Abstract
Loss of pollen-S function in Prunus self-compatible mutants has recently been associated with deletions or insertions in S-haplotype-specific F-box (SFB) genes. We have studied two self-compatible cultivars of apricot (Prunus armeniaca), Currot (SCSC) and Canino (S2SC), sharing the naturally occurring self-compatible (SC)-haplotype. Sequence analysis showed that whereas the SC-RNase is unaltered, a 358-bp insertion is found in the SFBC gene, resulting in the expression of a truncated protein. The alteration of this gene is associated with self-incompatibility (SI) breakdown, supporting previous evidence that points to SFB being the pollen-S gene of the Prunus SI S-locus. On the other hand, PCR analysis of progenies derived from Canino showed that pollen grains carrying the S2-haplotype were also able to overcome the incompatibility barrier. However, alterations in the SFB2 gene or evidence of pollen-S duplications were not detected. A new class of F-box genes encoding a previously uncharacterized protein with high sequence similarity (approximately 62%) to Prunus SFB proteins was identified in this work, but the available data rules them out of producing S-heteroallelic pollen and thus the cause of the pollen-part mutation. These results suggest that cv Canino has an additional mutation, not linked to the S-locus, which causes a loss of pollen-S activity when present in pollen. As a whole, these findings support the proposal that the S-locus products besides other S-locus independent factors are required for gametophytic SI in Prunus.
Gametophytic self-incompatibility (GSI) is a widespread mechanism in flowering plants often controlled by a single multiallelic locus, termed the S-locus, that prevents inbreeding and promotes out-crossing (de Nettancourt, 2001). In the Solanaceae, Scrophulariaceae, and Rosaceae, the S-locus is considered to contain at least two linked genes; one encodes glycoproteins with ribonuclease (S-RNase) activity in the pistils (McClure et al., 1989; Boskovic and Tobutt, 1996; Xue et al., 1996) and the other is an F-box pollen-expressed gene (named SLF or S-haplotype-specific F-box [SFB]; Lai et al., 2002; Entani et al., 2003; Ushijima et al., 2003; Sijacic et al., 2004).
In the Solanaceae, Scrophulariaceae, and Rosaceae, active S-RNases expressed in the style are essential for rejection of haploid pollen when the S-allele of pollen matches either S-allele of the diploid pistil (Huang et al., 1994; Xue et al., 1996; Sassa et al., 1997). The recent identification of S-linked F-box genes as the pollen-S determinants of the GSI system in Antirrhinum (Lai et al., 2002), Prunus (Entani et al., 2003; Ushijima et al., 2003), and Petunia (Sijacic et al., 2004) has thrown some light on the underlying mechanism. F-box proteins are involved in the ubiquitin/26S proteasome proteolytic pathway (Deshaies, 1999), suggesting that the function of S-linked F-box genes might be to inactivate nonself S-RNases by proteolytic degradation following their entry into the pollen tube (Ushijima et al., 2003; Ikeda et al., 2004). Consistent with this hypothesis is the finding that the Antirrhinum pollen-expressed F-box protein AhSLF2 interacts with both self and nonself S-RNases (Qiao et al., 2004). These findings are compatible with the inhibitor model of self-incompatibility (SI), which proposes that all S-RNases, regardless of their S-haplotype, enter into the pollen tube (Luu et al., 2000), and once there all of them are inhibited except the cognate S-RNase that degrades self-pollen RNA. How the cognate S-RNase is specifically protected from degradation remains unknown.
Spontaneous and induced self-compatible mutants have been used extensively to study the molecular basis of the GSI mechanism. Stylar-part mutations in the S-locus have been reported in Solanaceae (Royo et al., 1994) and Rosaceae (Sassa et al., 1997), revealing that RNase activity of S-RNases is needed to inhibit pollen growth. Numerous pollen-part mutants (PPMs) have been described in the Solanaceae, and most of them are consistent with competitive interaction in which S-heteroallelic pollen, containing two different S-alleles, fails to function in SI (Thompson et al., 1991; Golz et al., 1999, 2001). In Prunus, the recent analysis of self-compatible mutant haplotypes has shown that SFB genes are defective, providing additional evidence that SFBs are the pollen-S genes in GSI in Prunus (Ushijima et al., 2004; Sonneveld et al., 2005) and suggesting that mutations in or deletions of SFBs, as well as S-allele duplications, can cause pollen function breakdown in the Rosaceae. Finally, breakdown of SI has also been associated with mutations that may affect additional S-locus external factors in sweet cherries (Prunus avium; Wünsch and Hormaza, 2004).
The degree of natural self-fertility varies between Prunus species. Peaches (Prunus persica) are normally self-pollinated, apricots (Prunus armeniaca) partly self-compatible, and almonds (Prunus dulcis) and sweet cherries mostly self-incompatible (Watkins, 1976). Among the apricots, many early Spanish cultivars that are genetically closely related are self-compatible (Burgos et al., 1993). Two of these self-compatible apricots, Currot (SCSC) and Canino (S2SC), were analyzed in this work. We show that loss of pollen-S function in the naturally occurring SC-haplotype is associated with an insertion in SFBC that cosegregates with self-compatibility. In addition, this study provides evidence that the loss of function of an additional factor not linked to the S-locus is also involved in the breakdown of SI in the PPM Canino. Finally, as a result of the pollen-S gene duplication analysis in both cultivars, an F-box gene with high sequence similarity to Prunus SFB proteins that is not tightly linked to the S-locus has been identified in apricot.
RESULTS
Genetic Analysis of the Self-Compatible cv Currot (SCSC)
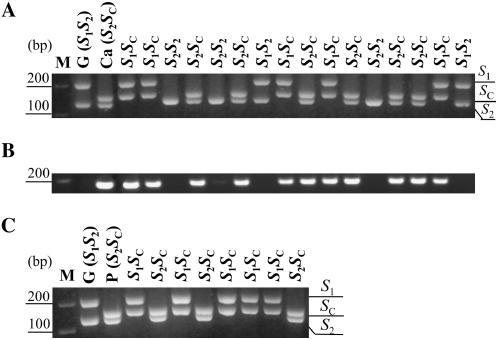
Based on evidence obtained from stylar RNase analysis, PCR, and pollen-tube growth tests on controlled crosses, the self-compatible apricot cv Currot (SCSC) was suggested to be homozygous for the naturally occurring SC-haplotype (Alburquerque et al., 2002; Vilanova et al., 2005). S-RNase PCR-typing of the cross population Goldrich (S1S2) × Currot (SCSC) showed, as expected from the Currot S-genotype, that the SC-RNase is present in all the progeny (Supplemental Fig. S1A) and grouped into two heterozygote classes, S1SC and S2SC, with an observed proportion of 31:39 that fits with the expected segregation ratio 1:1 (Table I). To also analyze this progeny for the SC-RNase pollen counterpart, we cloned and sequenced an SFBC fragment PCR amplified with consensus primers from Currot. Subsequently, we designed specific primers (RFBc-F/RFBc-R) to identify SFBC by PCR. Supplemental Figure S1B shows that SFBC is also present in all the Goldrich × Currot seedlings. The cosegregation of the SC-RNase and SFBC in the Goldrich (S1S2) × Canino (S2SC) population (see Fig. 3, A and B) provided evidence of genetic linkage between the two genes.
Table I.
Segregation of the S-RNase alleles in the progenies of controlled field crosses and self-pollinations
S-genotypes were determined by PCR. Observed S-RNase genotypes, expected segregation ratios, and χ2 values obtained for each population are indicated.
| Seed Parent (S-Genotype) | Pollen Parent (S-Genotype) |
S-Genotypes Observed
|
Total | Expected Segregation Ratio | χ2d (P Value) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| S1SC | S2SC | S1S2 | S2S2 | S1S1 | SCSC | |||||
| Goldrich (S1S2) | Currot (SCSC) | 31 | 39 | – | – | – | – | 70 | 1:1 | 0.91 (0.339) |
| Goldrich (S1S2) | Canino (S2SC) | 66 | 55 | 28 | 22 | – | – | 171 | 2:2:1:1b | 2.98 (0.394) |
| Canino (S2SC) | Canino (S2SC) | – | 53 | – | 11 | – | 35 | 99 | 3:1:2b | 2.20 (0.333) |
| GC-8 (S2SC)a | GC-8 (S2SC) | – | 14 | – | 4 | – | 6 | 24 | 3:1:2b | 0.83 (0.659) |
| GC-10 (S2SC)a | GC-10 (S2SC) | – | 14 | – | – | – | 10 | 24 | 1:1c | 0.67 (0.414) |
| GC-80 (S1SC)a | GC-80 (S1SC) | 15 | – | – | – | 3 | 6 | 24 | 3:1:2b | 1.50 (0.472) |
| GC-86 (S1SC)a | GC-86 (S1SC) | 13 | – | – | – | – | 11 | 24 | 1:1c | 0.17 (0.683) |
Seedlings derived from the cross Goldrich (S1S2) × Canino (S2SC).
Expected ratios for a single mutation unlinked to the S-locus.
Expected ratios for nonmutated GC seedlings.
Observed ratios do not differ significantly from expected at P < 0.05 in any case.
Figure 3.
