Abstract
A new ubiquitin-processing protease (Ubp-M) has been identified in mammalian cells that is phosphorylated at the onset of mitosis and dephosphorylated during the metaphase/anaphase transition. The carboxyl-terminal domain of this 823-aa protein can be phosphorylated in vitro with either extracts of mitotic cells or purified cdc-2/cyclin B complexes. Recombinant Ubp-M is able to deubiquitinate histone H2A in vitro, and the phosphorylated form is also enzymatically active. Wild-type Ubp-M, transiently expressed as green fluorescent protein-fusion proteins, localizes in the cytoplasm of cultured cells, but mutant forms, lacking an active-site cysteine, associate closely with mitotic chromosomes during all stages of cell division and remain within the nucleus during the postmitotic period. Cells transfected with plasmids containing mutant Ubp-M genes stop dividing and eventually undergo apoptosis. Ubp-M may deubiquitinate one or more critical proteins that are involved in the condensation of mitotic chromosomes, possibly acting selectively on histones H2A and H2B, the major ubiquitinated proteins of chromatin.
A large number of critical regulatory proteins are modified covalently by the enzymatic attachment of ubiquitin to side chains of lysines, a modification that often leads to their proteolytic degradation (1, 2). Proteins that regulate the cell cycle, such as cyclin B and p27, and either activate or inhibit specific protein kinases are ubiquitinated by multiprotein complexes, known as the anaphase-promoting complexes or cyclosomes (3, 4).
It is generally accepted that the covalently bound polyubiquitin chains, linked together as polymers through internal isopeptide bonds, are recognized by one or more subunits of the 26S proteosome as the first step in the pathway that eventually leads to degradation of the ubiquitinated protein (5–7). Although the ubiquitins themselves are 76-aa polypeptides, they are not degraded by the proteosomes, but instead are recycled through the actions of enzymes known as ubiquitin-processing proteases (Ubps) or deubiquitinating enzymes (DUBs). The enzymes and protein cofactors that attach ubiquitin to polypeptides have been studied extensively (8, 9), but our understanding of how the ubiquitin-processing enzymes function lags far behind. A number of proteins are ubiquitinated and then degraded during mitosis, but it is not known how, when, or where deubiquitinating enzymes act during the mitotic cycle. Furthermore, many studies indicate that the recycling of ubiquitin from degraded proteins by deubiquitinating enzymes is not their sole function.
Yeast are believed to have 20 or more proteins that may have ubiquitin-processing activity (10), but it remains unresolved how individual Ubps recognize specific substrates or how they themselves are regulated. The enzymes that attach ubiquitin to protein substrates, known as ubiquitin-conjugating enzymes, share a common active site and have unique domains that determine substrate specificity and intracellular localization (11–13); it is likely that ubiquitin-processing enzymes have similar properties, but, so far, none have been defined.
In addition to their role in protein degradation during mitosis (14) and in the processing of growth regulatory factors that are activated by cytokines (15, 16), recent studies suggest that deubiquitinating enzymes are involved in regulating transcriptional activity of chromatin, both in yeast, in collaboration with the silent information regulator (SIR) proteins (17), and in Drosophila, in which a mutant form of a ubiquitin-processing enzyme (D-Ubp-64E) modifies position–effect variegation (18). In both cases ubiquitin processing is necessary, but it is uncertain how the removal of attached ubiquitins influences gene expression or contributes to chromatin stability.
It also has been evident for more than two decades that histones H2A and H2B are monoubiquitinated (19), but this posttranslational modification does not lead to enhanced proteolytic degradation (20). Normally, between 5 and 10% of nuclear histones H2A and H2B are ubiquitinated during interphase, but a study of precisely synchronized slime mold cells found that both histones were completely deubiquitinated during metaphase, which coincided with complete condensation of the mitotic chromatin (21). As the cells progressed to anaphase, within minutes later, both histones were reubiquitinated.
We have identified a new deubiquitinating enzyme (Ubp-M) that is phosphorylated and then dephosphorylated at critical points in the mitotic cycle, and its properties and behavior suggest that it may play some role in regulating mitotic chromatin, possibly by deubiquitinating histones or other relevant substrates.
MATERIALS AND METHODS
Identification of Ubp-M.
