT and yeast strain PR1 and Dr. John Cooper for helpful discussions. This work was supported by the National Science Foundation, Grant MCB 9603807.
ABBREVIATIONS
- Ni-NTA
nickel-nitrilotriacetate
- YPD
yeast extract/peptose/dextrose
References
- 1.Cook R K, Blake W T, Rubenstein P A. J Biol Chem. 1992;267:9430–9436. [PubMed] [Google Scholar]
- 2.Solomon L R, Rubenstein P A. J Biol Chem. 1987;262:11382–11388. [PubMed] [Google Scholar]
- 3.Solomon T L, Solomon L R, Gay L S, Rubenstein P A. J Biol Chem. 1988;263:19662–19669. [PubMed] [Google Scholar]
- 4.Taniguchi S, Sagara J, Kakunaga T. J Biochem. 1988;103:707–713. doi: 10.1093/oxfordjournals.jbchem.a122333. [DOI] [PubMed] [Google Scholar]
- 5.Feng L, Kim E, Lee W L, Miller C J, Kuang B, Reisler E, Rubenstein P A. J Biol Chem. 1997;272:16829–16837. doi: 10.1074/jbc.272.27.16829. [DOI] [PubMed] [Google Scholar]
- 6.Frankel S, Condeelis J, Leinwand L. J Biol Chem. 1990;265:17980–17987. [PubMed] [Google Scholar]
- 7.Lehrer S S, Kerwar G. Biochemistry. 1972;11:1211–1217. doi: 10.1021/bi00757a015. [DOI] [PubMed] [Google Scholar]
- 8.Frankel S, Sohn R, Leinwand L. Proc Natl Acad Sci USA. 1991;88:1192–1196. doi: 10.1073/pnas.88.4.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McNally E, Sohn R, Frankel S, Leinwand L. Methods Enzymol. 1991;196:368–389. doi: 10.1016/0076-6879(91)96033-n. [DOI] [PubMed] [Google Scholar]
- 10.Gao Y, Thomas J O, Chow R L, Lee G H, Cowan N J. Cell. 1992;69:1043–1050. doi: 10.1016/0092-8674(92)90622-j. [DOI] [PubMed] [Google Scholar]
- 11.Melki R, Vainberg I E, Chow R L, Cowan N J. J Cell Biol. 1993;122:1301–1310. doi: 10.1083/jcb.122.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian G L, Vainberg I E, Tap W D, Lewis S A, Cowan N J. Nature (London) 1995;375:250–253. doi: 10.1038/375250a0. [DOI] [PubMed] [Google Scholar]
- 13.Ng R, Abelson J. Proc Natl Acad Sci USA. 1980;77:3912–3916. doi: 10.1073/pnas.77.7.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallwitz D, Sures I. Proc Natl Acad Sci USA. 1980;77:2546–2550. doi: 10.1073/pnas.77.5.2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wertman K F, Drubin D G, Botstein D. Genetics. 1992;132:337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hennessey E S, Drummond D R, Sparrow J C. Biochem J. 1993;282:657–671. doi: 10.1042/bj2910657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magdolen V, Drubin D G, Mages G, Bandlow W. FEBS Lett. 1993;316:41–47. doi: 10.1016/0014-5793(93)81733-g. [DOI] [PubMed] [Google Scholar]
- 18.Espinet C, de la Torre M A, Aldea M, Herrero E. Yeast. 1995;11:25–32. doi: 10.1002/yea.320110104. [DOI] [PubMed] [Google Scholar]
- 19.Boeke J D, Trueheart J, Natsoulis G, Fink G R. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 20.Rose M D, Winston F, Hieter P, editors. Methods in Yeast Genetics: A Laboratory Course Manual. Plainview, NY.: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 21.Kaiser C, Michaelis S, Mitchell A, editors. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 22.Buzan J M, Frieden C. Proc Natl Acad Sci USA. 1996;93:91–95. doi: 10.1073/pnas.93.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du J Y, Frieden C. Biochemistry. 1998;37:13276–13284. doi: 10.1021/bi981117r. [DOI] [PubMed] [Google Scholar]
- 24.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 25.Kilmartin J V, Adams A E M. J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novick P, Botstein D. Cell. 1985;40:405–416. doi: 10.1016/0092-8674(85)90154-0. [DOI] [PubMed] [Google Scholar]
- 27.Muneyuki E, Nishida E, Sutoh K, Sakai H. J Biochem (Tokyo) 1985;97:563–568. doi: 10.1093/oxfordjournals.jbchem.a135091. [DOI] [PubMed] [Google Scholar]
- 28.Aspenstrom P, Karlsson R. Eur J Biochem. 1991;200:35–41. doi: 10.1111/j.1432-1033.1991.tb21045.x. [DOI] [PubMed] [Google Scholar]
- 29.Van E J E, Hodges R S. Biochemistry. 1991;30:11676–11682. doi: 10.1021/bi00114a010. [DOI] [PubMed] [Google Scholar]
- 30.Bonafe N, Chaussepied P. Biophys J. 1995;68:S 35–S 43. [PMC free article] [PubMed] [Google Scholar]
- 31.Kunori S, Katoh T, Mogi Y, Morita F. J Biochem (Tokyo) 1995;118:1239–1247. doi: 10.1093/oxfordjournals.jbchem.a125013. [DOI] [PubMed] [Google Scholar]
- 32.Doyle T, Botstein D. Proc Natl Acad Sci USA. 1996;93:3886–3891. doi: 10.1073/pnas.93.9.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]