Segregation analysis of S-alleles of the Goldrich × Canino and Goldrich × Pepito progenies. A, PCR amplification of apricot genomic DNA with consensus primers (SRc-F/SRc-R) for the first S-RNase intron. B, SFBC-specific PCR amplification performed with RFBc-F/RFBc-R primers. Samples in A and B are as follows: (G) Goldrich (S1S2); (Ca) Canino (S2SC); and 16 seedlings derived from the Goldrich × Canino cross. C, S-RNase allele fragments PCR amplified with SRc-F/SRc-R primers in the Goldrich × Pepito progeny. Samples are as follows: (G) Goldrich (S1S2); (P) Pepito (S2SC); and eight seedlings derived from the Goldrich × Pepito cross.
Molecular Analysis of the Apricot SC-Haplotype
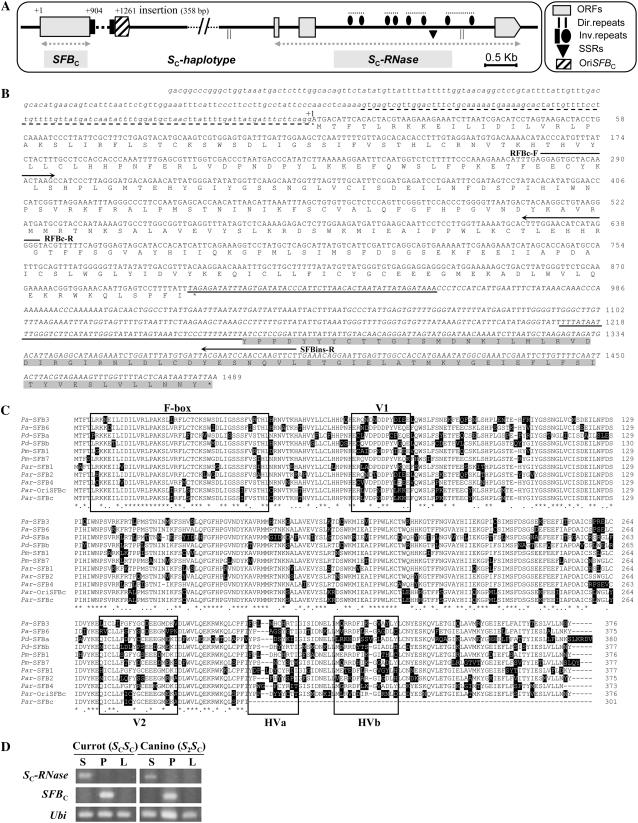
To test whether mutations or indels, affecting the putative SC-RNase or SC-haplotype-specific SFB genes, were the cause of SI breakdown in the SC-haplotype, we cloned and sequenced genomic DNA fragments, containing both genes and their adjacent regions, from the apricot self-compatible cv Currot (SCSC). Figure 1A shows the putative genomic structure of the SC-haplotype. Two fragments (approximately 3.0 and 5.0 kb, respectively) containing open reading frames (ORFs) with homology to the known SFB and S-RNase genes (SFBC and SC-RNase) were identified. Several direct and inverted repeats, along with microsatellites, were found, mainly in the SC-RNase intron. Physical distances between the genes and their relative orientations are not yet definitively established, but PCR segregation analysis suggests that they are genetically linked (see Fig. 3, A and B).
Figure 1.
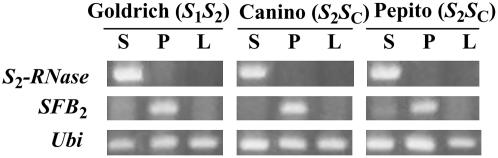
Genetic analysis of the apricot SC-haplotype. A, Genomic structure of the apricot S-locus region in the SC-haplotype. Gray boxes represent ORFs for the S-RNase and SFB alleles. The physical distance between both genes and their relative positions has not yet been established. Direct and inverted repeats and microsatellites identified in the genomic sequences are also indicated. The broken line represents the inserted fragment, black boxes the approximately 52-bp inverted repeats, and lined box the sequence corresponding to the OriSFBC below the insertion. B, Nucleotide and the deduced amino acid sequence of the SFBC allele. The first nucleotide of the translational initiation codon is indicated (+1). The nucleotide sequence indicated by lowercase corresponds to the 5′ flanking region of SFBC. The broken line is underlining the putative intron within the 5′ untranslated region found in Japanese apricot (Ushijima et al., 2004) and sweet cherry (Vaughan et al., 2006). A 358-bp DNA fragment inserted at position 904 to 1,261 produces a premature stop codon in translation. Inverted repeats located at both ends of the insertion are underlined. The nucleotide sequence below the stop codon is in italics. Residues highlighted in gray correspond to the original intact OriSFBC. Arrows indicate positions and directions of primers used in Supplemental Figure S1B, Figures 1D and 3B (RFBc-F/RFBc-R), and Figure 2 (RFBc-F/SFBins-R). C, Alignment of the deduced amino acid sequences of SFBs from apricot Par, sweet cherry Pa, Japanese apricot Pm, and almond Pd. Asterisks indicate conserved sites, dots conservative substitutions, and dashes gaps. Residues different from the consensus are highlighted in black boxes. F-box and (hyper)variable (V1, V2, HVa, and HVb) regions (Ikeda et al., 2004) are boxed. D, RT-PCR analysis of the SC-RNase and SFBC gene expression in pollen (P), styles (S), and leaves (L) of the apricot cultivars Currot (SCSC) and Canino (S2SC). Ubi, Ubiquitin (positive control) genes.
The putative apricot SC-RNase shows the typical features of Prunus T2-type RNases with five conserved domains (C1, C2, C3, RC4, and C5) and one hypervariable region (RHV; Supplemental Fig. S2). The positions of the two SC-RNase introns (260 and 2,680 bp in size, respectively) are also identical to those of the other Prunus S-RNases. Amino acid identity of the SC-RNase to other Prunus S-RNases ranged from 59.7% to 80.0%, preserving the typical high allelic sequence diversity of the S-RNases (Table II). The primary structure of the encoded apricot SC-RNase is intact and, furthermore, motifs surrounding the His residues necessary for the RNase activity in the C2 and C3 domains (Kawata et al., 1989) are present. The only conserved potential N-glycosylation site (NxS/T consensus sequence; Ishimizu et al., 1998) located at the Asn-142 according to the numbering for the apricot S1-RNase (AY587561) was also identified (Supplemental Fig. S2). Finally, reverse transcription (RT)-PCR analysis showed that the SC-RNase is specifically expressed in style tissues but not in pollen or leaves (Fig. 1D).
Table II.
Amino acid sequence identities (%) among Prunus S-locus genes
The upper half shows identities between the S-RNases and the lower half between the SFBs.
| Sweet Cherry
|
Almond
|
Japanese Apricot
|
Apricot
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pa-S3 | Pa-S6 | Pd-Sa | Pd-Sb | Pm-S1 | Pm-S7 | Par-S1 | Par-S2 | Par-S4 | Par-Sc | |
| Pa-S3 | – | 72.8 | 57.7 | 73.8 | 72.2 | 67.3 | 73.1 | 75.6 | 74.6 | 74.1 |
| Pa-S6 | 80.0 | – | 55.8 | 80.0 | 74.3 | 74.4 | 77.4 | 71.8 | 71.6 | 77.9 |
| Pd-Sa | 67.2 | 68.3 | – | 57.7 | 58.3 | 55.9 | 59.1 | 54.9 | 57.9 | 59.7 |
| Pd-Sb | 77.1 | 78.7 | 67.3 | – | 72.5 | 71.7 | 81.8 | 73.8 | 75.1 | 80.0 |
| Pm-S1 | 82.1 | 81.1 | 69.3 | 80.6 | – | 75.7 | 75.0 | 68.8 | 70.9 | 75.8 |
| Pm-S7 | 77.7 | 79.6 | 69.5 | 78.6 | 80.6 | – | 72.1 | 69.0 | 67.7 | 75.5 |
| Par-S1 | 77.1 | 80.6 | 67.6 | 75.3 | 81.1 | 78.6 | – | 77.5 | 73.7 | 77.3 |
| Par-S2 | 78.1 | 79.7 | 67.2 | 79.3 | 82.1 | 79.3 | 79.8 | – | 73.3 | 75.9 |
| Par-S4 | 79.8 | 80.6 | 70.8 | 79.6 | 83.0 | 82.5 | 80.1 | 79.0 | – | 75.5 |
| Par-Sc | 77.1 | 78.9 | 65.1 | 78.7 | 78.9 | 78.0 | 77.7 | 77.1 | 78.5 | – |
Sequence analysis revealed a 358-bp insertion disrupting the putative SFBC ORF at +904. At the ends of the inserted sequence are two approximately 52-bp inverted repeats (approximately 75% nucleotide identity) that are similar to the inverted terminal repeats (ITR) of transposable elements (Fig. 1B). Interestingly, BLASTN analysis (Altschul et al., 1990) revealed significant similarity between the insert and two sequences found in the EMBL/GenBank/DDBJ database, the peach bacterial artificial chromosome (BAC) clone 28F08 (AC154900) and a partial coding sequence of the Prunus salicina gene for Sn-RNase (AB093136; 82% and 77% nucleotide identities, respectively). Furthermore, the insertion also displayed significant similarity (52%) with the SC-RNase 5′ flanking region located between −404 and −740 from the translational start codon. The inserted sequence leads to a premature stop codon at +904 in the SFBC transcript (Fig. 1, A–C). Figure 1B also indicates the position of the putative intron associated with the 5′ untranslated region of Prunus SFB (Vaughan et al., 2006). Therefore, SFBC transcript encodes a putative truncated protein lacking the last 75 amino acid residues of the C-terminal half, including the HVa and HVb hypervariable domains (Fig. 1C). No progenitor allele is available, but the nucleotide sequence predicted for the hypothetical allele, which is supposed to be the original SFBC (OriSFBC, according to the nomenclature established by Ushijima et al. [2004]), lacking the insert, encodes a typical S-locus F-box protein (Ikeda et al., 2004), with one F-box domain and four (hyper)variable regions (V1, V2, HVa, and HVb; Fig. 1C). OriSFBC also shows the typical high sequence diversity to other Prunus SFBs with amino acid identities ranging from 65.1% to 78.9% (Table II). Gene expression analysis performed by RT-PCR showed that SFBC is expressed in pollen tissues but not in styles or leaves (Fig. 1D).