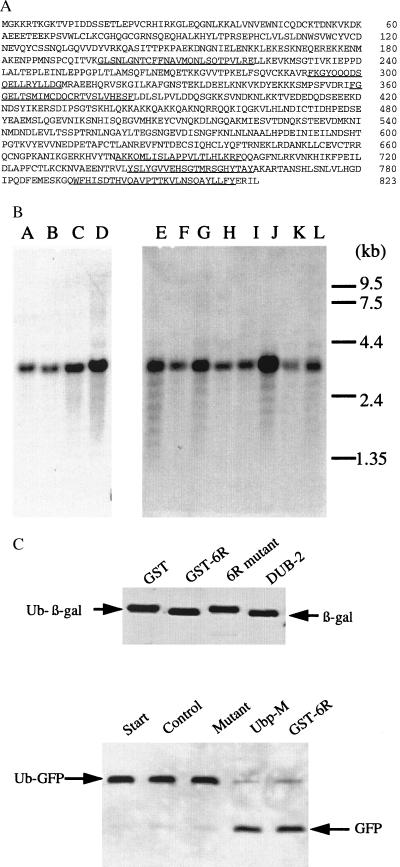
To clone the Ubp-M gene, two human cDNA libraries, a Jurkat λgt11 (a gift of S. Weissman, Yale School of Medicine, New Haven, CT) and a HeLa Uni-ZAP RX (Stratagene), were screened with an IgM mAb that was raised against a mixture of phosphoproteins derived from mitotic Chinese hamster ovary (CHO) cells. Seven positive clones were selected: four from the Jurkat library, with sizes of 2.1, 1.9, 1.6, and 1.3 kb, and three from HeLa, with sizes of 2.1, 2.0, and 1.4 kb. The DNA sequences of these clones indicated that all seven clones had the same sequence at the 3′ end but contained different-sized extensions to the 5′ end. A 3.0-kb transcript was demonstrated by Northern blot analysis in a variety of tissues. A number of probes were made from clones derived from the 5′ end to search for additional unique 5′ sequences, but all the new clones found were consistent with a single ORF that coded for the same polypeptide chain. Three of them contain the whole coding region.
Phosphorylation of Ubp-M During the Mitotic Cycle.
Synchronized cultures of CHO cells were prepared by exposing overnight cultures of CHO cells to nocodazole (0.25 μM) for 4 hours at 37°C and harvesting cells arrested in prometaphase by mechanical agitation. Cells were washed in medium and incubated in suspension for varying periods before sampling. Aliquots of cells were lysed in standard Laemmli buffer and analyzed by SDS/PAGE using 10% polyacrylamide gels. For immunoblots, gels were transferred to nitrocellulose paper (Schleicher & Schuell) overnight, then blocked in 3% nonfat milk in Tris-buffered saline at room temperature for 1 hr, incubated in primary and secondary antibodies for 45 min each, and recorded on x-ray film after treatment with the enhanced chemiluminescence system. A polyclonal antiserum, raised in chickens using a glutathione S-transferase (GST)-fusion peptide (pGEX-3X, Pharmacia) derived from the first 190 aa of the N-terminal domain of Ubp-M, was used to follow the change in mobility of Ubp-M as a function of the degree of phosphorylation.
In Vitro Phosphorylation of Ubp-M.
GST-Ubp-M was enriched on glutathione-Sepharose beads and then incubated at 37°C for 45 min with extracts prepared from synchronized CHO cells grown in suspension. After incubation, the beads were collected by centrifugation, lysed in SDS solubilizing buffer, and analyzed by immunoblotting with a monoclonal IgM antibody. Glutathione-Sepharose beads containing bound GST-Ubp-M also were incubated in a cdc-2/cyclin B complex (from New England Biolabs) and [γ-32P]ATP, with and without olomoucine (200 μM).
Deubiquitination Assays.
Deubiquitination assays were carried out either in cotransformed E. coli or with soluble extracts of recombinant proteins. For cotransformation assays, either a GST-Ubp-M construct or GST-DUB (a gift from Alan D’Andrea, Harvard Medical School, Cambridge) were cotransformed with pACYC184-Ub-Met-beta-gal (a gift from Mark Hochstrasser, University of Chicago) into MC 1061 Escherichia coli. Overnight cultures were induced with isopropyl β-d-thiogalactoside, and total lysates were analyzed by SDS/PAGE and immunoblotted with anti-β-galactosidase antisera. A ubiquitinated green fluorescent protein (GFP) fusion protein was made as an additional substrate, using PCR to generate a Ub-GFP chimeric gene that was inserted into pET-28a vector (Novagen) and expressed as a histidine-containing fusion protein.
A preparation of histone H2A/H2B dimer (a gift from E. Moudrianakis, Johns Hopkins University, Baltimore, MD) was used to follow the deubiquitinating activity of GST fusion proteins of both wild-type and mutant forms of Ubp-M that were purified with glutathione beads. Aliquots of histone dimer (10 μg) and GST-Ubp-M (2 μg) or its mutant form were incubated at 37°C for 30 and 45 min, and the products were analyzed by SDS/PAGE using 15% polyacrylamide gels.
Preparation of GFP-Fusion Proteins.
