Abstract
We have identified and cloned the cDNA for a 912-aa protein, rab11BP, that interacts with the GTP-containing active form of rab11, a GTP-binding protein that plays a critical role in receptor recycling. Although rab11BP is primarily cytosolic, a significant fraction colocalizes with rab11 in endosomal membranes of both the sorting and recycling subcompartments. In vitro binding of rab11 to native rab11BP requires partial denaturation of the latter to expose an internal binding site located between residues 334 and 504 that is apparently masked by the C-terminal portion of the protein, which includes six repeats known as WD40 domains. Within the cell, rab11BP must undergo a conformational change in which the rab11-binding site becomes exposed, because when coexpressed with rab11 in transfected cells the two proteins formed abundant complexes in association with membranes. Furthermore, although overexpression of rab11BP did not affect transferrin recycling, overexpression of a truncated form of the protein, rab11BP(1–504), that includes the rab11-binding site but lacks the WD40 domains inhibited recycling as strongly as does a dominant negative rab11 mutant protein that does not bind GTP. Strikingly, the inhibition caused by the truncated rab11BP was prevented completely when the cells also expressed a C-terminally deleted, nonprenylatable form of rab11 that, by itself, has no effect on recycling. We propose that rab11BP is an effector for rab11, whose association with this GTP-binding protein is dependent on the action of another membrane-associated factor that promotes the unmasking of the rab11-binding site in rab11BP.
Small GTP-binding proteins of the rab family have characteristic subcellular distributions, and specific rab proteins have been implicated in the control of vesicular transport steps between different subcellular compartments (see ref. 1). The precise mechanism of action of rab proteins has not yet been elucidated, although there is evidence that the active GTP-containing form of some rab proteins serves to stabilize SNARE complexes required for vesicle fusion (2–4). Several proteins have been identified (5–12), in most cases by using a yeast two-hybrid interaction screen, that are capable of binding to the active forms of rab proteins and may serve as their effectors in mediating specific transport steps. In general, these proteins do not show sequence similarity to each other and, in fact, may have diverse functional roles ranging from stabilizing the active form of the rab (9, 13, 14) to mediating interactions with the cytoskeleton (7).
At least five rab proteins (rabs 4, 5, 7, 9, and 11) have been shown to participate in endocytosis and/or transport of endocytosed material or receptors between endosomal compartments or between endosomes and the plasma membrane or the trans-Golgi network (TGN; see ref. 1). Rab11, in particular, has been shown to play a key role in plasma membrane receptor recycling, because expression of a form of this protein that cannot be activated by GTP binding inhibited transport of endocytosed transferrin receptors from sorting endosomes to a pericentriolar recycling compartment, from where receptors normally are returned to the cell surface (15, 16). In addition to its presence in sorting endosomes and the recycling compartment, rab11 has been localized to TGN membranes and post-Golgi secretory vesicles (17, 18) and is highly concentrated in the apical tubulovesicular system of parietal cells in the gastric epithelium (19).
To further our understanding of the mechanism of action of rab11, we sought to identify proteins that interact with its active GTP-binding form and, therefore, may serve as the downstream effectors that execute its function. Using a blotting assay, we have identified a rab11-binding protein, rab11BP, that interacts with the GTP form of rab11 only if the latter contains an intact effector domain. Moreover, we present evidence that rab11BP, together with rab11, participates in the regulation of transferrin receptor recycling.
MATERIALS AND METHODS
DNA Cloning and Construction of Expression Plasmids Encoding Wild-Type or Mutant rab11 or rab11BP.
Wild-type and mutant human rab11 cDNAs (16) were cloned into the bacterial expression vector pET-11d (Novagen), and the bacterially expressed rab11 proteins were purified from lysates by ammonium sulfate precipitation, followed by fast protein liquid chromatography on a Superdex HR-75 gel-filtration matrix (Pharmacia). The full-length rab11BP cDNA coding region and various subfragments of it were cloned into pcDNA3 (Invitrogen), downstream of a hemagglutinin epitope tag, or into pMAL-C2 (New England Biolabs) downstream of sequences encoding the maltose-binding protein moiety, used to purify the hybrid proteins made in bacteria on an amylose-containing resin. Transfection of cultured cells was carried out as described (16).
Cell Fractionation.
Confluent monolayers of cultured cells (MDCK, 293T, or 3T3) were harvested by scraping in a buffer containing 25 mM Hepes/150 mM NaCl/5 mM MgCl2/3 mM DTT/1 μg/ml leupeptin/5 μg/ml Trasylol. Cells were broken by sonication, and sedimentable membrane (P100) and cytosolic supernatant fractions (S100) were obtained by centrifugation (60 min at 100,000 × g; Beckman 100.3 rotor).
Rat liver subcellular fractions were prepared by the procedure of Fuchs et al. (20), and the distribution of various markers was determined by immunoblotting.
Filter-Blotting Assay and Purification of rab11BP.
Gel-electrophoresis/filter-binding overlay assays, including the competition experiments with rab proteins charged with nonradioactive nucleotides, were performed essentially as described previously (21).