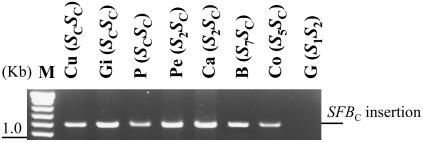
The presence of the fragment inserted in SFBC was confirmed by PCR amplification in all apricot self-compatible cultivars sharing the SC-haplotype used in this study (Fig. 2). Because the SC-RNase sequence is intact and an insertion leading to a premature stop codon in translation is found in the SFBC, the SC-haplotype is believed to be a PPM. As PPMs generated in the Solanaceae are mostly associated with S-locus duplications (Brewbaker and Natarajan, 1960; Golz et al., 1999, 2001), we tested this possibility in Currot using flow cytometry and DNA-blot analysis. In the flow cytometry analysis, the peaks of nuclei isolated from Currot and the control diploid plant (Goldrich) were coincident, indicating that Currot is a diploid (data not shown). In addition, DNA-blot analysis conducted with several restriction enzymes provided no evidence of genetic duplication (see below).
Figure 2.
Genomic PCR amplification of several apricot cultivars for the SFBC insertion. The primers used (RFBc-F/SFBins-R) were designed from the consensus sequence of the Prunus SFB alleles (Romero et al., 2004). Samples are as follows: (Cu) Currot (SCSC), (Gi) Ginesta (SCSC), (P) Palau (SCSC), (Pe) Pepito (S2SC), (Ca) Canino (S2SC), (B) Beliana (S7SC), (Co) Colorao (S5SC), and (G) Goldrich (S1S2).
Genetic Analysis of the Self-Compatible cv Canino
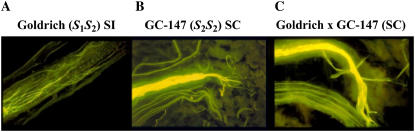
The S-genotype of the self-compatible cv Canino (S2SC) was previously determined by analysis of stylar RNases (Alburquerque et al., 2002) and PCR (Vilanova et al., 2005). Initially, the origin of self-compatibility in this cultivar was thought to be associated with the naturally occurring SC-haplotype. Nevertheless, segregation analysis of S-haplotypes performed in different controlled crosses did not agree with this hypothesis. Figure 3A shows the S-RNase genotyping of the Goldrich (S1S2) × Canino (S2SC) population determined by PCR amplification. The S-genotypes of this population fell into four classes (S1SC, S1S2, S2S2, and S2SC; Table I), instead of the two expected (S1SC and S2SC). Therefore, two unexpected S-genotype classes were obtained in this cross, S1S2 and S2S2, as pollen tubes carrying the S2-haplotype from Canino were expected to be incompatible on Goldrich styles. Similarly, self-pollination of Canino produced three S-genotype classes (S2SC, S2S2, and SCSC), including the unexpected S2S2 seedlings (Table I) instead of the two expected (S2SC and SCSC). Pepito (S2SC), a genetically related cultivar with the same S-genotype as Canino (S2SC), was also tested for the SI breakdown not associated with the SC-haplotype. However, in this case, only the expected S1SC and S2SC genotypes derived from the Goldrich (S1S2) × Pepito (S2SC) cross were observed (Fig. 3C). No pollen contamination was found by the detection of unrelated S-haplotypes in these populations. After self-pollination in the field, the four S-genotype classes obtained in the Goldrich (S1S2) × Canino (S2SC) cross were shown to be self-compatible, setting at least 5% fruit in most seedlings and 2% fruit in nearly all seedlings (Table III). It is noteworthy that the average fruit set decreases slightly in the S1S2 and S2S2 genotypes (Table III). To confirm self-compatibility, two selected trees (GC [for Goldrich (S1S2) × Canino (S2SC) cross]-35 and GC-147) with a fruit set >10% and belonging to the S1S2 and S2S2 classes, respectively, were subjected to pollen-growth tests. Figure 4 shows that self-pollen growth of GC-147 (S2S2) as well as cross-pollination with the self-incompatible cv Goldrich (S1S2) were successful. Similar results were obtained with GC-35 (S1S2; data not shown).
Table III.
Evaluation of self-compatibility in seedlings derived from the cross Goldrich (S1S2) × Canino (S2SC) by recording fruit set after field self-pollination
| S-Genotype | No. of Seedlings Evaluated | No. and Percentage of Seedlings with Fruit Set >2% | No. and Percentage of Seedlings with Fruit Set >5% | Average Fruit Set (%) |
|---|---|---|---|---|
| S1SC | 47 | 46 (98%) | 42 (89%) | 19.2 |
| S2SC | 38 | 36 (95%) | 34 (89%) | 18.0 |
| S1S2 | 16 | 15 (94%) | 12 (75%) | 11.7 |
| S2S2 | 14 | 13 (93%) | 10 (71%) | 8.3 |
| Total | 115 | 110 | 98 |
Figure 4.
Pollen-tube growth after controlled pollination in two apricot selections. Photographs show the style of pistils. A, Self-pollination in Goldrich (S1S2). B, Self-pollination in seedling GC-147 (S2S2). C, Cross-pollination between Goldrich and seedling GC-147 (S2S2). Pollen tubes of self-incompatible cv Goldrich are arrested at the upper part of the pistil (A), but those of the two compatible pollinations complete their development in the pistil and reach the ovary tissue (B and C).
SI of cv Goldrich, previously determined by controlled cross-pollinations (Egea and Burgos, 1996), has been confirmed by pollen-growth tests in this work (Fig. 4A). Thus, regardless of the SC-haplotype, SI breakdown in the Goldrich (S1S2) × Canino (S2SC) cross is due to an alteration in the pollen of Canino. This hypothesis is also supported by the S-genotype segregation resulting from the Canino self-pollination. To test whether the pollen-part mutation in Canino is linked or not to the S-locus, we performed χ2 tests of the segregation ratios observed in these two populations. The S-haplotypes of the Goldrich (S1S2) × Canino (S2SC) progeny segregated into four classes (S1SC:S2SC:S1S2:S2S2) comprising 66:55:28:22 individuals, respectively (Table I). In addition, self-pollination of Canino (S2SC) produced three S-genotype classes (S2SC:S2S2:SCSC) comprising 53:11:35 individuals, respectively (Table I). These observed ratios fit with the expected ratios for a model based on the pollen parent being heterozygous for a mutation affecting pollen function that is unlinked to the S-locus (2:2:1:1 and 3:1:2, respectively; Table IV) with χ2 values of 2.98 and 2.20 (P = 0.394 and P = 0.333; Table I). On the contrary, if we consider pollen-part mutations linked in coupling to the S-locus, the expected ratios (1:1:1:1 and 2:1:1, respectively) do not fit with the observed data, with χ2 values of 31.31 and 12.13, respectively (P < 0.0001 and P = 0.002). Furthermore, S-haplotype segregation data of progenies derived from the self-pollination of some S1SC and S2SC GC seedlings revealed that both S-genotypes are able to produce homozygous (S1S1 or S2S2) individuals (Table I). This result also seems to indicate that the loss of pollen-S function of Canino is not tightly linked to the S-locus and therefore is inherited independently of the S-haplotype. In these cases, segregations fit with the expected ratio for a single mutation unlinked to the S-locus (3:1:2; Table I). In agreement with this hypothesis, some GC seedlings are not able to produce homozygous individuals, supposedly because they did not inherit the mutation, and their segregations fit with the expected ratio (1:1) for a nonmutated seedling (Table I). To confirm these results, the progeny derived from selfing 25 S1SC and 20 S2SC GC seedlings were tested for the presence of S1S1 and S2S2 genotypes, respectively. Fourteen S1SC and 13 S2SC GC seedlings produced these progeny, showing that the S-locus-independent PPM segregates with a 1:1 ratio (χ2 values of 0.36 and 1.80, P = 0.548 and P = 0.180) in the GC population.
Table IV.
Expected gamete and seedling genotypes formed from the outcross Goldrich (S1S2) × Canino (S2SC) and the selfing of Canino (S2SC) considering Canino heterozygous for a pollen-part mutation unlinked to the S-locus (Mm)
| Female Goldrich (S1S2MM)/Male Canino (S2SCMm) | S2Ma | S2m | SCM | SCma |
|---|---|---|---|---|
| S1M | Xb | S1S2Mm | S1SCMM | S1SCMm |
| S2M | X | S2S2Mm | S2SCMM | S2SCMm |
| Female/Male Canino (S2SC Mm) | S2Ma | S2m | SCM | SCma |
| S2M | X | S2S2Mm | S2SCMM | S2SCMm |
| S2m | X | S2S2mm | S2SCMm | S2SCmm |
| SCM | X | S2SCMm | SCSCMM | SCSCMm |
| SCm | X | S2SCmm | SCSCMm | SCSCmm |
If the pollen-part mutation was linked in coupling with S2, the S2M and SCm gametes from Canino would not be formed.