Fusion proteins containing GFP were made by inserting the Ubp-M gene at the 3′ end of the GFP gene using the pEGFP-C1 vector (CLONTECH) under the control of the cytomegalovirus promoter. A Flag peptide tag was added to the N-terminal end of Ubp-M by PCR and subcloned into pBluescript (KS), and the products were confirmed by sequencing.
To generate mutant clones of Ubp-M, in which the active-site cysteine was replaced by serine as the only change, flag-tagged Ubp-M gene was subcloned into pAlter-Max vector (Promega), and a mutagenesis oligonucleotide (5′-AACTGCATTGAAGAAGCTTGTGTTTCCCAAATT-3′) was synthesized, to change C205 to S, and introduced by HindIII at the desired mutation site, using Altered Sites II Mammalian Mutagenesis System.
Localization of GFP-Fusion Proteins.
GFP-Ubp-M constructs were transiently transfected into several cell lines (293T, CHO, COS-7) by lipofection or calcium phosphate coprecipitation using the manufacturer’s protocols. Cells were grown on glass coverslips and fixed in 4% paraformaldehyde/0.1% glutaraldehyde/0.25% Triton X-100/PBS at room temperature for 15–30 min, blocked in 5% BSA/PBS, washed in distilled water, mounted in a gelatin-based medium that contained 0.05% 4′,6-diamidino-2-phenylindole (DAPI), and examined and photographed with a Nikon Mikrophat FXA.
To determine the fraction of cells that were expressing GFP-fusion proteins at different times after transfection, we photographed populations of living cells that were grown on grid-marked coverslips, using phase-contrast microscopy, and then determined the number of cells expressing GFP by fluorescence microscopy. About 500 cells were counted for each construct. Comparable counts were made on fixed cells, but the fraction of cells showing positive fluorescence was lower because of the tendency of fixed cells to wash off the coverslips during processing.
RESULTS AND DISCUSSION
We have characterized a new ubiquitin-processing enzyme that was identified by screening two mammalian cDNA libraries with an IgM mAb raised against an enriched mixture of mitotic phosphoproteins. Antibody-positive clones coded for a unique polypeptide chain of 823 aa that was identified as a Ubp, based on the peptide segments it shares with many other known processing enzymes (see Fig. 1A). Northern blots of different tissue extracts showed that a 3-kb transcript, coding for this protein, is present in all tissues examined (Fig. 1B). To confirm that Ubp-M was indeed an active processing protease, different recombinant forms of the enzyme were incubated with two ubiquitinated substrates, ubiquitin β-galactosidase, a standard Ubp substrate, and ubiquitinated GFP. Both substrates were cleaved by wild-type Ubp-M, but mutant forms, in which the active-site cysteine was replaced by serine, were inactive (Fig. 1C). Both assays are useful ways to measure ubiquitin-processing activity, and we assume that they provide some indication of the ability of the enzyme to act on naturally occurring ubiquitinated proteins. Later studies, described below, revealed that Ubp-M is able to cleave a potentially significant physiological substrate.
Figure 1.
(A) Deduced amino acid sequence of Ubp-M derived from cDNA clones isolated from Jurkat and Hela cell libraries. Sequences that are highly homologous to sequences of known ubiquitin-processing proteases are underlined. (B) Northern blot of human tissues using a full-length Ubp-M probe. Lanes A–L were loaded with extracts from fetal brain, fetal lung, fetal liver, fetal kidney, heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas. (C) Deubiquitinating activity of Ubp-M. (Upper) Enzymatically active Ubp-M (GST-6R) increases the mobility of β-galactosidase by removing an 8.0-kDa ubiquitin, an activity shared by DUB-2, a deubiquitinating enzyme provided by Alan D’Andrea (15). Mutant Ubp-M (6R mutant) is inactive. (Lower) The mobility of ubiquitinated GFP (Ub-GFP) is also increased by two forms of Ubp-M, soluble Ubp-M or a GST-fusion protein (GST-6R).
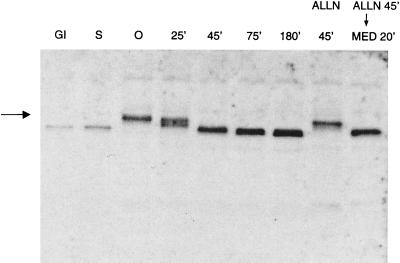
Ubp-M is phosphorylated in cultured CHO cells during the G2/M transition and is maximally phosphorylated in cells harvested in prometaphase after nocodazole treatment. We determined this by analyzing the mobility of Ubp-M as a function of the mitotic cycle, using a polyclonal antibody that was generated against a GST-Ubp-M fusion protein containing a 190-aa fragment derived from the amino terminus. As shown in Fig. 2, Ubp-M abruptly shifts to dephosphorylated states as the cells pass from metaphase to anaphase, at a time that also coincides with the degradation of cyclin B and the separation of sister chromatids that signal the onset of anaphase. The fully phosphorylated form of Ubp-M persists in cells arrested in metaphase by ALLN, a proteolytic inhibitor that acts on proteosomes (22), but it is rapidly dephosphorylated if the ALLN block is relieved and the cells are allowed to progress to anaphase. This demonstrates that the dephosphorylation of Ubp-M is tightly coupled to the events that accompany anaphase.