Rab11BP was purified from a high-speed supernatant (40,000 rpm, 2 hr in a Beckman Ti60 rotor) prepared from a bovine brain homogenate (1.5 kg in 2 vol of 50 mM Mes, pH 7.0/0.5 mM MgCl2/3 mM NaN3/1 mM EGTA/2 mM EDTA/1 mM DTT/2.5 μg/ml antipain/2.5 μg/ml pepstatin/5 μg/ml Trasylol/0.5 mM phenylmethylsulfonyl fluoride). A 25–50% saturation ammonium sulfate (AS) precipitate obtained from the supernatant was submitted to a sequence of chromatographic steps on Q Sepharose, phenyl-Sepharose, and mono S, and gel filtration was on HR 10/30 Superose-12. Fractions from the last two columns were assayed by a slot-blotting procedure in which the nitrocellulose filters containing the protein fractions were incubated in a buffer containing 8 M urea before washing and probing. An aliquot (150 μg) of the pooled final fractions was fractionated by SDS/PAGE (10%), and, after transfer to nitrocellulose, the major band at 130 kDa, which was positive in the filter-overlay assay, was excised for proteolytic fragmentation and peptide microsequencing (22, 23). The sequences of 8 peptides were obtained: (i) L(L or I)TPEPDIVASTK, (ii) Q(L or I)TAANFCQNGK, (iii) IWALK, (iv) NAFDYFNNMR, (v) LPTGINPLTLHIMR, (vi) AHNAVVTSAIFAPNPSLML(S)LDVQ, (vii) ASFSHDFNYLV(S)(G)xED(K)YVYI, and (viii) GQSDQATASPVTAGTEL(S)NIP(g)L(L)AI(d)(q)VL. The underlined peptide sequences were chosen to design degenerate primers used for PCR-based cloning.
Cloning of rab11BP.
cDNA was synthesized using bovine brain poly(A)+ RNA as template and oligo(dT) as primer and was amplified by PCR using the degenerate primers with 5′ EcoRI sites added. The PCR product (≈950 bp), purified and cloned using the TA Cloning Kit (Invitrogen), contained an ORF of 309 codons. This encoded in its interior peptide 7, in which x corresponded to a serine, and a peptide, LITAANFCQNGK, very similar to peptide 2. The cDNA fragment was used to screen a “stretched” bovine cDNA library (CLONTECH), and three lambda-phage clones containing overlapping inserts were identified that, in aggregate, encompassed an ORF encoding the C-terminal 823 aa of a protein that contained all of the sequenced peptides. The remaining portion of the cDNA ORF was obtained by a 5′ RACE procedure with a kit from CLONTECH and bovine brain mRNA as template. This segment contained an initiation codon immediately preceded by a termination codon. The amino acid sequence derived from the overlapping cDNA segments comprised 912 residues.
A rab11BP cDNA encompassing the entire coding region was assembled by sequential ligations of appropriate restriction fragments and cloned into the pcDNA3 expression vector (Invitrogen).
Antibodies.
A rabbit antiserum to rab11BP was prepared against the peptide GQSDQATASPVTAGTEK (residues 88–103, with an added K). Antibodies to rab11BP and to rab11 (16) were affinity-purified by using the respective recombinant proteins immobilized on nitrocellulose. Antibodies to endolyn (24), cathepsin D (25), and the Na,K-ATPase (26) had been described previously. Antibodies to mannosidase II and TGN38 were gifts of M. Farquhar (University of California at San Diego) and J. Bonifacino (National Institute of Child Health and Human Development), respectively.
RESULTS
Identification of a Protein That Binds Rab11-GTP.
Using a blotting assay (21) with [α-32P]GTP-charged rab11 as a probe, we searched for rab11-binding proteins in lysates from MDCK, 293T, and NIH 3T3 cells. This revealed (Fig. 1A, lanes a–c) the presence of an rab11-binding protein with an Mr of ≈130,000 in all three cell lines. The binding protein, designated rab11BP, was primarily cytosolic (Fig. 1A, lanes d–f) but was also detectable in a sedimentable membrane fraction (Fig. 1A, lane e). Competition experiments showed that rab11BP bound specifically to the GTP-containing form of rab11, because an excess of rab11 charged with nonradioactive GTP eliminated the binding of the radioactive probe (Fig. 1A, lane g), whereas an excess of rab11 charged with nonradioactive GDP had no effect (Fig. 1A, lane h). A screening of cytosolic fractions from several organs using the blotting assay showed that rab11BP is particularly abundant in brain tissue (data not shown). A brain cytosolic subfraction obtained by precipitation with ammonium sulfate between 25 and 45% saturation was highly enriched in rab11BP and, therefore, was used for further characterization and purification of the protein.
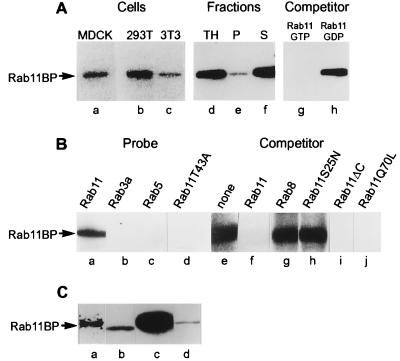
Figure 1.