Pollen incompatibility.
Molecular Analysis of the Self-Compatible cv Canino (S2SC)
To test whether the growth of Canino pollen tubes carrying the S2-haplotype was due to mutations or indels affecting the S2-haplotype, we cloned and sequenced genomic DNA fragments containing the SFB2 and S2-RNase genes and their flanking regions from apricot cv Canino (S2SC) and cv Pepito (S2SC). The nucleotide sequence of the S2-haplotype genomic region containing both genes in the self-incompatible cv Goldrich (S1S2) was used as reference (Romero et al., 2004). No changes were detected in the nucleotide sequences of the two S2-haplotype fragments cloned (approximately 1.9 and approximately 2.2 kb, respectively) containing the ORFs of the SFB2 and S2-RNase genes among the three cultivars. Furthermore, gene expression analysis based on RT-PCR showed that both genes are expressed specifically in pollen and styles, respectively, in the three cultivars (Fig. 5). Taken together, these findings indicate that neither the coding sequence nor the promoter functionality is altered in the Canino SFB2 and S2-RNase genes.
Figure 5.
RT-PCR analysis of the S2-RNase and SFB2 gene expression in pollen (P), styles (S), and leaves (L) of the apricot cultivars Goldrich (S1S2), Canino (S2SC), and Pepito (S2SC). Ubi, Ubiquitin (positive control) genes.
Mutations that affect the pollen response are frequently associated with duplicated S-alleles in the Solanaceae (Golz et al., 2001). Thus, duplication of the S-locus F-box genes in the Canino S2-haplotype might also be the cause of pollen function breakdown. Genetic and molecular approaches were carried out to test this possibility. First, as previously described for Currot, flow cytometry determined that Canino is diploid, excluding polyploidy as a possible cause of self-compatibility (data not shown). Subsequently, to look for evidence of genetic duplication, SFB-fragments PCR amplified with consensus primers (SFBc-F and SFBc-R) from genomic DNA of Canino (S2SC) and the self-compatible seedling GC-5 (S2S2) were cloned. Clones were analyzed for cleavage amplified polymorphism sequence (CAPS) digesting with the restriction enzymes EcoRI-NdeI, EcoRI-KpnI, and MvaI that recognize specific sites for cleavage in SFB2 and SFBC. Forty-three clones obtained from Canino exhibited the restriction patterns predicted by the SFB2 or SFBC sequences and 26 obtained from GC-5 exhibited the pattern predicted by SFB2. Sequencing of six clones from each group showed that they indeed encoded SFBC and SFB2. A different restriction EcoRI-NdeI pattern was found in three clones, named FB1 (F-box like), two from Canino and one from GC-5 (Supplemental Fig. S3, A and B). Digestions with KpnI and MvaI confirmed these results (data not shown). The sequences obtained from these three clones were identical to each other and significantly similar to the S-locus F-box genes. Specific genomic PCR amplification of this fragment showed that it is present in all the cultivars and GC seedlings analyzed, but, interestingly, it could not be amplified from BAC clones spanning an approximately 180-kb region containing the Goldrich S-locus, suggesting that it is located outside of the S-locus (Supplemental Fig. S3C). The genomic region adjacent to the FB1 gene was cloned and sequenced in Canino and Goldrich to obtain the complete F-box gene coding sequence. One and two allelic variants with no introns were found in Canino and Goldrich, respectively, and designated ParFB1, ParFB2, and ParFB3 (P. armeniaca F-box protein 1, 2, and 3). The insertion of guanosine single nucleotides in the ParFB1 and ParFB3 (at +608 and +499, respectively) coding sequences produce frame shifts leading to premature stop codons. On the contrary, ParFB2 coding sequence was not disrupted and encoded a complete protein of 378 amino acids. ParFB amino acid-deduced sequences show the N-terminal F-box domain observed in all Prunus SFB proteins (including those encoded that are supposed to be the original ParFB1 [OriParFB1] and ParFB3 [OriParFB3] lacking the inserted guanosines; Supplemental Fig. S4, A and B). Amino acid sequence identity among ParFB proteins is very high (>99%); hence, no typical SFB (hyper)variable regions were found, but the level of identity between ParFB2 and SFB proteins varies from 59% to 64%. RT-PCR assays (Supplemental Fig. S4C) using ParFB-specific primers showed that ParFB is strongly expressed in pollen but also in leaves and styles. Differential intensities of the bands may reflect stronger expression in pollen tissues, although quantitative analysis will need to be done to confirm this observation.
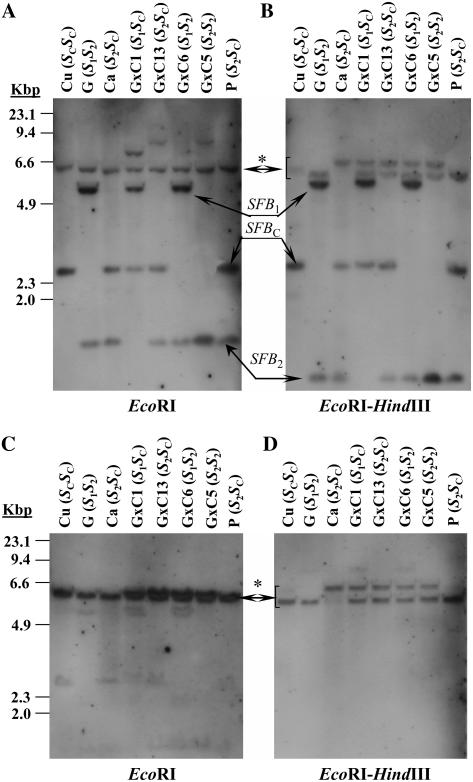
To complement the CAPS analysis, we studied the presence of genetic duplications by genomic DNA-blot analysis performed on four cultivars [Canino (S2SC), Currot (SCSC), Goldrich (S1S2), and Pepito (S2SC)] and four Goldrich × Canino seedlings using a 306-bp SFB1 DNA fragment as a probe. Digestions with EcoRI and EcoRI-HindIII restriction enzymes resulted in single restriction fragments corresponding to SFB1-, SFB2-, and SFBC-alleles (Fig. 6, A and B), according to the S-genotypes previously reported for each selection by genomic PCR (Vilanova et al., 2005). SFB2 and SFBC cross hybridized slightly weaker to the SFB1 probe than SFB1. In addition, extra bands common to all the analyzed selections were detected in the two digestions. In the EcoRI digest, the estimated size of the common band was approximately 6.2 kb (Fig. 6A), but in the EcoRI-HindIII digest the extra bands detected showed different sizes among the cultivars (approximately 5.9 kb for Goldrich and Pepito, approximately 6.1 kb for Currot, and approximately 6.6 kb for Canino). Moreover, the four seedlings share both approximately 5.9-kb and approximately 6.6-kb restriction fragments in agreement with Goldrich and Canino being homozygous for these bands (Fig. 6B). The rehybridization of the filters with a 707-bp ParFB1 probe showed that these common bands correspond to ParFB alleles, because they hybridized specifically to this probe and the hybridization signals corresponding to SFB-alleles were strongly reduced or undetected (Fig. 6, C and D). Results from genomic DNA-blot analysis suggest that all selections analyzed carry ParFB homologous fragments.
Figure 6.
DNA-blot analysis of apricot self-compatible and incompatible cultivars. A and B, Blots of genomic DNA hybridized with an SFB1 probe. C and D, Blots of genomic DNA rehybridized with a ParFB1 probe. Samples were Cu, Currot (SCSC); G, Goldrich (S1S2); Ca, Canino (S2SC); GC-1, seedling GC-1 (S1SC); GC-13, seedling GC-13 (S2SC); GC-6, seedling GC-6 (S1S2); GC-5, seedling GC-5 (S2S2); and P, Pepito (S2SC). DNAs were digested with two restriction enzymes, EcoRI (A and C) and EcoRI-HindIII (B and D).
DISCUSSION
An Insertional Mutation of the Specific SC-Haplotype F-Box Gene Is Associated with Self-Compatibility in Apricot
The presence of the SC-RNase has been determined by stylar RNase analysis and PCR in all apricot self-compatible cultivars studied to date (Burgos et al., 1998; Alburquerque et al., 2002; Vilanova et al., 2005). Moreover, it is absent in all self-incompatible cultivars analyzed, supporting the hypothesis of Burgos et al. (1998) that the SC-RNase is associated with an SC-haplotype. S-genotyping and self-compatibility determination of seedlings resulting from different crosses also confirmed this hypothesis and demonstrated that the SC-RNase cosegregates with self-compatibility (Burgos et al., 1998; Alburquerque et al., 2002).
Results obtained in this work are consistent with SFBC disruption being the cause of SI breakdown associated with the SC-haplotype. Indeed, sequence analysis showed that the SC-RNase gene encodes a likely functional S-RNase, whereas a 358-bp insertion was found in the middle of the putative SFBC coding region. This insertion is located just upstream from the HVa hypervariable region and leads to the production of transcripts of a defective SFBC protein lacking the HVa and HVb hypervariable regions, suggesting that the SC-haplotype is a PPM. RT-PCR analysis showed that SFBC is expressed in pollen tissues, but no data are available about the production of its truncated protein. However, even if the truncated protein is produced, it is unlikely to be functional since the two C-terminal half hypervariable regions are lacking, and both are suggested to be essential for the function of SFB (Ikeda et al., 2004). This feature is common to two PPMs recently reported in Japanese apricot (Prunus mume), where a 6.8-kb insertion found in the middle of the SFBf coding region leads to a frame shift and a premature stop codon (Ushijima et al., 2004), and sweet cherries, where the frame shift mutation is caused by a 4-bp deletion upstream from the HVa coding region of SFB4′ (Ushijima et al., 2004; Sonneveld et al., 2005).
