Abstract
Propagation of mouse embryonic stem (ES) cells in vitro requires exogenous leukemia inhibitory factor (LIF) or related cytokines. Potential downstream effectors of the LIF signal in ES cells include kinases of the Src, Jak, and mitogen-activated protein families and the signal transducer and transcriptional activator STAT3. Activation of nuclear STAT3 and the ability of ES cells to grow as undifferentiated clones were monitored during LIF withdrawal. A correlation was found between levels of STAT3 activity and maintenance of an undifferentiated phenotype at clonal density. In contrast, variation in STAT3 activity did not affect cell proliferation. The requirement for STAT3 was analyzed by targeted mutagenesis in ES cell lines exhibiting different degrees of LIF dependency. An insertional mutation was devised that abrogated Stat3 gene expression but could be reversed by Cre recombination-mediated excision. ES cells heterozygous for the Stat3 mutation could be isolated only from E14 cells, the line least dependent on LIF for self-renewal. Targeted clones isolated from other ES cell lines were invariably trisomic for chromosome 11, which carries the Stat3 locus, and retained normal levels of activated STAT3. Cre-regulated reduction of Stat3 gene copy number in targeted, euploid E14 clones resulted in dose-dependent losses of STAT3 activity and the efficiency of self-renewal without commensurate changes in cell cycle progression. These results demonstrate an essential role for a critical amount of STAT3 in the maintenance of an undifferentiated ES cell phenotype.
Keywords: leukemia inhibitory factor, cytokine signaling, JAK-STAT pathway, embryonic stem cell differentiation
Murine embryonic stem (ES) cells are permanent pluripotent cell lines derived by culture of preimplantation embryos (1–3). Various ES cell lines may show different characteristics during their establishment and maintenance (4); however, they all require leukemia inhibitory factor (LIF) for self-renewal. LIF initially was identified as a cytokine capable of inducing the differentiation of M1 myeloid leukemia cells (5) and later was shown to have a strong differentiation-inhibiting activity on ES cells (6–8). LIF, IL-6, and IL-11, ciliary neurotrophic factor, cardiotrophin 1, and oncostatin M belong to a family of cytokines characterized by their use of composite receptors that share the signal transducing chain, gp130 (reviewed in ref. 9). Ligand-induced assembly of gp130 triggers the activation of associated Jak tyrosine kinases and results in the activation of overlapping intracellular signaling cascades, which can account for the pleiotropic effects of this group of cytokines (10). Jak-mediated phosphorylation of specific tyrosine residues in the gp130 cytoplasmic domain creates docking sites for Src homology 2-containing STATs (signal transducer and activator of transcription), which are, in turn, phosphorylated at a single regulatory tyrosine residue, dimerize, and translocate to the nucleus to induce specific sets of genes (11). Among the seven members of the STAT family, STAT3 is the major mediator of gp130 signals (12–15), being involved in induction of acute-phase response genes in hepatic cells (16), transcriptional activation and self-renewal in ES cells (17, 18), LIF- and IL-6-dependent differentiation of M1 cells (19–21), as well as a variety of responses in vivo (22–24).
Recently, it has become clear that STAT3 plays an important role in embryonic cell growth. Targeted disruption of the Stat3 gene in mice resulted in early embryonic lethality (25). In addition, ectopic overexpression of dominant-negative versions of STAT3 in ES cells led to loss of pluripotency and enhanced cell differentiation (17, 18). We present direct genetic and biochemical evidence that STAT3 function in ES cells is linked to the maintenance of a stem-cell phenotype independent of cell proliferation. By modifying Stat3 gene expression through conditional mutagenesis of one of its alleles, we demonstrate that a minimal dose of STAT3 is required for ES cell propagation and pluripotency.
MATERIALS AND METHODS
Cloning and Conditional Mutation of the Murine Stat3 Gene in ES Cells.
An 18.5-kb genomic clone containing STAT3 exons 12–24 was isolated from a 129/SvJ genomic library (Stratagene) by using a STAT3 cDNA probe (12). A Stat3 targeting construct was assembled by cloning an 8-kb HindIII genomic fragment (exons 15–22) into pLitmus29 (New England Biolabs) and inserting (i) a loxP and a HindIII site into a ClaI site in intron 15, and (ii) a floxed (loxP-flanked) TK-neor cassette, obtained from the neor and thymidine kinase (TK) units from pPNT (26) subcloned into the BamHI site of pLoxLox (P. Orban, Institute for Genetics, Cologne, Germany), into a SphI site in intron 21.
To generate Stat3-mutant ES cells, linearized targeting vector (40 μg) was electroporated into 1.6 × 106 cells (240 V and 500 μF, Bio-Rad gene pulser). Resistant clones selected in 0.4 mg/ml G418 (Life Technologies, Grand Island, NY) were screened for homologous recombination by PCR (Expand, Boehringer Mannheim). For transient expression of the Cre enzyme, 30 μg of pMC-Cre (H. Gu, National Institutes of Health, Bethesda, MD) supercoiled plasmid were electroporated into 2 × 107 cells (240 V, 960 μF). Cells (106) were seeded onto a gelatinized 10-cm plate and replated at clonal density after 24 h. Fifty to 100 random clones were assayed by PCR for Cre-mediated recombination and for clonal purity. PCR primers were: 30_20, 5′-GCACGAACACAAAAGTGATGAACATGGAGGAGTC-3′; L2, 5′-GCAGCCCAAGCTTGTATTCTATAGTGTCACCTA-3′; 1R, 5′-GAAAAGCGCCTCCCCTACCCGGTAGAATTGAC-3′; 2, 5′-CACACAAGCCATCAAACTCTGGTCTCCAACAGAA-3′; L2rev, 5′-CGATTTAGGTGACACTATAGAATACAAGCTTGGGCTG-3′; and L2WT, 5′-CTTTCTTCAGACTCCTGGCACTGCTCACTCAG-3′.