Identification and cDNA cloning of a 130-kDa protein (rab11BP) that specifically interacts with rab11-GTP in a filter-overlay assay. (A) Lanes a–c, presence of rab11BP in MDCK (a), 293T (b), and 3T3 (c) cells. Cell lysates (100 μg of protein) were analyzed by the filter-overlay assay using [α-32P]GTP-rab11 as a probe. Lanes d–f, though primarily cytosolic (f), a fraction of rab11BP is membrane-associated (e). A total MDCK cell lysate (TH, lane d) and aliquots of sedimentable (P, lane e) and soluble subfractions (S, lane f) were analyzed by the blotting assay for the presence of rab11BP. Lanes g and h, only the active GTP-containing form of rab11 binds to rab11BP. Filter-overlay assays with MDCK cell lysates and [α-32P]GTP-rab11 as probe were carried out in the presence of a 30-fold excess of rab11 charged with nonradioactive GTP (g) or GDP (h). (B) Specificity of the rab11–rab11BP interaction. When used as probes charged with [α-32P]GTP, rab 3a (b), rab5 (c), and a rab11 effector domain mutant (rab11T43A) (d) fail to bind to rab11BP. Aliquots (100 μg protein) of a rat brain cytosolic subfraction obtained by ammonium sulfate precipitation (25–45% saturation) were analyzed by the blotting assay with the indicated [α-32P]GTP-charged probes. Lanes e–h, when used as competitors in the blotting assay with the brain cytosolic subfraction, rab8 and the dominant negative rab11 mutant (rab11S25N) do not diminish binding of [α-32P]GTP-rab11 to rab11BP. The various competitors were incubated for charging with cold GTP and used in the blotting assay at a 30-fold excess over the concentration of the labeled [32P]GTP-rab11 probe. (C) The cloned cDNA encodes a protein that comigrates with rab11BP and binds rab11-GTP. Lane a, an immunoprecipitate obtained from a partially purified brain cytosolic extract with an antibody to a peptide (residues 88–103) within the amino acid sequence derived from the cloned cDNA was used for the filter-overlay assay with [α-32P]GTP-rab11 as a probe. The protein in the immunoprecipitate that bound the probe (a) had the same electrophoretic mobility as rab11BP (d) in the original bovine brain protein fraction. Lanes b and c, aliquots of lysates of 293T cells transfected with the pcDNA vector alone (b) or the vector containing the cloned cDNA (c) were analyzed by the filter-overlay assay. The amount of extract analyzed in lane b, to reveal the endogenous human rab11BP, was 30 times greater than that from the transfected cells used in lane c.
Rab11BP appears to be specific for rab11, because in direct probing (Fig. 1B, lanes a–c) or in competition experiments (Fig. 1B, lanes e–g), it did not bind to the GTP-containing forms of rab3a, rab5, and rab8. The specificity of the rab11-rab11BP interaction was also examined using rab11 mutant proteins. As expected from the competition experiment in Fig. 1A (lanes g and h), a mutant protein unable to bind GTP (rab11S25N) could not compete with [α-32P]GTP-charged rab11 for binding to rab11BP (Fig. 1B, lane h). More importantly, a form of rab11 altered in its effector domain but capable of binding GTP (rab11T43A) did not bind to rab11BP (Fig. 1B, lane d). On the other hand, two other mutant forms of rab11, one that is C-terminally truncated (rab11ΔC:Q212STOP) and another expected to lack GTPase activity (rab11Q70L), when charged with GTP competed effectively with wild-type rab11-GTP for binding to rab11BP (Fig. 1B, lanes i and j). These observations indicate that rab11BP has some of the key properties expected of a functional effector of rab11. It should be noted, however, that the blotting assay used to identify rab11BP entails the denaturation of the protein during an SDS gel electrophoresis step. Only a very weak binding to rab11 could be demonstrated when the nondenatured protein in a cytosolic protein subfraction was immobilized directly on a nitrocellulose filter without subsequent denaturation (not shown).
Cloning of the rab11BP cDNA.
Rab11BP was purified from bovine brain cytosol by a sequence of chromatographic separations, using a simplified slot-blot assay to measure the [α-32P]GTP-rab11 binding capacity of immobilized, urea-denatured, protein fractions. After a final separation by SDS gel electrophoresis and transfer to nitrocellulose, the band at an Mr of 130,000 was excised and used for proteolytic fragmentation and microsequencing analysis (22, 23). The sequences of eight peptides (listed in Materials and Methods) were determined, which enabled us to obtain, by reverse transcription–PCR with degenerate primers, an amplified cDNA segment of 950 bp. This was used for subsequent library screening and a 5′ RACE procedure to obtain the entire coding region of a cDNA that encompasses 912 codons and includes all of the sequenced peptides (Fig. 2A).
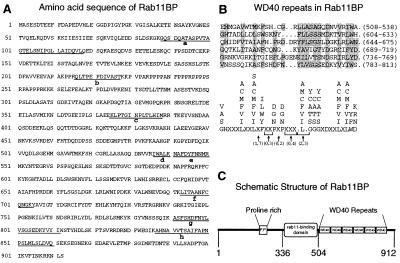
Figure 2.