The association between the self-compatibility phenotype and the loss of function of the haplotype-specific SFBC gene in apricot supports previous evidence that SFB is the pollen-S gene in Prunus (Entani et al., 2003; Ushijima et al., 2003, 2004; Sonneveld et al., 2005). In contrast to Prunus, all well-characterized PPMs in the Solanaceae result from duplications of an S-allele leading to competitive interaction and the breakdown of the pollen SI response (Brewbaker and Natarajan, 1960; de Nettancourt, 2001). S-heteroallelic pollen is also the cause of self-fertility in some petunia tetraploids (Stout and Chandler, 1941). Apricot is a diploid, although some tetraploids have been reported (Layne et al., 1996). Collectively, our data suggest that duplication of pollen S-alleles can be ruled out in the apricot PPMs studied in this work, reinforcing the conclusion that breakdown of pollen function in the apricot SC-haplotype is due to the defective SFBC protein.
In accordance with the original inhibitor model, defective SFBs have been suggested to lose the specific S-RNase interaction domain but retain the general interaction domain marking all S-RNases, including the cognate S-RNase, for subsequent degradation by the 26S proteasome pathway (Ushijima et al., 2004). However, Sonneveld et al. (2005) provided evidence of a self-compatible SFB knockout mutant in sweet cherry, suggesting that SFB cannot have the role of general S-RNase inactivator, otherwise SFB loss of function would result in universal incompatible pollen lacking the S-RNase inhibitor mechanism. Results obtained by these authors support a specific role of the SFB proteins in the protection of the cognate S-RNase from the general inactivation mechanism in agreement with the two-component inhibitor model proposed by Luu et al. (2000). The analysis of the apricot SC-haplotype has not provided evidence to support either of these two proposals.
A common evolutionary origin has been proposed for the Solanaceae, Rosaceae, and Scrophulariaceae S-RNase-based GSI systems (Igic and Kohn, 2001). Nevertheless, recent findings suggest differences at least between the pollen-S factor of the Rosaceae and Solanaceae. First, Prunus SFB sequence allelic diversity is significantly higher than that found in Antirrhinum AhSLF or Petunia PiSLF genes (Zhou et al., 2003; Ikeda et al., 2004; Sijacic et al., 2004). Second, recent analysis of tetraploid sour cherry selections has shown that S-heteroallelic pollen retains its SI phenotype in Prunus (Hauck et al., 2006), unlike the phenomenon reported in the Solanaceae (Stout and Chandler, 1941) and other Rosaceae (Crane and Lawrence, 1931) where polyploidy is a direct cause of self-compatibility as a result of competitive interaction. Finally, the absence of pollen-S deletions in the analyzed Solanaceae PPMs prompted Golz et al. (2001) to suggest that pollen-S is required for pollen viability. However, several loss-of-pollen-S-function self-compatible mutants have been found in Prunus besides those of apricot reported in this work, including a complete deletion of the sweet cherry SFB3 (Sonneveld et al., 2005).
Loss of Function of an S-Locus External Factor and SFBC Allele Are Coresponsible for SI Breakdown in Canino (S2SC)
In the cross-pollination Goldrich (S1S2) × Canino (S2SC), Canino pollen tubes carrying the S2-haplotype were supposed to be arrested in the styles of the self-incompatible cv Goldrich (Egea and Burgos, 1996; Alburquerque et al., 2002). However, S-genotypes containing the S2-haplotype derived from Canino (S1S2 and S2S2) were identified in the progeny. Furthermore, S-genotyping of Canino self-pollination progeny also showed homozygous S2S2 genotypes, reinforcing the previous observation and suggesting that a loss of pollen-S function is independent of the SC-haplotype in Canino. As described above, the loss of pollen-S function has been recently associated with deletions and mutations of the SFB genes in sweet cherry and Japanese apricot self-compatible selections (Ushijima et al., 2004; Sonneveld et al., 2005). However, no mutations, insertions, or deletions were found in the sequence analysis of the SFB2 and S2-RNase alleles of Canino (S2SC), and their transcripts were detected specifically in pollen and styles, respectively.
Moreover, segregation ratios observed in the two controlled pollinations performed with Canino (Table I) fit with a model in which the pollen-part mutation is not linked to the S-locus. Indeed, all S-genotypes were found to be self-compatible by field self-pollination (Table III), and the analysis of self-pollination progeny derived from S2SC and S1SC genotypes showed the presence of S1 and S2 homozygotes (Table I). Thus, the pollen-part mutation seems to affect both S1 and S2 haplotypes, indicating that it is inherited independently of the S-locus. Interestingly, the number of homozygous S2 genotypes obtained in both controlled crosses is lower than that expected for a mutation not linked to the S-locus (Table I). The average fruit set recorded from the progeny is consistent with that finding and seems to be lower in those S-genotypes containing the S2-haplotype derived from Canino (S1S2 and S2S2; Table III). A similar deviation was observed by Wünsch and Hormaza (2004) in the self-pollination of the sweet cherry PPM Cristobalina. These authors suggest several reasons to explain this deviation, such as differences in the pollen competitive capacity to grow through the style, postzygotic selection against homozygous embryos, or linkage between the dominant allele of the mutated factor and the S-locus. An additional possibility may be that the pollen-part mutation does not only break the SI rejection mechanism but also reduces pollen viability.
Pollen-part mutations can also arise from S-allele duplications located in a centric fragment, in a non-S chromosome or linked to the S-locus (Golz et al., 2001). In fact, S-genotype segregations observed in the Goldrich (S1S2) × Canino (S2SC) cross and Canino (S2SC) self-pollination might result from the loss of function of an additional factor outside the S-locus or from the duplicated pollen-S gene not linked in coupling with the S-locus. However, Canino was confirmed as diploid (see above), and neither CAPS nor DNA-blot analysis provided evidence of pollen-S genetic duplication. As discussed later, the ParFB SFB-like gene identified in this work was found in both self-compatible and self-incompatible cultivars, and, therefore, it cannot be considered as a duplicated pollen-S gene involved in the SI breakdown. Together, these results seem to rule out competitive interaction resulting from S-heteroallelic pollen; hence, apart from the mutated SC-haplotype, SI breakdown in Canino could be due to a defective additional factor outside the S-locus.
Genetic evidence of factors required for SI besides the specificity determinants has been widely reported in the Solanaceae (McClure et al., 2000). Indeed, similar to the case described in this work, Ai et al. (1991) showed that the self-compatible cultivar of Petunia hybrida Strawberry Daddy (SOSX) accumulates a nonfunctional S-allele (SO) and a recessive mutation in an additional factor necessary for SI. Moreover, mutations in modifier loci not related to the S-locus affecting the function of the pollen-S factor have also been suggested to explain SI breakdown in Solanum tuberosum (Thompson et al., 1991), Petunia axillaris (Tsukamoto et al., 2003), and more recently in sweet cherry (Wünsch and Hormaza, 2004). The hypothetical Canino defective factor does not seem to affect the expression of S-locus genes or result in sterility, suggesting that it belongs to group 2 of the modifier genes required for pollen rejection but with no wider role in pollination (McClure et al., 2000). If this is the case, according to the original RNase-mediated SI inhibitor model (Thompson and Kirch, 1992), this factor could be involved in a nonspecific interaction preventing ubiquitination of the S-RNases, and, therefore, its loss of function would facilitate a breakdown of SI. However, other possibilities cannot be ruled out, such as the requirement of this factor for the uptake of the S-RNases into the pollen tubes. Further research will be necessary to determine the SI function affected by the pollen-part mutation in Canino.
ParFB, a New Type of F-Box Gene with Sequence Similarity to SFB
S-locus pollen-expressed F-box genes with sequence similarity to SFB (or SLF) have been identified in Prunus (Entani et al., 2003; Ushijima et al., 2003), Anthirrinum (Zhou et al., 2003), and Petunia (Wang et al., 2003). In all these cases, amino acid sequence identity shared to SFB (or SLF) is less than 60% (Tsukamoto et al., 2005). The SLFL F-box proteins found in the S-locus genomic region of Japanese apricot and almond (Entani et al., 2003; Ushijima et al., 2003) exhibit high sequence identity among them (>90%) and low amino acid identities (approximately 25%) with the Prunus SFB. More recently, PaF1, an SFB-like gene probably not linked to the S-locus, has been reported in P. axillaris. PaF1 does not show any allelic polymorphism and, unexpectedly, PaF1 (P. axillaris F-box protein 1) shares approximately 90% sequence identity with PaSLF proteins (Tsukamoto et al., 2005). The function of all these SFB-like genes is presently unknown.