ES Cell Lines and ES Cell Differentiation Assay.
ES cell lines were propagated by standard methods (27) on monolayers of mitomycin C-treated fibroblasts derived from mouse embryos carrying the PGK-neor gene (28). E14 (29), J1 (30), R1 (31), and W4 (W. Auerbach, New York University) were derived, respectively, from 129/OlaHsd, 129/SvJae, (129/Sv x 129/SvJ)F1, and 129/SvEv mice. The supernatant of 720-LIF-D cells (C. Simon, University of Chicago) secreting recombinant mouse LIF was used to supplement the ES cell cultures at 1,000 units/ml. LIF units were calibrated with recombinant murine LIF (ESGRO, Life Technologies) for support of undifferentiated growth of R1 cells. For differentiation assays, 103 cells per well were seeded onto gelatin-coated 6-well plates. After 6–7 days, undifferentiated colonies were identified by staining for alkaline phosphatase expression (Kit 86R, Sigma-Aldrich). Total colony number was measured by automated colony counting (Quantity One, Bio-Rad).
Karyotype Analysis.
Mitotically arrested ES cells were harvested for chromosome counting and G-banding as described (32). For fluorescence in situ hybridization, a mouse chromosome 11 painting probe (Oncor) was hybridized to metaphase spreads, as described (33, 34).
Proliferation Assay.
ES cells from subconfluent cultures were seeded at 104 (see Fig. 2A) or 3 × 103 (see Fig. 6B) cells per well in 96-well gelatin-coated plates. Growth medium was replaced every 24 h, and [3H]thymidine (ICN Biomedicals, 2.5 μCi/ml) was added during the last 24 h of culture.
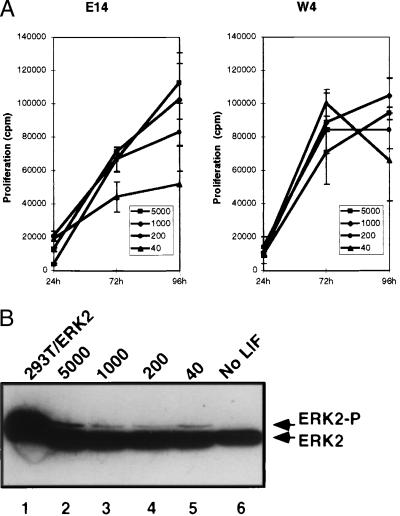
Figure 2.
LIF concentration has minimal effect on ES cell proliferation rate or MAPK activation. (A) Effect of LIF concentration on cell proliferation. [3H]Thymidine incorporation (mean and SD of triplicate samples) was measured during the final 24 h of growth of E14 (Left) and W4 cells (Right) harvested after 24, 72, and 96 h of culture. (B) Immunoblot of phosphorylation-dependent ERK2 migration shift (ERK2-P) in W4 cells cultured at different concentrations of LIF (lanes 2–5) or maintained in the absence of LIF for 12 h (lane 6). W4 cell lysate (120 μg, lanes 2–6) and 20 μg of total cell lysate from 293T cells cotransfected with the epidermal growth factor receptor and ERK2 (lane 1) were immunoblotted for ERK2.
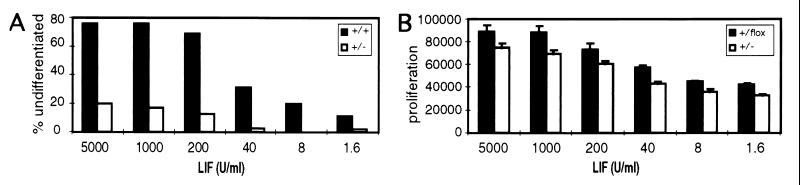
Figure 6.
STAT3 hemizygous cells are impaired for undifferentiated clonal growth. (A) Efficiency of clonal stem cell growth of wild-type E14 cells (filled bars) or Stat3+/− hemizygous cells (empty bars). The proportion of alkaline phosphatase-positive (undifferentiated) colonies was measured after 6 days of growth at the indicated concentrations of LIF (units/ml). Data are average of duplicate assays. (B) Dose-response effect of LIF on Stat3+/flox (filled bars) or Stat3+/− (empty bars) cell proliferation. [3H]Thymidine incorporation (cpm, mean and SD of triplicate samples) was measured on 4-day cultures.
Electrophoretic Mobility Shift Assays.
Nuclear extracts were prepared from 2–4 × 106 cells as described (35) except cells were lysed in 0.25% Nonidet P-40. Gel mobility shift assays were performed as described by using 5 μg of protein and a labeled, double-stranded oligonucleotide containing a high-affinity STAT binding site (36).
Protein Extracts and Immunoreactions.
Immunoprecipitation and Western blot analysis were performed by standard procedures (37). The antibodies used were: anti-p91C against STAT1 (C. Schindler, Columbia University, New York), LI-18 against Jak1 (M. Seidel, Ligand Pharmaceuticals, La Jolla, CA), ST3–5G7 against STAT3 (Zymed), anti-ERK2 C14-G (Santa Cruz Biotechnology), and antiphosphotyrosine antibodies 4G10 and PY20 (Upstate Biotechnology, Lake Placid, NY and Transduction Laboratories, Lexington, KY, respectively). Equal loading of total cell lysates was checked by Ponceau S staining of the filter before antibody probing.