Structure of Rab11BP. (A) Amino acid sequence of bovine rab11BP derived from the cDNA. The sequences of peptides originally identified by microsequencing in the purified protein are underlined. A proline-rich region extends from residues 210 to 256. The protein has a calculated molecular mass of 101,290 Da and a pI of 5.18. The nucleotide sequence that encodes the bovine rab11BP has been submitted to GenBank with accession no. AF117897. (B) Rab11BP contains six WD40 repeats in the region extending from residues 508 to 813. (a) The six repeats, whose positions are indicated by the numbers within the parentheses at the right of each sequence, are shown in an alignment to maximize their agreement (shaded residues) with the loose consensus sequence for WD40 repeats defined by Neer et al. (30), which is presented in b. (C) Schematic representation of the structure of rab11BP. In addition to the WD40 repeats and the proline-rich region, the figure indicates the location of the rab11-binding domain, as defined by the experiments in Fig. 3.
Several criteria were used to establish that the cDNA we cloned actually corresponds to that of rab11BP, despite the discrepancy between the derived molecular mass (101.29 kDa) of the protein encoded in the cDNA and the molecular mass (≈130 kDa) estimated from the electrophoretic mobility of rab11BP in SDS gels. First, an antibody produced against a peptide (GQSDQATASPVTAGTE) that is encoded in the cloned cDNA (residues 88–103 in Fig. 2A) immunoprecipitated a protein from a brain cytosolic subfraction that, in the blot assay, bound [α-32P]GTP-rab11 (Fig. 1C, lane a) and had the same electrophoretic mobility (≈130 kDa) as bovine brain rab11BP (Fig. 1C, lane d). Second, expression of the complete cDNA in 293T cells yielded a protein that in the blotting assay bound rab11GTP (Fig. 1C, lane c) and also had the same electrophoretic mobility as the bovine brain protein (Fig. 1C, lane d), which is slightly lower than that of the endogenous human product in 293T cells (Fig. 1C, lane b).
Rab11BP is a novel protein whose sequence (Fig. 2A) is not represented in available databases. A region of rab11BP comprising the C-terminal ≈400 aa contains six WD40-like repeats (Fig. 2B) and, therefore, shows significant homology to other WD40-containing proteins, in particular, 67% identity and 80% similarity over a region of approximately 200 aa to a Caenorhabditis elegans protein (GenBank accession no. Z92837), as well as a lower similarity to the yeast αCOP (27) and the yeast and mammalian β′COPs (28, 29). WD40 domains are segments of approximately 42–43 aa in length that fit a loose consensus sequence (Fig. 2B) and are thought to mediate protein–protein interactions (30, 31). No significant similarities with other proteins were observed for the amino-terminal half of rab11BP.
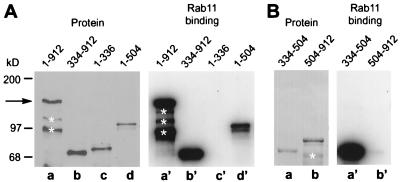
Analysis of the capacity to interact with rab11 of various rab11BP deletion constructs expressed either in mammalian cells or in bacteria (for protein segments that did not accumulate in transfected mammalian cells) indicated that the rab11-binding domain is located between residues 334 and 504 and not in the distal WD40-containing region. Thus, a bacterial fusion protein containing those residues linked to the maltose-binding protein effectively bound 32P-labeled GTPrab11 (Fig. 3B, lane a′), whereas proteins encompassing residues 1–336 or 504–912 did not (Fig. 3 A, lane c′, and B, lane b′, respectively). Moreover, proteins encompassing the first 504 residues or extending from residues 334 to 912 interacted strongly with rab11 (Fig. 3A, lane d′ and b′, respectively). It is noteworthy that all rab11BP deletion proteins containing the amino-terminal region (1–336) showed an anomalously slow electrophoretic migration (Fig. 3A, lanes c and d) that was not manifested by proteins containing only the distal regions of the rab11BP (Fig. 3 A, lane b, and B, lanes a and b) and was sufficient to account for the anomalous electrophoretic behavior of the full-length protein. A striking feature of the amino-terminal segment of rab11BP is the presence of a region of high proline content (22 proline residues between amino acids 210 and 256; Fig. 2A). The domain structure of rab11BP is illustrated in Fig. 2C.
Figure 3.
The rab11-binding region in rab11BP is located between residues 334 and 504. (A) HA-tagged intact rab11BP (lane a, 1–912) or the specific HA-tagged deleted forms of the protein indicated above each lane (b–d) were expressed in transfected 293T cells, and the cell lysates were analyzed by immunoblotting with the anti-HA antibody (Left, a–d) or, for [32P]GTP-rab11 binding, by the overlay assay (Right, a′–d′). The white asterisks indicate degradation products of rab11BP(1–912). (B) Purified fusion proteins containing the Escherichia coli maltose-binding protein linked to rab11BP segments extending from residues 334 to 504 (a and a′) or from 504 to 912 (b and b′) were fractionated by SDS gel electrophoresis and transferred to nitrocellulose. The Poinceau red-stained protein pattern (Left) and [32P]GTP-rab11 binding pattern (Right) are shown.
In Vivo Association of rab11BP and rab11.