Figure 2.
Changes in the degree of phosphorylation of Ubp-M during the mitotic cycle. The mobility of Ubp-M decreases dramatically in cells in prometaphase (0 time) as a result of phosphorylation. The fully phosphorylated form is marked by the arrow. Multiple bands of increased mobility appear in cells undergoing the metaphase/anaphase transition (25 min). Cells in anaphase (45 min), telophase (75 min), and the immediate postmitotic period (180 min) show only the dephosphorylated form, as do cells in G1 and S phase. Cells arrested in metaphase by ALLN show persistence of the phosphorylated form (ALLN 45 min), which is dephosphorylated in cells incubated in medium after ALLN is removed (MED 20 min). Ubp-M is identified by immunoblotting of cell extracts prepared as described (18). Extracts from equivalent numbers of cells were electrophoresed in lanes 3–9.
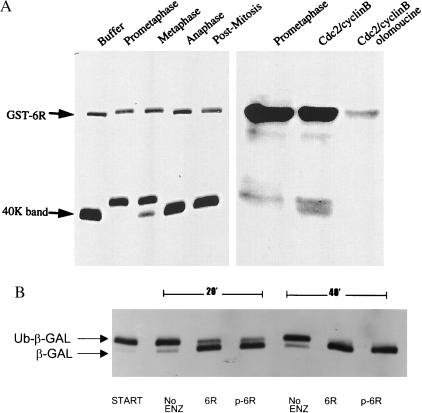
To determine which segments of Ubp-M are phosphorylated, we expressed different recombinant forms of Ubp-M in E. coli and found that both the complete molecule (residues 1–823) and a carboxyl-terminal fragment (residues 504–823), which was generated in E. coli via an internal initiation site, were phosphorylated by extracts obtained from cells isolated in either prophase or metaphase (Fig. 3A). Extracts obtained from cells taken after anaphase or in the immediate postmitotic period were inactive. We also found that recombinant Ubp-M was phosphorylated by purified cdc-2/cyclin B complexes, an action that was blocked by olomoucine, an inhibitor of the cdc-2 kinase. To determine whether phosphorylation of Ubp-M affected its capacity to deubiquitinate substrates, we incubated recombinant forms, before and after in vitro phosphorylation, with Ub-β-galactosidase (Fig. 3B) and found that phosphorylated recombinant Ubp-M was fully active against both substrates.
Figure 3.
(A) Phosphorylation of Ubp-M in vitro. (Left) A GST-fusion protein of Ubp-M (GST-6R) and a truncated carboxyl-terminal domain (40K band) were incubated together with soluble extracts prepared from CHO cells isolated at different stages of mitotic division. Extracts of cells in prometaphase and metaphase decreased the mobilities of the bands because of an increase in phosphorylation. Extracts from cells in anaphase, postmitosis, and buffer alone showed no effect. SDS/PAGE gels were analyzed by immunoblotting with an IgM mAb that reacts with a carboxyl-terminal domain of Ubp-M. (Right) 32P incorporation into Ubp-M (GST-6R) after incubation with [γ-32P]ATP and extracts from cells isolated in prometaphase or after incubation with cdc-2/cyclin B preparations. The addition of olomoucine blocked the phosphorylation of Ubp-M by the cdc-2/cyclin B complex. (B) Effect of phosphorylation on deubiquitinating activity. Ubiquitinated β-galactosidase (Ub-β-Gal) shifted to a lower-molecular-weight form, the result of deubiquitination, after incubation with both nonphosphorylated recombinant Ubp-M (6R) and phosphorylated preparations (p-6R). Deubiquitination was partial after a 20-min incubation at 37°C and complete after 40 min.