RESULTS
Various ES Cell Lines Exhibit Differing LIF Dependency.
Previous studies have shown constitutive activation of STAT3 in ES cells cultured in the presence of LIF (38), suggesting a role for STAT3 in LIF signaling. We surveyed different ES cell lines for LIF dependence and STAT3 activity. E14, J1, R1, and W4 cells that had been cultured in 1,000 units/ml of LIF were plated at single-cell density in media containing varying concentrations of LIF, and stem cell colonies were identified morphologically after 7 days. E14 showed higher undifferentiated cell plating efficiency than W4 under normal culture conditions (1,000 units/ml) and up to 40% higher efficiency at the lowest concentration of LIF tested (Fig. 1A). R1 and J1 cells exhibited an intermediate efficiency (not shown). Although all ES lines tested depended on LIF concentration for plating efficiency, E14 exhibited the lowest LIF dependency.
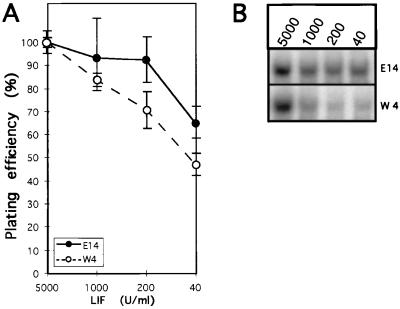
Figure 1.
Levels of nuclear STAT3 activity correlate with the efficiency of stem cell colony formation. (A) Differentiation inhibiting activity of LIF on ES cell lines E14 and W4. Number of undifferentiated colonies (mean and range of duplicate cultures) was normalized for each cell line to that observed at the highest LIF concentration (5,000 units/ml). At 5,000 units LIF/ml, the proportion of undifferentiated over total number of colonies was approximately 80%. (B) Electrophoretic mobility shift assay analysis of nuclear STAT3 binding activity in E14 and W4 cells grown for 7 days at the indicated concentrations of LIF.
We next measured the level of nuclear STAT3 binding activity present in E14 and W4 cells cultured for 7 days at different concentrations of LIF. LIF deprivation caused a reduction of nuclear STAT3 activity in both cases (Fig. 1B); however, each line exhibited a different degree of LIF dependency. At low LIF concentrations, W4 cells contained less than half the STAT3 activity detected in E14, similar to the decreased plating efficiency of this cell line under similar culture conditions. J1 and R1 cells showed 80% and 70%, respectively, of the level of activated STAT3 detected in E14 cells (not shown). These results showed a correlation in ES cell lines between the maintenance of their pluripotency, as measured by their ability to form stem cell colonies at low density, and the stability of nuclear STAT3 levels in response to different concentrations of LIF.
STAT3-Mediated Signaling Is Not Associated with ES Cell Proliferation.
The previous finding suggested that STAT3 plays a critical role in ES cell growth or survival. We tested its role in growth by measuring the proliferation of E14 and W4 cells growing at different LIF concentrations over a period of 4 days. W4 cells displayed a higher proliferation rate than E14 and started to arrest after 3 days, probably because of high cell density (Fig. 2A). E14 cell proliferation correlated somewhat with the concentration of added LIF, although there was only a 2-fold difference in proliferation between the highest and lowest LIF concentrations. In contrast, no significant differences in growth rate were detected for W4 at different LIF concentrations, despite its showing the highest LIF dependency for STAT3 activation and pluripotency.
LIF has been reported to activate the extracellular signal-regulated kinase (ERK) pathway in different cell types, including ES cells (10), which often is associated with cell proliferation (39). We examined activation of the ERK pathway as a function of LIF growth conditions. Total cell extracts prepared from W4 cells cultured at different concentrations of LIF were analyzed by using an anti-ERK2 antibody. No activated ERK2 was detected in cells maintained in the absence of LIF for 12 h (Fig. 2B, lane 6). However, the amount of activated ERK2 present in growing cells was independent of the concentration of LIF in the media (Fig. 2B, lanes 2–5). These results showed that the dose of LIF did not have an immediate impact on cell proliferation rate or ERK signaling, implying that the critical parameter mediated by STAT3 during stem cell growth is not cell cycle progression.
Only E14 Withstand Inactivation of One Stat3 Allele.
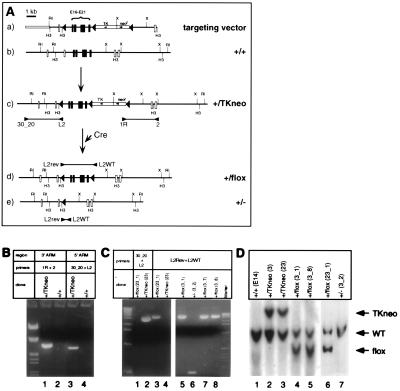
To investigate the requirement of STAT3 for ES cell self-renewal we adopted a conditional mutagenesis approach that would allow modification of Stat3 gene copy number. A replacement targeting vector was designed to introduce a loxP site into intron 15 of the Stat3 gene by homologous recombination and a selectable marker (TK-neor) flanked by loxP sites into intron 21 (Fig. 3Aa). Because the insertion of strong transcription units into introns often results in partial or complete loss of gene expression (40), homologous recombinants would be expected to exhibit lower levels of STAT3 protein. The targeting construct was designed so that the mutation could be either reversed by Cre-mediated excision of the TK-neor cassette (Fig. 3Ad), or completed by Cre-dependent removal of exons 16–21 (Fig. 3Ae). Because exons 19–21 encode the essential Src homology 2 domain of STAT3, their removal would result in the production of an inactive protein.