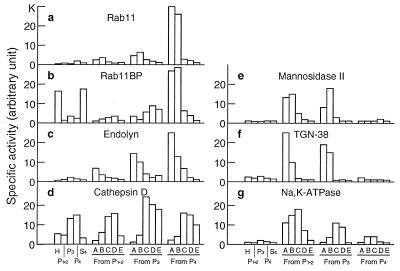
That a functional interaction between rab11 and rab11BP takes place in vivo, in spite of the fact that in vitro the interaction requires the unfolding of rab11BP, first was suggested by the finding that rab11 was colocalized with the membrane-bound subfraction of rab11BP molecules in an endosomal compartment, by both cell fractionation and immunofluorescence studies (Figs. 4 and 5). Thus, both proteins were found at their highest concentrations (Fig. 4 a and b, respectively) in an endosomal rat liver subfraction containing the highest specific activity of the endosomal membrane marker, endolyn (Fig. 4c) (24). In striking contrast, both rab11 and rab11BP were present at very low specific activities in Golgi fractions characterized by the presence of mannosidase II (Fig. 4e), in fractions highly enriched in the TGN marker, TGN38 (Fig. 4f), and in lysosomes, characterized by the presence of cathepsin D (Fig. 4d), as well as in a plasma membrane fraction, identified by its content of Na,K-ATPase (Fig. 4g).
Figure 4.
Colocalization of rab11 and rab11BP in subcellular fractions enriched in endosomes. A rat liver homogenate was fractionated (20) to obtain three sedimentable fractions (P1–2, P3, P4) enriched in nuclei and mitochondria (P1–2), smaller particulate components of the cytoplasm (P3 and P4), and a final supernatant (S). The sedimentable fractions were subfractionated by flotation in discontinuous sucrose gradients to obtain from each four subfractions (A, B, C, and D) and a pellet. Aliquots (100 μg protein) of each fraction and subfraction were analyzed by Western blotting with antibodies specific for rab11 or rab11BP, or for the marker proteins indicated in each chart. Bound antibodies were detected with 125I-labeled protein A and quantified by PhosphorImager analysis. The bars represent the specific activities for each protein in the various fractions and subfractions indicated on the abscissa at the bottom of the figure.
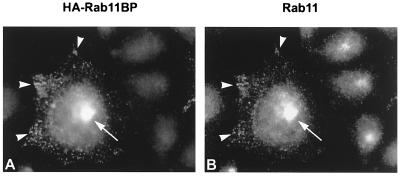
Figure 5.
Immunofluorescence colocalization of rab11 and membrane-associated rab11BP in endosomes and the pericentriolar recycling compartment. Permanent transformants of Chinese hamster ovary cells that express the human transferrin receptor (TRVb-1 cells) were transfected with a plasmid-encoding HA-tagged rab11BP. The cells grown on coverslips were permeabilized with 0.05% saponin in PBS for 4 min to release soluble cytosolic components and processed for double-label immunofluorescence with primary mouse mAbs to the HA epitope (A) and rabbit antibodies to rab11 (B), followed by Texas red-conjugated donkey anti-mouse antibody and fluorescein-conjugated donkey anti-rabbit antibody, respectively. The arrows and arrowheads point to the pericentriolar compartment and to peripheral-sorting endosomes in a transfected cell, respectively. Note that a trio of untransfected cells in B shows intense staining of the pericentriolar compartment with anti-rab11 antibodies.
Double-label immunofluorescence of Chinese hamster ovary cells that expressed an epitope-tagged form of rab11BP and were treated with saponin to remove cytosolic components, showed that membrane-associated rab11BP (Fig. 5A) colocalized with endogenous rab11 (Fig. 5B) in peripheral (arrowheads) as well as pericentriolar vesicular structures (arrows), which previously have been shown (15, 16) to correspond to sorting endosomes and to the pericentriolar recycling compartment, respectively.
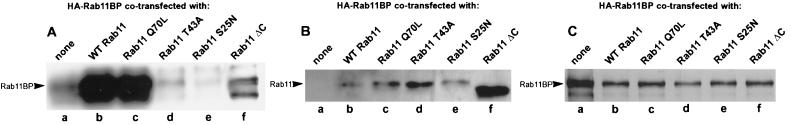
The in vivo association of rab11 and rab11BP, and its dependence on the activation of rab11 and on the integrity of the rab11 effector domain, was demonstrated in cells coexpressing exogenous hemagglutinin-tagged rab11BP (HA-rab11BP) and various rab11 mutant proteins (Fig. 6). Western blot analysis, using antibodies to the epitope tag, of immunoprecipitates obtained with anti-rab11 antibodies from cell extracts prepared with a nonionic detergent showed (Fig. 6A) that in vivo rab11BP formed abundant complexes with the wild type (Fig. 6A, lane b) and the constitutively active (rab11Q70L) protein (Fig. 6A, lane c), whereas it failed to interact with the effector domain mutant (rab11T43A) and the GDP form (rab11 S25N) of rab11 (Fig. 6A, lanes d and e, respectively). These observations are consistent with the behavior of the same rab11 mutant proteins in the blotting assay (Figs. 1B). On the other hand, in transfected cells rab11BP interacted only very poorly with the C-terminally truncated form of rab11 (rab11ΔC), which cannot be prenylated and, therefore, does not become associated with membranes (Fig. 6A, lane f), even though in the blotting assay rab11ΔC efficiently recognized denatured rab11BP (Fig. 1B). This suggests that in vivo intact rab11 and rab11BP only interact with each other when associated with membranes. Thus, it would appear that the membrane-associated form of rab11BP adopts a conformation in which the normally sequestered rab11-binding site is exposed and able to interact with rab11.