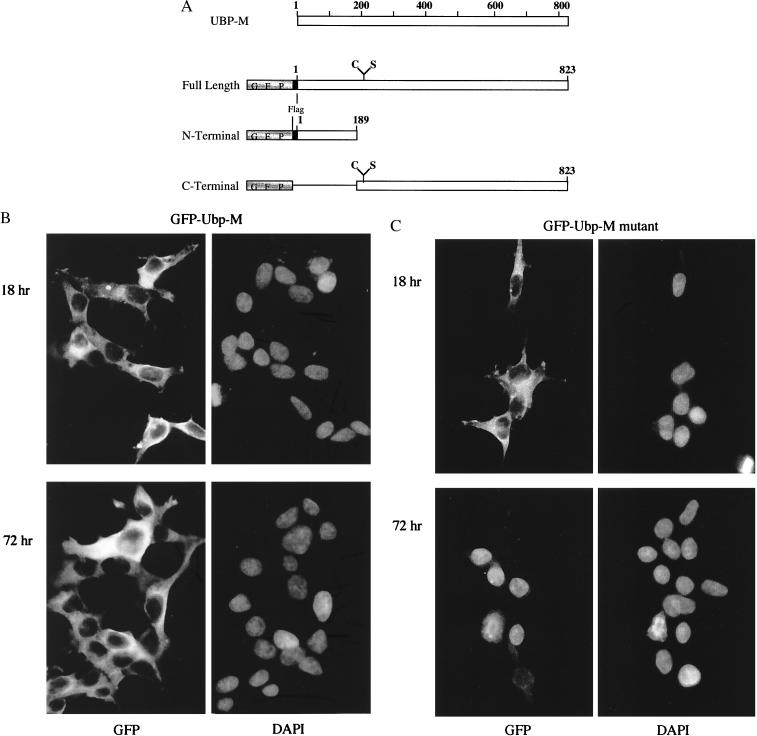
To explore the site of action of Ubp-M in cultured cells as a function of the cell cycle, we transfected cells with plasmids that code for different versions of the enzyme and compared the localization of enzymatically active GFP-labeled fusion proteins with mutant counterparts, in which the active-site cysteine was replaced by serine. The constructs used in these experiments are depicted in Fig. 4A. Both native and mutant forms of Ubp-M were expressed in HEK-293T cells with comparable efficiency, and, in each case, the transcribed protein was found in the cytoplasm of the transfected cells within the first 18 hr after transfection, as shown in Fig. 4 B and C (Upper). Cells transfected with enzymatically active versions of Ubp-M continued to divide and express the enzyme, and it remained within the cytoplasm of interphase cells. Fig. 4B (Lower) shows the localization of GFP-Ubp-M in cells cultured for 72 hr after transfection. A strikingly different localization was found in cells transfected with mutant Ubp-M at the same time period. Fewer cells continued to express mutant Ubp-M as the incubation proceeded, and those that were labeled had most of the GFP-Ubp-M mutant form within their nuclei (Fig. 4C Lower), a localization not seen in any of the cells that were expressing the enzymatically active protein.
Figure 4.
Localization of GFP-fusion proteins of Ubp-M in cells transfected with GFP-Ubp-M fusion genes. (A) Constructs of GFP-fusion genes with different domains of Ubp-M that were transfected into cultured cells. DNA fragments containing the catalytic domain contained codons for either cysteine or serine at position 205. (B) Enzymatically active GFP-Ubp-M localizes to the cytoplasm of cells fixed either 18 hr or 72 hr after transfection. The nuclei of the same cells are stained with DAPI. (C) Inactive mutant GFP-Ubp-M (C-S) also localizes to the cytoplasm of cells fixed 18 hr after transfection, but, by 72 hr, most of the GFP staining is nuclear. The nuclei of the same cells also are stained by DAPI. Cells were processed for fluorescence microscopy as described.
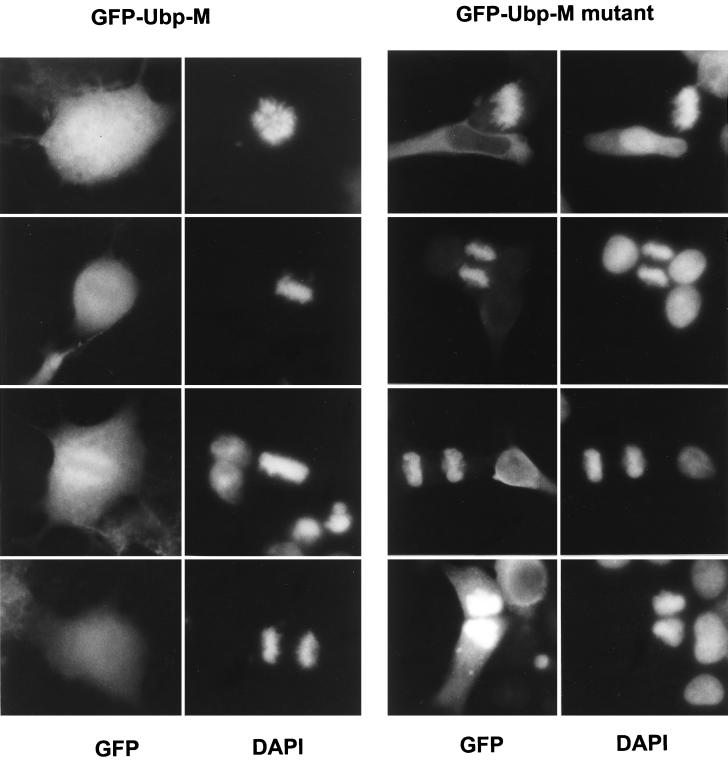
Another difference was seen when individual mitotic cells were examined after transfection with the mutant gene. Mutant Ubp-M appeared to coat each of the mitotic chromosomes (Fig. 5), and it remained associated with the chromatin as the cells completed cytokinesis and entered telophase. Enzymatically active Ubp-M was distributed more diffusely within transfected mitotic cells (Fig. 5), although in some cases it did appear to be concentrated in the vicinity of the chromosomes but it was never seen coating the chromosomes, nor was it associated with postmitotic chromatin. Cells transfected with a number of other GFP-fusion proteins, including GFP alone and GFP linked to peptides derived from the amino-terminal segment of Ubp-M, showed no association with mitotic chromosomes.