In this work, we have identified a new type of F-box gene with sequence similarity to SFB, ParFB. Three ParFB variants have been cloned and sequenced from two different apricot cultivars and they show very low allelic sequence polymorphism (approximately 99% nucleotide sequence similarity), similar to Prunus SLFL genes (Entani et al., 2003; Ushijima et al., 2003). Likewise, predicted amino acid sequences of these three F-box proteins exhibit low percentage identity (approximately 25%) with Prunus SLFL (Entani et al., 2003; Ushijima et al., 2003) and S-locus linked F-box proteins identified in Petunia inflata (Wang et al., 2003), but a high sequence identity with Prunus SFB (approximately 62% on average). Interestingly, two insertions comprising five and two amino acids were found in the middle of the putative ParFB regions corresponding to the SFB hydrophilic V1 and V2 domains, respectively. Similarly, a gap spanning three amino acids was found in the putative region of ParFB corresponding to HVb in the S-locus F-box gene. The V1, V2, and HVb domains of SFB have been shown to be under positive selection, suggesting that these regions may be responsible for the discrimination between self and non-self S-RNases (Ikeda et al., 2004). Thus, it seems that whatever is the role of ParFB proteins, functions related to these putative domains could be changed or lost with regard to SFB proteins.
The isolation of a full-length ParFB coding sequence from the self-incompatible cv Goldrich (S1S2) allows to discard its involvement in SI breakdown through competitive interaction. Moreover, the PPM Canino is supposed to be homozygous for the disrupted variant ParFB1 since it has been the only variant found in this cultivar. If this is the case, ParFB would not be the mutated additional factor outside the S-locus that causes SI breakdown in Canino. Otherwise, the number of individuals belonging to each S-genotype obtained in the cross pollination Goldrich (S1S2) × Canino (S2SC) would have been the same (Table I). On the other hand, ParFB genes share high sequence similarity with the pollen-S candidate SFB genes and are strongly expressed in pollen tissues, but, at the same time, show very low allelic diversity, and gene-expression is not pollen-specific. In addition, the analysis of their deduced amino acid sequences suggests that putative domains corresponding to those of SFB proteins involved in the specific recognition of the style counterpart are altered. Therefore, although the function of the ParFB proteins remains to be investigated, these results would suggest a nonspecific role, if any, in GSI.
Overall, findings of this study could be summarized in two main conclusions. On one side, the association between the partial loss-of-function mutation of the SFBC and the Currot self-compatibility phenotype reinforces previous evidence suggesting SFB as the pollen-S candidate in Prunus. On the other, genetic evidence has been provided supporting the involvement of an S-locus external factor in the S-RNase-based GSI breakdown of the self-compatible cv Canino. In addition, the ParFB gene identified in this work encodes a previously uncharacterized protein that shares high amino acid sequence identity to SFB but whose function is still unknown. Further research will be necessary to reveal the precise role of ParFB. In that respect, the study of natural self-compatible mutants has been shown as a useful tool to dissect the basis of GSI mechanism in Prunus where transgenic approaches present clear difficulties.
MATERIALS AND METHODS
Plant Material
Eight apricot (Prunus armeniaca) cultivars, Goldrich (S1S2), Pepito (S2SC), Colorao (S5SC; Burgos et al., 1998), Currot (SCSC), Canino (S2SC), Beliana (S7SC; Alburquerque et al., 2002), Ginesta (SCSC), and Palau (SCSC; Vilanova et al., 2005), and six progenies, consisting of eight to 171 seedlings, derived from the crosses Goldrich (S1S2) × Canino (S2SC; designated as GC-), Goldrich (S1S2) × Pepito (S2SC), and Goldrich (S1S2) × Currot (SCSC) as well as from the self-pollination of Canino (S2SC) and the seedlings GC-8, GC-10, GC-80, and GC-86, were used in this study. All these trees are maintained at the collections of the Instituto Valenciano de Investigaciones Agrarias (IVIA) in Valencia (Spain) and at the Departamento de Mejora de Frutales (CEBAS-CSIC) in Murcia (Spain).
DNA Extraction
Five grams or two discs of leaves of each selection were collected and stored at −80°C before DNA isolation. Genomic DNA was extracted from leaf samples following the method of Doyle and Doyle (1987). DNA quantification was performed by comparison with a λ DNA Mr marker (Promega).
Cloning and Sequencing of the SC-RNase and SFBC Genes from cv Currot (SCSC)
A fragment of the SC-RNase was PCR amplified with the primers SRc-F (5′-CTC GCT TTC CTT GTT CTT GC-3′; Romero et al., 2004) and Pru-C5 (5′-TAC CAC TTC ATG TAA CAA CTG AG-3′; Tao et al., 1999) using Currot (SCSC) genomic DNA as template. Cycling conditions were as follows: an initial denaturing step of 94°C for 1 min; 7 cycles of 94°C for 5 s and 68°C for 3 min; 30 cycles of 94°C for 5 s, 60°C for 1 min, and 68°C for 3 min; and a final extension of 68°C for 10 min (GeneAmpPCR System 9700; Perkin-Elmer). Similarly, an amplification product containing a fragment of the SFBC gene was generated with primers SFBc-F (5′-TCG ACA TCC TAG TAA GAC TAC CTG C-3′) and SFBc-R (5′-ATT TCT TCA CTG CCT GAA TCG-3′; Romero et al., 2004) also using Currot (SCSC) genomic DNA as template. In this case, amplifications were carried out using a temperature profile with an initial denaturing of 94°C for 2 min; 30 cycles of 94°C for 30 s, 55°C for 60 s, and 72°C for 1 min 30 s; and a final extension of 72°C for 10 min (GeneAmpPCR System 9700; Perkin-Elmer).
PCR products were electrophoresed in 0.8% (w/v) agarose gel. Molecular sizes of the amplified fragments were estimated using a 100-bp ladder (Invitrogen). To obtain the complete sequences, adjacent 5′ and 3′ ends were isolated using the Universal Genome Walker kit (CLONTECH). Finally, these fragments were purified from the agarose gels using the QIAquick Gel Extraction kit (Qiagen) and cloned into the pGEM T-Easy vector (Promega). DNA sequences from four independent clones were determined automatically using an ABI PRISM 377 DNA Sequencer (Applied Biosystems) and the BigDye Terminator v1.1 Cycle Sequencing kit (Applied Biosystems) following the manufacturer's instructions.
Cloning and Sequencing of the S2-RNase and SFB2 Genes from cv Canino (S2SC) and cv Pepito (S2SC)
Specific primers designed from the S2-haplotype sequence identified in apricot (Romero et al., 2004) were used for amplification of genomic fragments containing the complete S2-RNase [Sf-Hap2 (5′-CGC TAG AAA TCA AAG CCA CAG-3′)/Sr-Hap2 (5′-GGC GTA AGC AAG TGG AAA AG -3′)] and SFB2 coding sequences [FBf-Hap2 (5′-GCC CAA TTA CTT GGT CAC TG-3′)/FBr-Hap2 (5′-CAC CCA CTT GAC TTG TCA GC-3′)] using Canino (S2SC) and Pepito (S2SC) genomic DNA as templates. PCR conditions and methods for isolating, cloning, and sequencing these bands were the same used for the SC-RNase.
Sequence Analysis, Alignments, and Homology Searches
Sequences were assembled by BioEdit software (Hall, 1999). Primers were designed using the online program Primer3 (Rozen and Skaletsky, 2000). The analysis of the S-loci structure in the SC-haplotype was performed using the following software: EMBOSS (Rice et al., 2000), TROLL (Castelo et al., 2002), GeneScan (Burge and Karlin, 1997), and GenomeScan (Yeh et al., 2001). Alignment of the nucleotide and amino acidic sequences was carried out with ClustalW (Thompson et al., 1994) and ClustalX (Thompson et al., 1997) programs, respectively. Homology searches were performed with BLASTX and BLASTN (Altschul et al., 1990).
The following sequences were used for the alignment of Figure 1: EMBL/DDJB/GenBank accession numbers Par-SFB1 (AY587563), Par-SFB2 (AY587562), Par-SFB4 (AY587565; Romero et al., 2004), Par-SFBC (DQ422946; this work), Pa-SFB3 (AB096857), Pa-SFB6 (AB096858; Yamane et al., 2003), Pm-SFB1 (AB092621), Pm-SFB7 (AB092622; Entani et al., 2003), Pd-SFBa (AB092966), and Pd-SFBb (AB092967; Ushijima et al., 2003).
RT-PCR
Total RNA was extracted from leaves, pollen grains, and styles of the cultivars Currot (SCSC), Canino (S2SC), Pepito (S2SC), and Goldrich (S1S2) following the method of Salzman et al. (1999) with modifications. All RNAs were treated with DNase I (Roche Diagnostics). The cDNAs were synthesized by ThermoScript RT-PCR system (Invitrogen) with oligo-d(T) primer. The obtained cDNAs were used as a template for PCRs with gene-specific primer sets RSc-F (5′-GTG TTT CAT TAT GAG CAC TAG ATC-3′) and RSc-R (5′-TTA ATG TCA ACG TTA TTC CAG C-3′) for SC-RNase, RS2-F (5′-ATG AGC ACT GGT GAT GGA AC-3′) and RS2-R (5′-ACG GAG TGC AGG ATC AGT TC-3′) for S2-RNase, RFBc-F (5′-GAG GAG TGC TAC AAA CTA AGC-3′) and RFBc-R (5′-ACC CCT ATG ATG TTC CAA AG-3′) for SFBC, RFB2-F (5′-TGA ACG TCA GAA CGA CAC TG-3′) and RFB2-R (5′-ACC CTT ATA ATG CTG CCA AG-3′) for SFB2, FB-F (5′-TAC GAA AAC TAC GAG GAC TAC-3′) and FB-R (5′-AAG CAT CAT CTT TGT GGA CG-3′) for ParFBs, and UBI-F (5′-CTC CTC TGA CAC CAT CGA CAA-3′) and UBI-R (5′-CAT AGG TCA ACC CAC ACT TG-3′) for the ubiquitin gene (Entani et al., 2003). PCRs were performed as described by Romero et al. (2004).