Figure 3.
Targeted conditional mutation of the Stat3 gene. (A) Diagram of the two-step strategy to generate a Cre-loxP conditional mutation in the Stat3 gene. (a) Map of the targeting construct including two contiguous fragments of the Stat3 gene used as recombination arms, neomycin resistance (neor), and thymidine kinase (TK) cassettes flanked by loxP sequences (solid triangles), and an additional loxP site flanking the exons to be excised (exons 16–21, solid boxes). (b) Map of an 18.5-kb genomic clone for STAT3 containing exons 12–24 (boxes). (c) Partial map of the Stat3 locus after insertion of the targeting vector. Diagnostic PCR products spanning the recombination points are indicated. (d) Partial map of a floxed Stat3 gene (conditional mutant). (e) Partial map of an inactive Stat3 gene after Cre-mediated recombination. H3, HindIII; RI, EcoRI; X, XbaI; E, exon. (B) PCR analysis of genomic DNA from a representative targeted clone (+/TKneo, lanes 1 and 3) compared with wild-type cells (+/+, lanes 2 and 4). (C) PCR analysis of Cre-recombined clones (+/flox, lanes 3, 5, 7, and 8; +/−, lane 6). Clonal purity was verified by the absence of PCR products with the primer pair 30_20-L2 (lane 1). (D) Southern blot analysis of HindIII-digested genomic DNA from wild-type cells (lane 1), targeted clones (lanes 2 and 3) and Cre-recombined clones (lanes 4–7, probed with an 840-bp Stat3 cDNA) fragment encompassing exons 15–22.
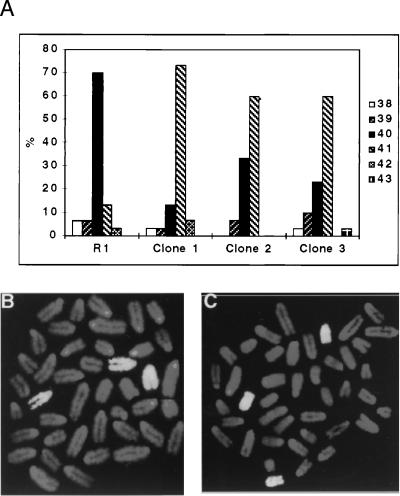
Four ES cell lines were analyzed by Stat3 gene targeting experiments (W4, R1, J1, and E14, Table 1). After electroporation with the targeting vector, 400–800 G418-resistant colonies of each cell line were screened in a total of eight experiments. Depending on the cell line and experiment, recombination at the Stat3 locus occurred with a frequency of 1.5–42%. Both 5′ and 3′ integration junctions were analyzed to identify clones that had integrated correctly the targeting vector and that had incorporated the loxP site distal to the selection cassette (Fig. 3Ac), which occurred in 5–10% of the homologous recombinants. We analyzed cytogenetically 55 targeted clones from the four cell lines (irrespective of the presence of the distal loxP site) and found that none of the W4, R1, or J1 clones tested was euploid (Table 1). Although up to 80% of the cells used for the electroporation displayed a normal karyotype (40 chromosomes) before transfection, the modal chromosome number of targeted W4, R1, and J1 clones was 41–43, with a minority of euploid metaphases (Fig. 4A).
Table 1.
Summary of gene targeting efficiencies in different ES cell lines
| ESC line | Integration frequency* | No. clones analyzed | Homologous recombinants† | Euploid recombinants‡ |
|---|---|---|---|---|
| W4 | 250 | 716 | 1/26 | 0 (19) |
| R1 | 400 | 817 | 1/63 | 0 (12) |
| J1 | 1,500 | 444 | 1/3.2 | 0 (14) |
| E14 | 1,500 | 672 | 1/2.4 | 42 (12) |
G418r clones/107 cells.
Frequency of integration events at the Stat3 locus among G418r colonies.
Percentage of euploid targeted clones and the number assayed.
Figure 4.
Targeted clones are aneuploid and carry trisomy 11. (A) Karyotype of parental ES cells and representative Stat3-targeted clones (as percentage of 30 metaphase spreads). (B and C) Fluorescence in situ hybridization painting of chromosome 11 on metaphase spreads of representative aneuploid, Stat3-targeted R1 (B) and W4 (C) clones shows selective chromosomal duplication.
Preliminary G-banding analysis of targeted clones indicated that chromosome 11 was consistently duplicated (not shown). Fluorescent in situ hybridization carried out on metaphase chromosomes of representative clones, using a probe specific for mouse chromosome 11, confirmed that targeted cells with a single disrupted Stat3 allele carried trisomy 11 (Fig. 4 B and C). Because the Stat3 gene is located on chromosome 11 (41, 42), trisomy 11 might be directly related to Stat3 gene disruption. Consistent with this notion, trisomic clones with a targeted Stat 3 allele did not have reduced levels of nuclear STAT3 activity compared with euploid cells (not shown), suggesting that the maintenance of a critical level of STAT3 was essential for ES cell self-renewal.
In contrast to the results obtained with W4, R1, and J1 cells, almost half of the E14 targeted clones retained a normal, euploid karyotype (Table 1). E14 thus seems to be not only less dependent on LIF concentration for activation of the STAT3 pathway and for growth at clonal density, but also capable of withstanding reduction of Stat3 gene dosage, which may reflect their ability to activate higher levels of STAT3 protein at the conditions of LIF used for routine culture and targeting (1,000 units/ml LIF, see Fig. 1B).