Figure 5.
Mutant Ubp-M (C-S) localizes to mitotic chromosomes and postmitotic chromatin. (Right) The mutant form of Ubp-M (GFP-Ubp-M C-S) stains chromosomes at all stages of mitosis. Examples of cells fixed in prometaphase, metaphase, anaphase, and telophase are shown. DAPI overlaps the staining of mitotic nuclei but also stains nuclei of nontransfected cells. (Left) Wild-type Ubp-M (GFP-Ubp-M) localizes diffusely throughout mitotic cells. In some cases it appears to be concentrated around metaphase chromosomes, but it does not stain individual chromosomes and does not remain in the postmitotic nucleus.
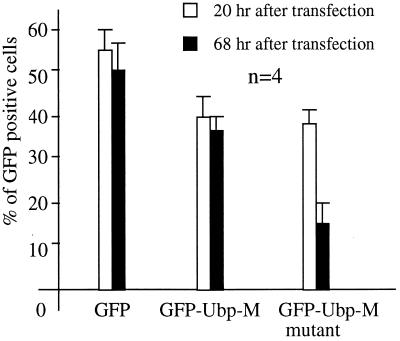
Transfection efficiencies of HEK-293T cells by plasmids containing GFP alone or different forms of Ubp-M were comparable when cell cultures were analyzed 20 hr after transfection (Fig. 6), but, as the cultures progressed, the number of cells expressing the mutant form of Ubp-M decreased significantly. The percentages of cells transfected by plasmids containing the different GFP constructs are presented in Table 1. In cultures exposed to plasmids containing the gene coding for GFP alone approximately 55% of the cells expressed GFP at the end of the transfection period (20 hr). A slightly smaller fraction (40%) expressed the GFP-Ubp-M constructs containing both mutant and native forms. After 68 hr of incubation the fraction of cells that continued to express the mutant form of GFP-Ubp-M decreased significantly, dropping from 40% at the end of the transfection period to approximately 16% after 68 hr. The proportion of cells expressing GFP alone or the enzymatically active form of GFP-Ubp-M remained essentially constant at the end of the same period, and the nontransfected cells continued to grow normally.
Figure 6.
The fraction of cells expressing GFP-fusion proteins at different times after transfection. The results of four experiments are shown.
Table 1.
Percentage GFP-positive cells
| Plasmid | 20 hr after transfection
|
68 hr after transfection
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | Average | 1 | 2 | 3 | 4 | Average | |
| GFP control | 63.5 | 54.3 | 48.9 | 56.0 | 55.7 ± 4.1 | 65.0 | 46.1 | 39.5 | 55.6 | 51.6 ± 7.8 |
| Ubp-M | 52.3 | 31.0 | 43.7 | 39.0 | 41.5 ± 6.5 | 42.2 | 32.0 | 38.3 | 38.1 | 37.7 ± 2.8 |
| Ubp-M mutant | 47.8 | 39.7 | 39.7 | 33.7 | 40.2 ± 3.8 | 19.1 | 25.5 | 10.9 | 10.2 | 16.4 ± 5.9 |
We presume that mutant GFP-Ubp-M blocks cell growth by interfering with the actions of the endogenous Ubp-M enzyme, rather than acting as a nonspecific toxic agent, because expression of the active enzyme has no apparent effect on the transfected cells. Initially, cells transformed with mutant Ubp-M grew normally, and their morphological appearance was unchanged, but, as the incubation continued, many transfected cells appeared to undergo apoptosis.