Genomic PCRs for S-Genotyping and SFBC-Specific Amplification
S-genotyping of populations and cultivars was performed by PCR amplification of the S-RNase first intron with the primer pair SRc-F (5′-CTC GCT TTC CTT GTT CTT GC-3′; Romero et al., 2004) and SRc-R (5′-GGC CAT TGT TGC ACA AAT TG-3′; Vilanova et al., 2005), following the protocol described by Vilanova et al. (2005).
SFBC-specific PCR amplification from genomic DNA was performed with the primer pair RFBc-F/RFBc-R. Two additional primers, RFBc-F and SFBins-R (5′-TCA AGA ACT TGG TTG GAT TCG-3′), designed from the consensus sequence of the Prunus SFB alleles (Romero et al., 2004), were used to amplify the SFBC insertion from genomic DNA of several apricot cultivars.
Pollination and Pollen-Tube Growth Test
One hundred and fifteen trees derived from the cross Goldrich (S1S2) × Canino (S2SC) were tested for self-compatibility by self-pollination in the field. Before anthesis, insect-proof bags were put over several branches, containing approximately 200 to 250 flower buds in total per tree, to prevent cross pollination. Subsequent fruit set was recorded and fruits collected about 3 months later. Following the same procedure, Canino was self-pollinated to obtain a population of study.
Pollen-tube growth tests were performed for self-pollinations of Goldrich (S1S2) and two seedlings derived from the cross Goldrich (S1S2) × Canino (S2SC), GC-35 (S1S2), and GC-147 (S2S2). Moreover, cross-pollinations between Goldrich, as female parent, and GC-35 or GC-147, as male parents, were also evaluated. Branches with about 30 flowers in balloon stage, for each combination, were placed in beakers with a 5% (w/v) Suc solution and the flowers were emasculated immediately. Branches were maintained in a chamber at a controlled temperature of 24°C. Pistils were self- or cross-pollinated 24 h later, and 72 h after pollination pistils were harvested in a 5% formaldehyde fixing solution at 40%, 5% acetic acid, and 90% ethanol at 70%. Pistils were washed in distilled water three times, for 1 h each, to remove the FAA and then placed in the autoclave for 30 min at 100 KP in 5% (w/v) sodium sulfite solution to soften the pistils. The pistils were then stained for 24 h with 0.1% (v/v) aniline blue in 0.1 n potassium phosphate, pH 8.7. The epidermis was removed and the pistils squashed for observation with an Olympus BH2 microscope with a BH2-RFL-T2 UV light source, using an Osram HBO 100 W/2 high-pressure mercury lamp.
Ploidy Level Determination
Ploidy level was determined using the Partec CyStain UV precise P reagent kit for nuclei extraction and DNA staining of nuclear DNA from plant tissues. Approximately 0.5 cm2 leaf tissue was chopped using a sharp razor blade in 400 μL of extraction buffer and filtered through a Partec 50-μm CellTrics disposable filter. Samples were then incubated for 60 s in the staining solution and analyzed in the Partec flow cytometer Ploidy Analyzer PA in the blue fluorescence channel.
Genomic DNA-Blot Analysis
Genomic DNA was digested with two restriction enzymes (EcoRI and HindIII; Roche Diagnostics), electrophoresed on 0.8% agarose gels, and blotted onto Hybond N nylon membranes (Amersham Biosciences). Blots were performed using approximately 3 μg of DNA per track. An SFB1 alkali-labile digoxygenin DIG-labeled (306 bp long) probe was synthesized by PCR from the apricot BAC DNA clone 102/D9 (Vilanova et al., 2003) using the primer pair SFB-5F (5′-TAG GAC CCC TCC AAT GAG C-3′) and SFBc-R (5′-ATT TCT TCA CTG CCT GAA TCG-3′). The SFB1 probe corresponds to a Prunus SFB-conserved region located between the V1 and V2 variable domains. Filters were hybridized overnight with the DIG-labeled probe at 65°C and washed at low-stringency conditions (twice at room temperature in 2× SSC, 0.1% SDS for 5 min, and twice at 68°C in 0.5× SSC, 0.1% SDS for 15 min). Hybridization signals were detected following the manufacturer's recommendations (Roche Diagnostics) using Kodak XAR films. Filters were then washed for removing the probe and rehybridized following the same procedure with a ParFB1 707-bp-long DIG-labeled probe synthesized by PCR from the apricot DNA clone C14 using the primer pair SRc-F (5′-CTC GCT TTC CTT GTT CTT GC-3′) and SFBc-R (5′-ATT TCT TCA CTG CCT GAA TCG-3′). Detection was performed as described above.
CAPS Analysis
SFB coding sequence fragments were nonspecifically PCR amplified with the consensus primers SFBc-F and SFBc-R (Romero et al., 2004) using Canino (S2SC) and GC-5 (S2S2) genomic DNA as templates. PCR products were cloned and subsequently digested. PCR conditions and methods for isolating and cloning these fragments into the pGEM T-Easy vector (Promega) were the same as described above for SFBC. Clones were digested with the restriction enzymes EcoRI-NdeI, EcoRI-KpnI, and MvaI (Roche Diagnostics). Restriction products were electrophoresed on 2% agarose gels and stained with ethidium bromide.
Cloning and Sequencing of the ParFB Genes from Goldrich (S1S2) and Canino (S2SC)
ParFB fragments amplified from Canino and Goldrich using the primers FB-F and SFBc-R (Romero et al., 2004) were cloned into the pGEM T-Easy vector (Promega) as described above. Twelve clones from each cultivar were sequenced falling into three variants (ParFB1 from Canino; ParFB2 and ParFB3 from Goldrich). PCR conditions and methods for isolating, cloning, and sequencing these fragments and the adjacent 5′ and 3′ ends were the same used for SFBC.
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession numbers DQ422943, DQ422944, DQ422945, DQ422946, and DQ422947.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Segregation analysis of S-RNases of the Goldrich × Currot progeny.
Supplemental Figure S2. Alignment of the amino acidic sequences of S-RNases from several Prunus species carried out with ClustalX (Thompson et al., 1997). Sequences used were the following: EMBL/DDJB/GenBank accession numbers Par-S1 (AY587561), Par-S2 (AY587562), Par-S4 (AY587564), Par-SC (DQ422947), Pa-S3 (AB010306), Pa-S6 (AB010305), Pm-S1 (AB101438), Pm-S7 (AB101439), Pd-Sa (AB026836), and Pd-Sb (AB011469).
Supplemental Figure S3. Search of SFB genetic duplications in cv Canino (S2SC) by CAPS analysis with EcoRI-NdeI.
Supplemental Figure S4. Molecular characterization of ParFB alleles. Sequences used for the alignments were: Par-SFB1 (AY587563), Par-SFB2 (AY587562), Par-SFB4 (AY587565; Romero et al., 2004), Par-FB1 (DQ422943), Par-FB2 (DQ422944), and Par-FB3 (DQ422946; this work).
Supplementary Material
Acknowledgments
We thank Dr. Wim Deleu for his helpful revision of this manuscript, José Juarez for his help in the ploidy level determination, and to all the members of the Fruit Trees lab for their contributions. All experiments described in this article comply with the current laws of Spain.
This work was supported by the Ministerio de Ciencia y Tecnología of Spain (grant no. AGL2001–1122–C02–02; fellowship to S.V.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Carlos Romero (cromero@ivia.es).
The online version of this article contains Web-only data.