Expression of STAT3 Correlates with Gene Copy Number and Affects ES Cell Clonal Growth.
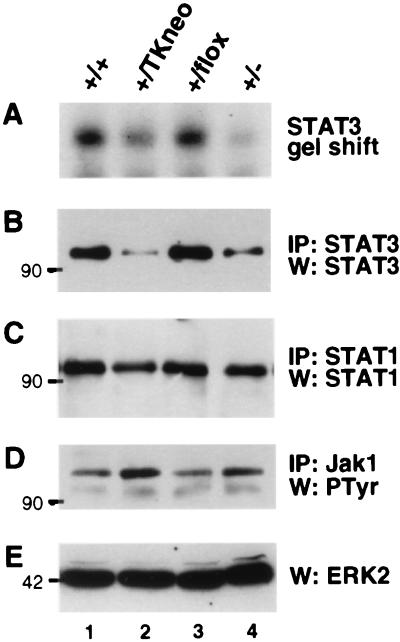
We took advantage of the ability of E14 to yield heterozygous Stat3 conditionally targeted cells to explore the phenotypic effect of reduced Stat3 gene dosage. Four targeted E14 clones (+/TKneo) were examined in which insertion of the TK-neor cassette reduced Stat3 gene expression. These clones were transiently transfected with a Cre recombinase expression vector, and isolated progeny were analyzed for the type of recombination that had occurred (Fig. 3 A d and e, C, and D). A panel of clones of the same lineage, having different modifications in one of the Stat3 alleles, was characterized for the effect of the mutation on LIF signaling. The amount of nuclear-activated STAT3 was reduced approximately by half in the Stat3+/− cells (Fig. 5 A and B, lane 4), as expected given the nature of the mutation. Insertion of the TK-neor cassette also resulted in impaired expression from the affected allele because the level of activated and total STAT3 protein was also significantly reduced (Fig. 5 A and B, lane 2). In contrast, the presence of single loxP sites in introns 15 and 21 had no significant effect on expression (Fig. 5 A and B, lane 3). The amount of STAT3 protein in total cell extracts, as assessed by immunoprecipitation with a STAT3 specific antibody, paralleled that of its nuclear activity (Fig. 5 A and B). However, LIF-dependent activation of Jak1 was unaffected by the genotype at the Stat3 locus, indicating that deficient signaling from the LIF receptor was not the cause of reduced activation of STAT3 (Fig. 5D). To test the specificity of the mutation, we examined the expression of STAT1, a factor also activated in ES cells in response to LIF (28). The levels of STAT1, as determined by immunoprecipitation, were not affected by any of the mutations (Fig. 5C). Reduction in STAT3 levels caused by the genomic mutations also had no effect on ERK activation, as revealed by immunoblot detection of ERK2 (Fig. 5E).
Figure 5.
Targeted mutation of the Stat3 gene reduces STAT3 levels. Extracts from wild type and the indicated E14-derived mutant ES cell clones were analyzed. (A) Electrophoretic mobility shift assay analysis nuclear STAT3. (B) STAT3 immunoblot after immunoprecipitation from 2 mg of whole-cell extract. (C) Supernatants from the STAT3 immunoprecipitation, blotted for STAT1 after immunoprecipitation with an anti-STAT1 antibody. (D) Antiphosphotyrosine blot of Jak1 immunoprecipitated from 2 mg of total cell extract. (E) ERK2 immunoblot showing phosphorylation-dependent ERK2 migration shift. Lines 3_1 and 3_2 (lanes 3 and 4, respectively) were subcloned in parallel from E14-derived line 3 (lane 2) after transient expression of Cre.
Because our previous observations suggested that STAT3 plays an essential role in the maintenance of a pluripotential stem cell phenotype in ES cells, we tested the ability of Stat3+/− mutants to form stem cell colonies in a plating efficiency assay. Heterozygous cells, which expressed about half the amount of STAT3 present in wild-type ES cells (Fig. 5 A and B), were impaired in clonal growth at all concentrations of LIF tested (Fig. 6A). At the highest concentration of LIF (5,000 units/ml), the number of Stat3+/− colonies was approximately 25% of that observed with wild-type cells. Although significant numbers of wild-type colonies survived at low concentrations of LIF (1.6–40 units/ml), virtually no undifferentiated heterozygous colonies were detected. To analyze the effect of STAT3 dose on cell proliferation, we measured the amount of [3H]thymidine incorporated by Stat3+/− and Stat3+/flox cells cultured for 4 days at different concentrations of LIF. In contrast to their drastically reduced ability for undifferentiated clonal growth, Stat3+/− cells proliferate at a rate only slightly lower (Fig. 6B) than that of Stat3+/flox cells expressing normal levels of STAT3 (Fig. 5 A and B, lane 3). [3H]Thymidine incorporation by Stat3+/− cells was 15–25% lower than controls at any concentration of LIF tested. This difference may be the result of a reduced number of viable Stat3+/− cells initially plated rather than of any intrinsic proliferative deficiency in Stat3+/− cells, given that propidium iodide flow cytometry analysis revealed no differences in cell cycle progression between mutant Stat3+/− cells and Stat3+/flox or wild-type E14 cells at any concentration of LIF in the range of 1,000 to 1.6 units/ml (data not shown).