Figure 6.
In vivo association of rab11 and rab11BP in cotransfected cells. Cultures of 293T cells were transfected with (a) a pcDNA3-based expression plasmid encoding HA-Rab11BP alone or together with plasmids encoding (b) WT Rab11, (c) Rab11Q70L (GTPase-deficient mutant), (d) Rab11T43A (effector domain mutant), (e) Rab11S25N (GDP-bound mutant), or (f) Rab11ΔC (a deletion mutant that lacks the last 6 amino acids and therefore cannot be prenylated). Forty hours posttransfection, complexes containing wild-type or rab11 mutant proteins were immunoprecipitated from detergent-solubilized cell lysates using immobilized polyclonal antibody to rab11. (A) Recoveries of HA-rab11BP in the immunoprecipitates analyzed by immunoblotting with a monoclonal anti-HA antibody. (B) Recoveries of rab11 or its mutants in the same immunoprecipitates analyzed with rabbit anti-rab11 antibodies. (C) Comparable levels of HA-Rab11BP are expressed in all the transfected cells. Equal aliquots of the cell lysates (2% of the total) were analyzed by immunoblotting with the anti-HA antibody.
Role of rabBP in Transferrin Recycling.
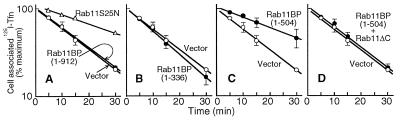
It has been shown previously (15, 16), and, for comparative purposes, is illustrated in Fig. 7A, that a dominant negative form of rab11 (rab11S25N), which does not bind GTP and is believed to competitively inhibit the activation of the endogenous wild-type protein, strongly inhibits transferrin recycling. This effect results from an inhibition of transport of interiorized receptors from sorting endosomes to the recycling compartment, which is dependent on activated rab11. Moreover, a GTPase-deficient form of rab11 was also found to inhibit recycling (15, 16), apparently because it blocks exit of transferrin receptors from the pericentriolar compartment (16). To determine whether rab11BP plays a role in the recycling pathway, we examined the effects of overexpression of this protein, and of various C-terminally truncated mutants of it, on transferrin recycling, as well as on the inhibition of recycling caused by expression of rab11 and its mutants. As shown in Fig. 7, expression of intact rab11BP(1–912) or of the mutant lacking the rab11-binding domain, rab11BP(1–336), had no effect on recycling (Fig. 7 A and B, respectively), whereas expression of the truncated protein, rab11BP(1–504), containing the rab11-binding region but lacking the WD40 domains, inhibited recycling as strongly as did the dominant negative rab11S25N mutant (compare Fig. 7 A and C). Thus, rab11BP(1–504) itself can be considered to be a dominant negative mutant capable of interacting with rab11 but defective in mediating its function. The strong inhibitory effect of rab11BP(1–504) was observed even though in the transfected cells it accumulated to lower levels than the intact protein or the rab11BP(1–336) variant (not shown). Because rab11ΔC—the C-terminally truncated form of rab11 that lacks the prenylation site required for its direct association with membranes—does not inhibit recycling (16) but can interact in vitro with rab11BP (Fig. 1B, lane i), we determined whether this mutant protein can abrogate the inhibitory effect of rab11BP(1–504). Indeed, when coexpressed with rab11BP(1–504), rab11ΔC completely prevented the marked inhibition caused by rab11BP(1–504) alone. No other rab11 mutant affected the inhibitory capacity of rab11BP(1–504) (not shown). Moreover, neither rab11BP(1–912) nor rab11BP(1–336), which, by themselves did not affect transferring recycling, altered the inhibitory capacity of the various rab11 mutant proteins (not shown).
Figure 7.
Inhibition of transferrin recycling by truncated rab11BP(1–504) and its relief by the nonprenylated rab11ΔC. Rates of transferrin recycling were assessed as described previously (16) in cultures of TRVb cells, a mutant line of Chinese hamster ovary cells lacking a functional endogenous transferrin receptor, that were cotransfected with a plasmid encoding the human transferrin receptor (A–D) and plasmids encoding the intact rab11BP (A), or its truncated variants, rab11BP (1–336) (B) and rab11BP (1–504) (C and D). A also shows the recycling kinetics (▵) for cells that express the receptor together with the rab11 dominant negative mutant (rab11S25N). In D, the cells express—in addition to the transferrin receptor and the truncated rab11BP(1–504)—the nonprenylatable rab11ΔC. In A–D, the recycling kinetics also are shown (○) for control cells that were cotransfected with the plasmid encoding the transferrin receptor and the pCDNA3 vector lacking an insert.
DISCUSSION
Using a blotting assay with radioactive rab11GTP as a probe, we have identified, purified, and cloned the cDNA for a protein of 912 aa, rab11BP, that specifically binds the active GTP-containing form of rab11 only if the latter contains an intact effector domain. Although a large pool of rab11BP molecules is present in the cytosol, a fraction is membrane-associated and is found in both sorting endosomes and the pericentriolar recycling compartment, where rab11 itself is also normally concentrated. The region within rab11BP that interacts with rab11 is contained within a polypeptide segment that extends from residues 334 to 504.