We do not know whether the expressed mutant proteins affected the production of endogenous Ubp-M in HEK-293T cells, but CHO and COS cells were especially sensitive to plasmids coding for mutant or antisense forms of Ubp-M genes. In contrast to HEK-293T cells, transfected COS cells became apoptotic within 24–48 hr after transfection with either mutant forms of Ubp-M or antisense GFP-Ubp-M, as analyzed by flow cytometry or nuclear morphology.
It is not entirely clear why mutant forms of the ubiquitin-processing enzyme bind to mitotic chromosomes and why they remain within the postmitotic nucleus, but one possible explanation deserves consideration. Enzymatically inactive Ubp-M, containing a single amino acid substitution at its active site, should be able to bind to ubiquitinated substrates, but, lacking the ability to cleave isopeptide bonds, may remain bound to unhydrolyzed substrates, similar to the way specific inhibitors bind to and occupy the active sites of other proteases. Among the ubiquitinated proteins on chromosomes that may be substrates for Ubp-M are histones 2A and 2B (14). Although only 5–10% of histone H2A is ubiquitinated in interphase cells, it is the most abundant ubiquitinated protein in cells, and, as part of the core nucleosome structure, it is distributed uniformly throughout the chromatin. The staining of mitotic chromosomes by the mutant form of GFP-Ubp-M mirrors precisely the expected distribution of the ubiquitinated histones.
It has been known for some time that the histones are deubiquitinated as dividing cells pass through metaphase, and they are reubiquitinated as cells complete division (21, 23), but the enzymes responsible for this rapid and cell cycle-specific turnover of ubiquitin have not been identified. We think it more than coincidental that deubiquitination of the histones occurs at the time that Ubp-M is phosphorylated, enzymatically active, and accessible to the mitotic chromosomes, and the timing of histone reubiquitination also coincides with the dephosphorylation of Ubp-M and its return to the postmitotic cytoplasm. The reubiquitination of histones at the completion of mitosis could be due to either the activation of ubiquitin conjugating enzymes or a reduced level of deubiquitinating activity, or a combination of both.
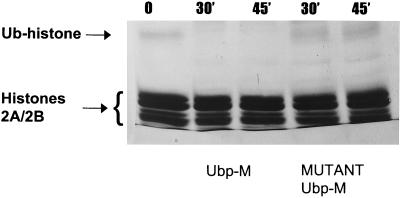
To determine whether recombinant Ubp-M is able to hydrolyze ubiquitinated histones, extracts of chromatin, enriched for histones H2A/H2B, were incubated in vitro with either active or mutant forms of Ubp-M and analyzed by SDS/PAGE, as shown in Fig. 7. Essentially all the detectable ubiquitinated H2A/H2B was lost after incubation with enzymatically active forms of Ubp-M, whereas the mutant enzyme showed no visible decrease. Although the capacity of Ubp-M to hydrolyze ubiquitinated histones in vitro is no guarantee that similar actions take place in vivo, the in vitro data are consistent with this possibility.
Figure 7.
Deubiquitination of histone H2A in vitro by Ubp-M. A small fraction of a histone H2A/H2B dimer preparation is ubiquitinated (Ub-histone) and has a lower mobility on SDS/PAGE gels. This fraction is lost after incubation at 37°C with active Ubp-M but not affected by mutant Ubp-M. Bands were stained by Coomassie blue.
Why does the mutant form of Ubp-M block cell division and eventually lead to programmed cell death?
Mutant Ubp-M could compete with the native deubiquitinating enzyme in many different ways, but its tight association with mitotic chromosomes suggests that it may interfere with cell viability by directly affecting chromatin functions. Recent crystallographic studies of nucleosomes (24) indicate that the amino acid of histone H2A that is normally ubiquitinated (lysine-119) is located close to a critical interface of the nucleosome, at which site a bulky ubiquitin molecule might be expected to influence intranucleosomal packing. If histone H2A is not deubiquitinated during mitosis, one might expect condensation of mitotic chromatin to be affected. Chromatin less able to pack into dense heterochromatin might be more sensitive to oxidative damage or nuclease digestion. If ubiquitinated nucleosomes remain exposed throughout mitosis, additional ubiquitins could be added to histone H2A, creating polyubiquitin chains that are signals for proteolytic degradation. It is also conceivable that ubiquitinated histones may influence other posttranslational modifications of nucleosomes, such as the enzymes responsible for acetylation and deacetylation of chromatin.
Other deubiquitinating enzymes reported to act on chromatin may also interact, either directly or indirectly, with specific histones. Yeast Ubp-3 associates with several proteins known as silent information regulators (SIRs 2, 3, and 4) that are both essential for gene silencing (17) and interact with the amino-terminal segments of histones H3 and H4 (25). Although these histones are not ubiquitinated, crystallographic studies show that H3 and H4 in mammalian chromatin interact directly with the ubiquitinated histones 2A and 2B (24).