DISCUSSION
Perhaps the most interesting function that LIF exhibits in vitro is the maintenance of ES cells in an undifferentiated, pluripotent state. Although this property has been widely exploited for the introduction of targeted mutations into ES cells and for the derivation of the corresponding mutant mice, very little is known about the underlying molecular mechanisms. LIF originally was discovered as a cytokine capable of inducing terminal differentiation and growth arrest of mouse leukemia cells, which might be considered the opposite function. Further, LIF and related IL-6-type cytokines are mitogenic for some cell types. This study demonstrates that STAT3, a major target of LIF, is essential in ES cells. A critical level of STAT3 is required for the maintenance of a pluripotent stem phenotype that is not required directly for mitogenesis.
ES cells subjected to Stat3 gene targeting experiments and heterozygous for an inactivating mutation of Stat3 were impaired in their ability to grow at clonal density. This behavior was observed in ES cell lines derived from various 129 mouse substrains, suggesting a general requirement for STAT3. Failure to recover targeted, euploid cell lines was not caused by a defect in gene replacement because insertion of the inactivating sequences (TK-neor cassette) in the Stat3 locus was obtained at moderate to high frequencies in all cell lines tested. Instead, the targeted clones invariably carried additional chromosomes. Cytogenetic analysis of representative clones revealed that chromosome 11, carrying the Stat3 gene, was the only duplicated chromosome common among the aneuploid clones. It is known that in vitro cultivation of ES cells can lead to accumulation of chromosome aberrations (43, 44); however, general chromosomal instability was not a likely cause for the increased frequency of trisomy 11 because all of the untargeted clones isolated in parallel had normal complements of 40 chromosomes (not shown). The association between disruption of the Stat3 gene and trisomy 11, along with the fact that the chromosomal instability inherent to ES cells in culture does not lead to biased karyotypic patterns (45), strongly indicate that the selective duplication of chromosome 11 conferred a selective advantage to ES cells sustaining Stat3 mutations. Taken together, these results suggest that maintenance of a normal number of functional Stat3 copies was the driving force behind the selection of cells carrying duplicated chromosome 11. The genes for LIF and oncostatin M also are located on the same chromosome (46, 47), and their amplification may exert additional selective pressure for trisomy 11.
E14, an ES cell previously used to generate Stat3-deficient mice (25), was unique among the lines analyzed in that it yielded normal, euploid clones hemizygous for the targeted disruption of Stat3. This observation indicates that E14 cells are more refractile to reduced Stat3 gene copy number, although not totally insensitive because more than half of the targeted clones carried trisomy 11. E14 cells also exhibited a more active STAT3 pathway, especially at low concentrations of LIF (Fig. 1B). Although the mechanism underlying the increase in activated STAT3 protein remains to be determined, it is clear that E14 cells are more sensitive to cytokine stimulation. It is likely that the higher STAT3 activity present in E14 cells made them less sensitive to reduction of Stat3 gene dosage, indicating that the requirement of LIF for clonal growth is mediated by a threshold level of nuclear STAT3. Gene targeting in lines other than E14 presumably brings STAT3 expression below the level required for ES cell survival.
The importance of STAT3 for ES cell pluripotency has been recently analyzed through the use of dominant negative STAT3 isoforms. ES cell transfectants overexpressing a STAT3-Y705F mutant exhibited a tendency toward differentiation after 1 month in culture (18), indicating a role for STAT3 function in maintaining an undifferentiated state. In a second study, episomal supertransfection or inducible overexpression of the same dominant negative STAT3 abrogated self-renewal and promoted differentiation of ES cells growing in LIF (17). Our results are consistent with the conclusion that inactivation of STAT3 function has a dramatic and rapid effect on ES cell differentiation. In addition, they reveal a critical role for STAT3 dosage in its biologic effects, because partial reduction of active STAT3 protein by LIF withdrawal or reduction of expression by half through gene targeting were detrimental to ES cell self-renewal.
Activation of the Ras-mitogen-activated protein kinase (MAPK) pathway by gp130 occurs in parallel with activation of the JAK-STAT pathway (10). Some evidence points to a role for the Ras-MAPK pathway transmitting mitogenic signals (48, 49). Although it has been suggested that the Ras-MAPK pathway has a dual role in ES cells, affecting proliferation as well as inhibiting differentiation (50), our data suggest that activation of this pathway alone is not sufficient for suppression of ES cell differentiation. Of the three parameters we measured in mass culture (nuclear STAT3 activation, proliferation, and MAPK activation), only STAT3 activity was linked to LIF-dependent changes self-renewal at clonal density. Together with the observation that a critical level of STAT3 was essential for ES cells in gene targeting or dominant negative experiments, we suggest that LIF inhibits differentiation pathways in ES cells through STAT3, whereas proliferation rate and MAPK activation are less sensitive to the abundance of STAT3.
ES cell lines hemizygous for a conditional mutation of Stat3 will provide a key reagent to further address signaling pathways and gene expression required for ES cell self-renewal. Disruption of Stat3 in vivo causes an early, postimplantation embryonic lethality (25), although the cause of lethality has not been determined. Maternal expression of LIF is required for fetal viability and may act at least in part on the embryo itself (51, 52), although its exact requirement is unclear (53). However, absence of maternal LIF causes lethality at an earlier stage than does loss of Stat3 (52), whereas disruption of the LIF receptor (54) or gp130 (55) results in lethality at a considerably later stage of development. STAT3 protein is known to be active during early postimplantation development, particularly in extra-embryonic tissue (56), suggesting roles for this transcription factor in addition to maintaining inner cell mass cell pluripotency. It will be important to determine the requirements of STAT3 during early embryogenesis as well as during later development, and mutant animals generated from ES cells carrying conditionally mutant Stat3 alleles will be instrumental in this process.