Several observations led us to suggest that in the cell rab11BP exists in two conformations, a cytosolic one that is not competent for rab11 binding, and a membrane-associated one in which its binding site for rab11 is exposed. Thus, native rab11BP isolated from brain cytosol could not bind rab11-GTP in a slot-blotting assay unless it was first partially denatured. However, in cotransfected cells expressing both proteins, a substantial amount of rab11BP could be recovered in a complex with wild-type rab11, or with the constitutively active rab11 mutant, by immunoprecipitation from nonionic detergent extracts of postnuclear supernatants. It appears that in the cytosolic form of rab11BP the rab11-binding site is masked by the C-terminal region of the protein that contains six WD40 repeats. This is inferred from the observation that overexpression of intact rab11BP(1–912) had no effect on transferrin recycling, whereas a truncated rab11BP(1–504) lacking the WD40 domains interfered with this process. This inhibition presumably results from binding of rab11BP(1–504) to the membrane-associated active form of rab11, and failure of the truncated protein to execute its effector function, which would require the deleted region. We have found that in transfected cells, rab11BP(1–504) becomes membrane-associated to a greater extent than does the intact protein (data not shown). This would be expected if during its normal function to associate with rab11, rab11BP must first undergo a conformation change induced by its interaction with another membrane component, which causes a displacement of the C-terminal region of rab11BP that exposes the rab11-binding site. Such an activation would not be required for rab11BP(1–504) to associate with rab11. That an association with the membrane is required for the activation of rab11BP can be concluded from our finding that a C-terminally deleted form of rab11 (rab11ΔC), which cannot be prenylated and therefore remains cytosolic in vivo, does not efficiently form a complex with the intact rab11BP, as do the wild-type and the constitutively active forms of rab11, which are membrane-associated. On the other hand, the nonprenylated rab11ΔC efficiently prevented the inhibition caused by the truncated cytosolic rab11BP(1–504), which would be expected if the nonprenylated rab11 associates with the truncated rab11BP in the cytosol and prevents it from complexing with and blocking the action of the active membrane-associated form of rab11. Indeed, we have observed (M.R. et al., unpublished observations) that coexpression of rab11ΔC with rab11BP(1–504) effectively suppresses the association of the latter with the membrane by sequestering it into the cytosol.
It is noteworthy that overexpression of the intact rab11BP(1–912) neither enhanced nor inhibited transferrin recycling. It is very likely that, if the normal function of this protein is carried out by the small fraction of molecules (≈5%) that is associated with endosomal membranes, then normally a sufficient excess of cytosolic rab11BP molecules is present, so that the increase of exogenous molecules resulting from transfection would have a negligible effect. Considering our proposal that a yet unidentified membrane-associated molecule is required to enable rab11BP to undergo the conformational change that allows it to bind rab11 and to carry out its function, it seems possible that the levels of the hypothetical rab11BP activator may, in fact, be rate-limiting for the function of rab11BP in transferrin recycling.
A direct demonstration of the role of rab11BP in receptor recycling and an understanding of the pathway of its activation would seem to require the use of an in vitro cytosolic protein-dependent system to study recycling, in which the activity of normal or mutant rab11BP molecules can be examined.
Acknowledgments
We thank Drs. P. de Camilli and R. Jahn for samples of the rab5 and rab3a proteins and Drs. M. Farquhar and J. Bonifacino for antibodies to α-mannosidase and TGN38, respectively. We are pleased to acknowledge the expert technical contributions of Ms. Irit Edelman-Novemsky, the help of Mss. Heide Plesken and Jody Culkin in preparing the illustrations, and the help of Ms. Myrna Cort in the preparation of the manuscript. We appreciate the valuable assistance Mr. Antonio J. D. Rocha throughout the work. This work was supported by National Institutes of Health Grant GM43583.
ABBREVIATIONS
- TGN
trans-Golgi network
- HA
hemagglutinin
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF117897).