References
- Ai Y, Kron E, Kao T-H (1991) S-alleles are retained and expressed in a self-compatible cultivar of Petunia hybrida. Mol Gen Genet 230: 353–358 [DOI] [PubMed] [Google Scholar]
- Alburquerque N, Egea J, Pérez-Tornero O, Burgos L (2002) Genotyping apricot cultivars for self-(in)compatibility by means of RNases associated with S alleles. Plant Breed 121: 343–347 [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Brewbaker JL, Natarajan AT (1960) Centric fragments and pollen part mutation of incompatibility alleles in Petunia. Genetics 45: 699–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boskovic R, Tobutt KR (1996) Correlation of stylar ribonuclease zymograms with incompatibility alleles in sweet cherry. Euphytica 90: 245–250 [Google Scholar]
- Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268: 78–94 [DOI] [PubMed] [Google Scholar]
- Burgos L, Berenguer T, Egea J (1993) Self- and cross-compatibility among apricot cultivars. Hort Sci 28: 148–150 [Google Scholar]
- Burgos L, Pérez-Tornero O, Ballester J, Olmos E (1998) Detection and inheritance of stylar ribonucleases associated with incompatibility alleles in apricot. Sex Plant Reprod 11: 153–158 [Google Scholar]
- Castelo AT, Martins W, Gao GR (2002) TROLL—tandem repeat occurrence locator. Bioinformatics 18: 634–636 [DOI] [PubMed] [Google Scholar]
- Crane MB, Lawrence WJC (1931) Sterility and incompatibility in diploid and polyploid fruits. J Genet 24: 97–107 [Google Scholar]
- de Nettancourt D (2001) Incompatibility and Incongruity in Wild and Cultivated Plants. Springer, Berlin
- Deshaies RJ (1999) SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol 15: 435–467 [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL (1987) A rapid isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19: 11–15 [Google Scholar]
- Egea J, Burgos L (1996) Detecting cross-incompatibility of three North American apricot cultivars and establishing the first incompatibility group in apricot. J Am Soc Hortic Sci 121: 1002–1005 [Google Scholar]
- Entani T, Iwano M, Shiba H, Che FS, Isogai A, Takayama S (2003) Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: identification of a pollen-expressed F-box gene with allelic diversity. Genes Cells 8: 203–213 [DOI] [PubMed] [Google Scholar]
- Golz JF, Oh HY, Su V, Kusaba M, Newbigin E (2001) Genetic analysis of Nicotiana pollen-part mutants is consistent with the presence of an S-ribonuclease inhibitor at the S locus. Proc Natl Acad Sci USA 98: 15372–15376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golz JF, Su V, Clarke AE, Newbigin E (1999) A molecular description of mutations affecting the pollen counterpart of the Nicotiana alata S locus. Genetics 152: 1123–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98 [Google Scholar]
- Hauck NR, Yamane H, Tao R, Iezzoni AF (2006) Accumulation of non-functional S-haplotypes results in the breakdown of gametophytic self-incompatibility in tetraploid Prunus. Genetics 172: 1191–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Lee HS, Karunanandaa B, Kao TH (1994) Ribonuclease activity of Petunia inflata S proteins is essential for rejection of self-pollen. Plant Cell 6: 1021–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igic B, Kohn JR (2001) Evolutionary relationships among self-incompatibility RNases. Proc Natl Acad Sci USA 98: 13167–13171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Igic B, Ushijima K, Yamane H, Hauck NR, Nakano R, Sassa H, Iezzoni AF, Kohn JR, Tao R (2004) Primary structural features of the S haplotype-specific F-box protein, SFB, in Prunus. Sex Plant Reprod 16: 235–243 [Google Scholar]
- Ishimizu T, Shinkawa T, Sakiyama F, Norioka S (1998) Primary structural features of rosaceous S-RNases associated with gametophytic self-incompatibility. Plant Mol Biol 37: 931–941 [DOI] [PubMed] [Google Scholar]
- Kawata Y, Sakiyama F, Hayashi F, Kyogoku Y (1989) Identification of two essential histidine residues of ribonuclease T2 from Aspergillus oryzae. Eur J Biochem 187: 255–262 [DOI] [PubMed] [Google Scholar]
- Lai Z, Ma W, Han B, Liang L, Zhang Y, Hong G, Xue Y (2002) An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol Biol 50: 29–42 [DOI] [PubMed] [Google Scholar]
- Layne REC, Bailey CH, Hough LF (1996) Apricots. In J Janick, JN Moore, eds, Fruit Breeding: Tree and Tropical Fruits, Vol 2. John Wiley & Sons, New York, p 89
- Luu DT, Qin XK, Morse D, Cappadocia M (2000) S-RNase uptake by compatible pollen tubes in gametophytic self-incompatibility. Nature 407: 649–651 [DOI] [PubMed] [Google Scholar]
- McClure BA, Cruz-García F, Beecher B, Sulaman W (2000) Factors affecting inter- and intra-specific pollen rejection in Nicotiana. Ann Bot (Lond) 85: 113–123 [Google Scholar]
- McClure BA, Haring V, Ebert PR, Anderson MA, Simpson RJ, Sakiyama F, Clarke AE (1989) Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature 342: 955–957 [DOI] [PubMed] [Google Scholar]
- Qiao H, Wang H, Zhao L, Zhou J, Huang J, Zhang Y, Xue Y (2004) The F-box protein AhSLF-S2 physically interacts with S-RNases that may be inhibited by the ubiquitin/26S proteasome pathway of protein degradation during compatible pollination in Antirrhinum. Plant Cell 16: 571–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A (2000) EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Romero C, Vilanova S, Burgos L, Martínez-Calvo J, Vicente M, Llácer G, Badenes ML (2004) Analysis of the S-locus structure in Prunus armeniaca L. Identification of S-haplotype specific S-RNase and F-box genes. Plant Mol Biol 56: 145–157 [DOI] [PubMed] [Google Scholar]
- Royo J, Kuntz C, Kowyama Y, Anderson M, Clarke AE (1994) Loss of a histidine residue at the active site of S-locus ribonuclease is associated with self-compatibility in Lycopersicon peruvianum. Proc Natl Acad Sci USA 91: 6511–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In S Krawetz, S Misener, eds, Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa, NJ, pp 365–386 [DOI] [PubMed]
- Salzman RA, Fujita T, Zhu-Salzman K, Hasegawa PM, Bressan RA (1999) An improved RNA isolation method for plant tissues containing high levels of phenolic compounds or carbohydrates. Plant Mol Biol Report 17: 11–17 [Google Scholar]
- Sassa H, Hirano H, Nishio T, Koba T (1997) Style-specific self-compatible mutation caused by deletion of the S-RNase gene in Japanese pear (Pyrus serotina). Plant J 12: 223–227 [Google Scholar]
- Sijacic P, Wang X, Skirpan AL, Wang Y, Dowd PE, McCubbin AG, Huang S, Kao T-h (2004) Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature 429: 302–305 [DOI] [PubMed] [Google Scholar]
- Sonneveld T, Tobbutt KR, Vaughan SP, Robbins TP (2005) Loss of pollen-S function in two self-compatible selections of Prunus avium is associated with deletion/mutation of an S haplotype-specific F-box gene. Plant Cell 17: 37–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout AB, Chandler C (1941) Change from self-incompatible to self-compatible accompanying change from diploid to tetraploidy. Science 94: 118. [DOI] [PubMed] [Google Scholar]
- Tao R, Yamane H, Sugiura A (1999) Molecular typing of S-alleles through identification, characterization and cDNA cloning for S-RNases in sweet cherry. J Am Soc Hortic Sci 124: 224–233 [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RD, Kirch HH (1992) The S locus of flowering plants: when self-rejection is self-interest. Trends Genet 8: 381–387 [DOI] [PubMed] [Google Scholar]
- Thompson RD, Uhrig H, Hermsen JGT, Salamini F, Kaufmann H (1991) Investigation of a self-compatible mutation in Solanum tuberosum clones inhibiting S-allele activity in pollen differentially. Mol Gen Genet 226: 283–288 [DOI] [PubMed] [Google Scholar]
- Tsukamoto T, Ando T, Takahashi K, Omori T, Watanabe H, Kokubun H, Marchesi E, Kao T-h (2003) Breakdown of self-incompatibility in a natural population of Petunia axillaris caused by loss of pollen function. Plant Physiol 131: 1903–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T, Ando T, Watanabe H, Marchesi E, Kao T (2005) Duplication of the S-locus F-box gene is associated with breakdown of pollen function in an S-haplotype identified in a natural population of self-incompatible Petunia axillaris. Plant Mol Biol 57: 141–153 [DOI] [PubMed] [Google Scholar]
- Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao R (2003) Structural and transcriptional analysis of the self-incompatibility locus of almond: identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell 15: 771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima K, Yamane H, Watari A, Kakehi E, Ikeda K, Hauck NR, Iezzoni AF, Tao R (2004) The S haplotype-specific F-box protein gene, SFB, is defective in self-compatible haplotypes of Prunus avium and P. mume. Plant J 39: 573–586 [DOI] [PubMed] [Google Scholar]
- Vaughan SP, Russell K, Sargent DJ, Tobutt KR (2006) Isolation of S-locus F-box alleles in Prunus avium and their application in a novel method to determine self-incompatibility genotype. Theor Appl Genet (in press) [DOI] [PubMed]
- Vilanova S, Romero C, Abernathy D, Abbott AG, Burgos L, Llácer G, Badenes ML (2003) Construction and application of a bacterial artificial chromosome (BAC) library of Prunus armeniaca L. for the identification of clones linked to the self-incompatibility locus. Mol Genet Genomics 269: 685–691 [DOI] [PubMed] [Google Scholar]
- Vilanova S, Romero C, Burgos L, Llácer G, Badenes ML (2005) Identification of self-(in)compatibility alleles in apricot (Prunus armeniaca L.) by PCR and sequence analysis. J Am Soc Hortic Sci 130: 893–898 [Google Scholar]
- Wang Y, Wang X, McCubbin AG, Kao T-h (2003) Genetic mapping and molecular characterization of the self-incompatibility (S) locus in Petunia inflata. Plant Mol Biol 53: 565–580 [DOI] [PubMed] [Google Scholar]
- Watkins R (1976) Cherry, plum, peach, apricot and almond. In NW Simmonds, ed, Evolution of Crop Plants. Longman, London, pp 242–247
- Wünsch A, Hormaza JI (2004) Genetic and molecular analysis in Cristobalina sweet cherry, a spontaneous self-compatible mutant. Sex Plant Reprod 17: 203–210 [Google Scholar]
- Xue Y, Carpenter R, Dickinson HG, Coen ES (1996) Origin of allelic diversity in Antirrhinum S locus RNases. Plant Cell 8: 805–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane H, Ikeda K, Ushijima K, Sassa H, Tao R (2003) A pollen-expressed gene for a novel protein with an F-box motif that is very tightly linked to a gene for S-RNase in two species of cherry, Prunus cerasus and P. avium. Plant Cell Physiol 44: 764–769 [DOI] [PubMed] [Google Scholar]
- Yeh RF, Lim LP, Burge CB (2001) Computational inference of homologous gene structures in the human genome. Genome Res 11: 803–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang F, Ma W, Zhang Y, Han B, Xue Y (2003) Structural and transcriptional analysis of S-locus F-box genes in Antirrhinum. Sex Plant Reprod 16: 165–177 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.