Acknowledgments
We thank J. Leiden, C. Simon, A. Auerbach, W. Auerbach, S. Ortega, M. Lafaille, and J. Benjamin for ES cells, plasmid constructs, and expert advice. We are grateful to H. Gu and P. Orban for the gift of plasmids, L.-T. Yang, J. Sap, C. Schindler, M. Seidel, and S. Guadagno for antibodies, J. Wider for image analysis, J. Hirst for flow cytometry and M. Zohouri, R. Gertner, and L. Pan for excellent technical assistance. This work was supported by grants from the National Institutes of Health (AI28900 and CA09161).
ABBREVIATIONS
- ES
embryonic stem
- LIF
leukemia inhibitory factor
- STAT
signal transducer and activator of transcription
- ERK
extracellular signal-regulated kinase
- MAPK
mitogen-activated protein kinase
- TK
thymidine kinase
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Rossant J, Papaioannou V E. Cell Differ. 1984;15:155–161. doi: 10.1016/0045-6039(84)90068-x. [DOI] [PubMed] [Google Scholar]
- 2.Evans M J, Kaufman M H. Nature (London) 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 3.Martin G R. Proc Natl Acad Sci USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawase E, Suemori H, Takahashi N, Okazaki K, Hashimoto K, Nakatsuji N. Int J Dev Biol. 1994;38:385–390. [PubMed] [Google Scholar]
- 5.Ichikawa Y. J Cell Physiol. 1970;76:175–184. doi: 10.1002/jcp.1040760207. [DOI] [PubMed] [Google Scholar]
- 6.Smith A G, Nichols J, Robertson M, Rathjen P D. Dev Biol. 1992;151:339–351. doi: 10.1016/0012-1606(92)90174-f. [DOI] [PubMed] [Google Scholar]
- 7.Williams R L, Hilton D J, Pease S, Willson T A, Stewart C L, Gearing D P, Wagner E F, Metcalf D, Nicola N A, Gough N M. Nature (London) 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 8.Moreau J F, Donaldson D D, Bennett F, Witek-Giannotti J, Clark S C, Wong G G. Nature (London) 1988;336:690–692. doi: 10.1038/336690a0. [DOI] [PubMed] [Google Scholar]
- 9.Taga T, Kishimoto T. Annu Rev Immunol. 1997;15:797–819. doi: 10.1146/annurev.immunol.15.1.797. [DOI] [PubMed] [Google Scholar]
- 10.Hirano T, Nakajima K, Hibi M. Cytokine Growth Factor Rev. 1997;8:241–252. doi: 10.1016/s1359-6101(98)80005-1. [DOI] [PubMed] [Google Scholar]
- 11.Darnell J E. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 12.Raz R, Durbin J E, Levy D E. J Biol Chem. 1994;269:24391–24395. [PubMed] [Google Scholar]
- 13.Zhong Z, Wen Z, Darnell J E. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 14.Akira S, Nishio Y, Inoue M, Wang X-J, Wei S, Matsusaka T, Yoshida K, Sudo T, Maruto M, Kishimoto T. Cell. 1994;77:63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 15.Lütticken C, Wegenka U M, Yuan J, Buschmann J, Schindler C, Ziemiecki A, Harpur A G, Wilks A F, Yasukawa K, Taga T, et al. Science. 1994;263:89–92. doi: 10.1126/science.8272872. [DOI] [PubMed] [Google Scholar]
- 16.Wegenka U M, Buschmann J, Lütticken C, Heinrich P C, Horn F. Mol Cell Biol. 1993;13:276–288. doi: 10.1128/mcb.13.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niwa H, Burdon T, Chambers I, Smith A. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boeuf H, Hauss C, Graeve F D, Baran N, Kedinger C. J Cell Biol. 1997;138:1207–1217. doi: 10.1083/jcb.138.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minami M, Inoue M, Wei S, Takeda K, Matsumoto M, Kishimoto T, Akira S. Proc Natl Acad Sci USA. 1996;93:3963–3966. doi: 10.1073/pnas.93.9.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakajima K, Yamanaka Y, Nakae K, Kojima H, Ichiba M, Kiuchi N, Kitaoka T, Fukada T, Hibi M, Hirano T. EMBO J. 1996;15:3651–3658. [PMC free article] [PubMed] [Google Scholar]
- 21.Yamanaka Y, Nakajima K, Fukada T, Hibi M, Hirano T. EMBO J. 1996;15:1557–1565. [PMC free article] [PubMed] [Google Scholar]
- 22.Cressman D E, Greenbaum L E, DeAngelis R A, Ciliberto G, Furth E E, Poli V, Taub R. Science. 1996;274:1379–1383. doi: 10.1126/science.274.5291.1379. [DOI] [PubMed] [Google Scholar]
- 23.Sierra-Honigmann M R, Nath A K, Murakami C, García-Cardeña G, Papapetropoulos A, Sessa W C, Madge L A, Schechner J S, Schwabb M B, Polverini P J, Flores-Riveros J R. Science. 1998;281:1683–1686. doi: 10.1126/science.281.5383.1683. [DOI] [PubMed] [Google Scholar]
- 24.Vaisse C, Halaas J L, Horvath C M, Darnell J E, Jr, Stoffel M, Friedman J M. Nat Genet. 1996;14:95–97. doi: 10.1038/ng0996-95. [DOI] [PubMed] [Google Scholar]