References
- 1.Novick P, Zerial M. Curr Opin Cell Biol. 1997;9:496–504. doi: 10.1016/s0955-0674(97)80025-7. [DOI] [PubMed] [Google Scholar]
- 2.Sogaard M, Tani K, Ye R R, Geromanos S, Tempst P, Kirchhausen T, Rothman J E, Sollner T. Cell. 1994;78:937–948. doi: 10.1016/0092-8674(94)90270-4. [DOI] [PubMed] [Google Scholar]
- 3.Lian J P, Stone S, Jiang Y, Lyons P, Ferro-Novick S. Nature (London) 1994;372:698–701. doi: 10.1038/372698a0. [DOI] [PubMed] [Google Scholar]
- 4.Lupashin V V, Waters M G. Science. 1997;276:1255–1258. doi: 10.1126/science.276.5316.1255. [DOI] [PubMed] [Google Scholar]
- 5.Shirataki H, Kaibuchi K, Sakoda T, Kishida S, Yamaguchi T, Wada K, Miyazaki M, Takai Y. Mol Cell Biol. 1993;13:2061–2068. doi: 10.1128/mcb.13.4.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brondyk W H, McKiernan C J, Fortner K A, Stabila P, Holz R W, Macara I G. Mol Cell Biol. 1995;15:1137–1143. doi: 10.1128/mcb.15.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Echard A, Jollivet F, Martinez O, Lacapere J-J, Rousselet A, Janoueix-Lerosey I, Goud B. Science. 1998;279:580–585. doi: 10.1126/science.279.5350.580. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Okamoto M, Schmitz F, Hofmann K, Sudhof T C. Nature (London) 1997;388:593–598. doi: 10.1038/41580. [DOI] [PubMed] [Google Scholar]
- 9.Diaz E, Schimmoller F, Pfeffer S R. J Cell Biol. 1997;138:283–290. doi: 10.1083/jcb.138.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stenmark H, Vitale G, Ullrich O, Zerial M. Cell. 1995;83:423–432. doi: 10.1016/0092-8674(95)90120-5. [DOI] [PubMed] [Google Scholar]
- 11.Horiuchi H, Lippe R, McBride H M, Rubino M, Woodman P, Stenmark H, Rybin V, Wilm M, Ashman K, Mann M, Zerial M. Cell. 1997;90:1149–1159. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- 12.Ren M, Zeng J, De Lemos-Chiarandini C, Rosenfeld M, Adesnik M, Sabatini D D. Proc Natl Acad Sci USA. 1996;93:5151–5155. doi: 10.1073/pnas.93.10.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rybin V, Ullrich O, Rubino M, Alexandrov K, Simon I, Seabras M C, Goody R, Zerial M. Nature (London) 1996;383:266–269. doi: 10.1038/383266a0. [DOI] [PubMed] [Google Scholar]
- 14.Kishida S, Shirataki H, Sasaki T, Kato M, Kailbuchi K, Takai Y. J Biol Chem. 1993;268:22259–22261. [PubMed] [Google Scholar]
- 15.Ullrich O, Reinsch S, Urbe S, Zerial M, Parton R G. J Cell Biol. 1996;135:913–924. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ren M, Xu G, Zeng J, De Lemos-Chiarandini C, Adesnik M, Sabatini D D. Proc Natl Acad Sci USA. 1998;95:6187–6192. doi: 10.1073/pnas.95.11.6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urbe S, Huber L A, Zerial M, Tooze S A, Parton R G. FEBS Lett. 1993;334:175–182. doi: 10.1016/0014-5793(93)81707-7. [DOI] [PubMed] [Google Scholar]
- 18.Chen W, Feng Y, Chen D, Wandinger-Ness A. Mol Biol Cell. 1998;9:3241–3257. doi: 10.1091/mbc.9.11.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldenring J R, Soroka C J, Shen K R, Tang L H, Rodriguez W, Vaughan H D, Stoch S A, Modlin I M. Am J Physiol. 1994;267:G187–G194. doi: 10.1152/ajpgi.1994.267.2.G187. [DOI] [PubMed] [Google Scholar]
- 20.Fuchs R, Male P, Mellman I. J Biol Chem. 1989;264:2212–2220. [PubMed] [Google Scholar]
- 21.Coutavas E, Ren M, Oppenheim J D, D’Eustachio P, Rush M G. Nature (London) 1993;366:585–587. doi: 10.1038/366585a0. [DOI] [PubMed] [Google Scholar]
- 22.Erdjument-Bromage H, Lui M, Sabatini D M, Snyder S H, Tempst P. Protein Sci. 1994;3:2435–2446. doi: 10.1002/pro.5560031227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lui M, Tempst P, Erdjument-Bromage H. Anal Biochem. 1996;241:156–166. doi: 10.1006/abio.1996.0393. [DOI] [PubMed] [Google Scholar]
- 24.Croze E, Ivanov I E, Kreibich G, Adesnik M, Sabatini D D, Rosenfeld M G. J Cell Biol. 1989;108:1597–1613. doi: 10.1083/jcb.108.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenfeld M, Kreibich G, Popov K, Kato K, Sabatini D D. J Cell Biol. 1982;93:135–143. doi: 10.1083/jcb.93.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desai-Yajnik V, Zeng J, Omori K, Sherman J, Morimoto T. Endocrinology. 1995;136:629–639. doi: 10.1210/endo.136.2.7835297. [DOI] [PubMed] [Google Scholar]
- 27.Letourneur F, Gaynor E C, Hennecke S, Démolliere C, Duden R, Emr S D, Riezman H, Cosson P. Cell. 1994;79:1199–1207. doi: 10.1016/0092-8674(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 28.Stenbeck G, Harter C, Brecht A, Herrmann D, Lottspeich F, Orci L, Wieland F T. EMBO J. 1993;12:2841–2845. doi: 10.1002/j.1460-2075.1993.tb05945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duden R, Hosobuchi M, Hamamoto S, Winey M, Byers B, Schekman R. J Biol Chem. 1994;269:24486–24495. [PubMed] [Google Scholar]
- 30.Neer E J, Schmidt C J, Nambudripad R, Smith T F. Nature (London) 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 31.van der Voorn L, Ploegh H L. FEBS Lett. 1992;2:131–134. doi: 10.1016/0014-5793(92)80751-2. [DOI] [PubMed] [Google Scholar]