Ubiquitin-processing enzymes are known to play an important role in the degradation of short-lived proteins, but the enzyme we describe here may be involved in ubiquitin turnover pathways that regulate mitotic events through other mechanisms, possibly including direct effects on the organization of mitotic chromatin through its capacity to regulate histone functions.
Acknowledgments
We thank Drs. Tian Xu, Arthur Horwich, Evangelos Moudrianakis, and Alan D’Andrea for helpful advice and Karen Muth for help with the manuscript. These studies were supported in part by a grant from Bayer Corporation of West Haven, CT, and a grant from the Lucille B. Markey Charitable Trust Foundation.
ABBREVIATIONS
- GFP
green fluorescent protein
- Ubp
ubiquitin-processing protease
- GST
glutathione S-transferase
- CHO
Chinese hamster ovary
- DAPI
4′,6-diamidino-2-phenylindole
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF126736).
References
- 1.Ciechanover A. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 2.Hochstrasser M. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 3.King R W, Peters J-M, Tugendreich S, Rolfe M, Hieter P, Kirschner M W. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 4.Sudakin, V. Cell6, 185–198.
- 5.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 6.Pickart C M. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 7.Fu H, Sadis S, Rubin D M, Glickman M, van Nocker S, Finley D, Vierstra R D. J Biol Chem. 1998;273:1970–1981. doi: 10.1074/jbc.273.4.1970. [DOI] [PubMed] [Google Scholar]
- 8.Tong H, Hateboer G, Perrakis A, Bernards R, Sixma T K. J Biol Chem. 1997;272:21381–21387. doi: 10.1074/jbc.272.34.21381. [DOI] [PubMed] [Google Scholar]
- 9.Haas A L, Siepmann T J. FASEB J. 1997;11:1257–1270. doi: 10.1096/fasebj.11.14.9409544. [DOI] [PubMed] [Google Scholar]
- 10.Wilkinson K D. FASEB J. 1997;11:1245–1256. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- 11.Gosink M M, Vierstra R D. Proc Natl Acad Sci USA. 1995;92:9117–9121. doi: 10.1073/pnas.92.20.9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaiser P, Mandl S, Schweiger M, Schneider R. FEBS Lett. 1995;377:193–196. doi: 10.1016/0014-5793(95)01323-7. [DOI] [PubMed] [Google Scholar]
- 13.Matuschewski K, Hauser H-P, Treier M, Jentsch S. J Biol Chem. 1996;271:2789–2794. doi: 10.1074/jbc.271.5.2789. [DOI] [PubMed] [Google Scholar]
- 14.Pagano M. FASEB J. 1997;11:1067–1075. doi: 10.1096/fasebj.11.13.9367342. [DOI] [PubMed] [Google Scholar]
- 15.Zhu Y, Carroll M, Papa F R, Hochstrasser M, D’Andrea A D. Proc Natl Acad Sci USA. 1996;93:3275–3279. doi: 10.1073/pnas.93.8.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Y, Lambert K, Corless C, Copeland N G, Gilbert D J, Jenkins N A, D’Andrea A D. J Biol Chem. 1997;272:51–57. doi: 10.1074/jbc.272.1.51. [DOI] [PubMed] [Google Scholar]
- 17.Moazed D, Johnson A D. Cell. 1996;86:667–677. doi: 10.1016/s0092-8674(00)80139-7. [DOI] [PubMed] [Google Scholar]
- 18.Henchoz S, De Rubertis F, Pauli D, Spierer P. Mol Cell Biol. 1996;16:5717–5725. doi: 10.1128/mcb.16.10.5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldknopf I L, Taylor C W, Baum R M, Yeoman L C, Olson M O J, Prestayko A W, Busch H. J Biol Chem. 1975;250:7182–7187. [PubMed] [Google Scholar]
- 20.Bradbury E M. BioEssays. 1992;14:9–16. doi: 10.1002/bies.950140103. [DOI] [PubMed] [Google Scholar]
- 21.Mueller R D, Yasuda H, Hatch C L, Bonner W M, Bradbury E M. J Biol Chem. 1985;260:5147–5153. [PubMed] [Google Scholar]
- 22.Sherwood S W, Kung A L, Roitelman J, Simoni R D, Schimke R T. Proc Natl Acad Sci USA. 1993;90:3353–3357. doi: 10.1073/pnas.90.8.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsui S I, Seon B K, Sandberg A A. Proc Natl Acad Sci USA. 1979;76:6386–6390. doi: 10.1073/pnas.76.12.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Nature (London) 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 25.Heckt A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]