- 25.Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. Proc Natl Acad Sci USA. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tybulewicz V J, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 27.Wurst W, Joyner A L. In: Production of Targeted Embryonic Stem Cell Clones. Joyner A L, editor. Oxford: Oxford Univ. Press; 1993. pp. 33–61. [Google Scholar]
- 28.Durbin J E, Hackenmiller R, Simon M C, Levy D E. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 29.Hooper M, Hardy K, Handyside A, Hunter S, Monk M. Nature (London) 1987;326:292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- 30.Li E, Bestor T H, Jaenisch R. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 31.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder J C. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 33.Cannizzaro L A, Shi G. Methods Mol Biol. 1997;75:313–322. doi: 10.1385/0-89603-441-0:313. [DOI] [PubMed] [Google Scholar]
- 34.Shi G, Cannizzaro L A. Cytogenet Cell Genet. 1996;75:180–185. doi: 10.1159/000134473. [DOI] [PubMed] [Google Scholar]
- 35.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silvennoinen O, Witthuhn B A, Quelle F W, Cleveland J L, Yi T, Ihle J N. Proc Natl Acad Sci USA. 1993;90:8429–8433. doi: 10.1073/pnas.90.18.8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silvennoinen O, Schindler C, Schlessinger J, Levy D E. Science. 1993;261:1736–1739. doi: 10.1126/science.8378775. [DOI] [PubMed] [Google Scholar]
- 38.Hocke G M, Cui M Z, Fey G H. Cytokine. 1995;7:491–502. doi: 10.1006/cyto.1995.0067. [DOI] [PubMed] [Google Scholar]
- 39.Seger R, Krebs E G. FASEB J. 1995;9:726–735. [PubMed] [Google Scholar]
- 40.Hartung S, Jaenisch R, Breindl M. Nature (London) 1986;320:365–367. doi: 10.1038/320365a0. [DOI] [PubMed] [Google Scholar]
- 41.Copeland N G, Gilbert D J, Schindler C, Zhong Z, Wen Z, Darnell J E, Jr, Mui A L, Miyajima A, Quelle F W, Ihle J N, Jenkins N A. Genomics. 1995;29:225–228. doi: 10.1006/geno.1995.1235. [DOI] [PubMed] [Google Scholar]
- 42.Shi W, Inoue M, Minami M, Takeda K, Matsumoto M, Matsuda Y, Kishimoto T, Akira S. Int Immunol. 1996;8:1205–1211. doi: 10.1093/intimm/8.8.1205. [DOI] [PubMed] [Google Scholar]
- 43.Robertson E J. In: Embryo-Derived Stem Cell Lines. Robertson E J, editor. Oxford: IRL; 1987. pp. 71–112. [Google Scholar]
- 44.Brown D G, Willington M A, Findlay I, Muggleton-Harris A L. In Vitro Cell Dev Biol. 1992;28A:773–778. doi: 10.1007/BF02631066. [DOI] [PubMed] [Google Scholar]
- 45.Longo L, Bygrave A, Grosveld F G, Pandolfi P P. Transgenic Res. 1997;6:321–328. doi: 10.1023/a:1018418914106. [DOI] [PubMed] [Google Scholar]
- 46.Kola I, Davey A, Gough N M. Growth Factors. 1990;2:235–240. doi: 10.3109/08977199009071509. [DOI] [PubMed] [Google Scholar]
- 47.Yoshimura A, Ichihara M, Kinjyo I, Moriyama M, Copeland N G, Gilbert D J, Jenins N A, Hara T, Miyajima A. EMBO J. 1996;15:1055–1063. [PMC free article] [PubMed] [Google Scholar]
- 48.Fukada T, Hibi M, Yamanaka Y, Takahashi-Tezuka M, Fujitani Y, Yamaguchi T, Nakajima K, Hirano T. Immunity. 1996;5:449–460. doi: 10.1016/s1074-7613(00)80501-4. [DOI] [PubMed] [Google Scholar]
- 49.Billadeau D, Jelinek D F, Shah N, LeBien T W, Van Ness B. Cancer Res. 1995;55:3640–3646. [PubMed] [Google Scholar]
- 50.Ernst M, Oates A, Dunn A R. J Biol Chem. 1996;271:30136–30143. doi: 10.1074/jbc.271.47.30136. [DOI] [PubMed] [Google Scholar]
- 51.Stewart C L, Cullinan E B. Dev Genet. 1997;21:91–101. doi: 10.1002/(SICI)1520-6408(1997)21:1<91::AID-DVG11>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 52.Stewart C L, Kaspar P, Brunet L J, Bhatt H, Gadi I, Kontgen F, Abbondanzo S J. Nature (London) 1992;359:76–79. doi: 10.1038/359076a0. [DOI] [PubMed] [Google Scholar]
- 53.Cheng L, Gearing D P, White L S, Compton D L, Schooley K, Donovan P J. Development (Cambridge, UK) 1994;120:3145–3153. doi: 10.1242/dev.120.11.3145. [DOI] [PubMed] [Google Scholar]
- 54.Ware C B, Horowitz M C, Renshaw B R, Hunt J S, Liggitt D, Koblar S A, Gliniak B C, McKenna H J, Papayannopoulou T, Thoma B, et al. Development (Cambridge, UK) 1995;121:1283–1299. doi: 10.1242/dev.121.5.1283. [DOI] [PubMed] [Google Scholar]
- 55.Yoshida K, Taga T, Saito M, Suematsu S, Kumanogoh A, Tanaka T, Fujiwara H, Hirata M, Yamagami T, Nakahata T, et al. Proc Natl Acad Sci USA. 1996;93:407–411. doi: 10.1073/pnas.93.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duncan S A, Zhong Z, Wen Z, Darnell J E. Dev Dyn. 1997;208:190–198. doi: 10.1002/(SICI)1097-0177(199702)208:2<190::AID-AJA6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]