Abstract
Although a wide range of structurally diverse small molecules can act as auxins, it is unclear whether all of these compounds act via the same mechanisms that have been characterized for 2,4-dichlorophenoxyacetic acid (2,4-D) and indole-3-acetic acid (IAA). To address this question, we used a novel member of the picolinate class of synthetic auxins that is structurally distinct from 2,4-D to screen for Arabidopsis (Arabidopsis thaliana) mutants that show chemically selective auxin resistance. We identified seven alleles at two distinct genetic loci that conferred significant resistance to picolinate auxins such as picloram, yet had minimal cross-resistance to 2,4-D or IAA. Double mutants had the same level and selectivity of resistance as single mutants. The sites of the mutations were identified by positional mapping as At4g11260 and At5g49980. At5g49980 is previously uncharacterized and encodes auxin signaling F-box protein 5, one of five homologs of TIR1 in the Arabidopsis genome. TIR1 is the recognition component of the Skp1-cullin-F-box complex associated with the ubiquitin-proteasome pathway involved in auxin signaling and has recently been shown to be a receptor for IAA and 2,4-D. At4g11260 encodes the tetratricopeptide protein SGT1b that has also been associated with Skp1-cullin-F-box-mediated ubiquitination in auxin signaling and other pathways. Complementation of mutant lines with their corresponding wild-type genes restored picolinate auxin sensitivity. These results show that chemical specificity in auxin signaling can be conferred by upstream components of the auxin response pathway. They also demonstrate the utility of genetic screens using structurally diverse chemistries to uncover novel pathway components.
The small molecule plant hormone auxin (indole-3-acetic acid [IAA]) regulates and influences many aspects of the growth and development of plants. Treatment of plants with exogenous IAA can result in a variety of physiological and morphological effects, including loss of apical dominance, effects on cell elongation and division, and adventitious root formation (Davies, 1995). A wide range of chemically diverse synthetic small molecules can also produce effects similar to treatment with the natural hormone (Woodward and Bartel, 2005). Some of these compounds have found use as herbicides, the most well known being 2,4-dichlorophenoxyacetic acid (2,4-D; Sterling and Hall, 1997). 2,4-D has proven to be a useful chemical probe of auxin action because it is a potent and stable xenobiotic compound that is not subject to the many endogenous homeostatic and metabolic mechanisms that can affect IAA (Ljung et al., 2002). Screens for Arabidopsis (Arabidopsis thaliana) mutants that are resistant to the action of 2,4-D have defined several genetic loci, including AXR1, AXR2, AXR3, AXR5, and AXR6 (Leyser et al., 1993, 1996; Nagpal et al., 2000; Hellmann et al., 2003; Yang et al., 2004), and mutations in these loci are cross-resistant to IAA, demonstrating the functional similarity of the two auxins. An additional locus, TIR1, has been identified from a screen using an auxin transport inhibitor and mutations at this locus were also found to confer resistance to both 2,4-D and IAA (Ruegger et al., 1998).
Investigation of these and other mutants has led to the understanding that auxin signal transduction involves the ubiquitin-proteasome pathway (Leyser, 2002; Dharmasiri and Estelle, 2004). IAA and 2,4-D stimulate the ubiquitination and degradation of the Aux/IAA family of transcriptional regulators that includes AXR2 (IAA7), AXR3 (IAA17), and AXR5 (IAA1). These proteins interact with DNA-binding auxin response factor transcription factors to regulate auxin-responsive genes (Guilfoyle and Hagen, 2001; Liscum and Reed, 2002). Their degradation results in derepression of auxin-regulated genes that in turn leads to the physiological and morphological events associated with auxin action. Specific ubiquitination of the proteins targeted for degradation is accomplished by a Skp1-cullin-F-box protein (SCF) ubiquitin E3 ligase complex (Gray et al., 2001; Risseeuw et al., 2003). The SCF components implicated in the Arabidopsis auxin response have been identified as ASK1, AXR6, CUL1, and TIR1, respectively (Gray et al., 1999). The cullin portion of the complex requires an additional subunit, the RING-finger protein RBX1 (Gray et al., 2002), and modification by the ubiquitin-related protein, RUB1, to be fully functional (del Pozo and Estelle, 1999). A variety of proteins responsible for the conjugation and deconjugation of RUB1 have been identified as affecting auxin response and these include AXR1 (Leyser et al., 1993; del Pozo et al., 2002). The functional SCF complex recruits an E2 ubiquitin ligase via the cullin subunit to ultimately catalyze the ubiquitination reaction of the target protein.
Proteins targeted for ubiquitination are recruited to the SCF complex via interaction with the F-box protein and this subunit is thought to confer a significant level of specificity to the action of the SCF complex (Leyser, 2002; Dharmasiri and Estelle, 2004). The first F-box protein implicated in 2,4-D and IAA-mediated SCF function was TIR1, a member of the Leu-rich repeat (LRR) subclass of the large F-box protein family in Arabidopsis (Ruegger et al., 1998; Gagne et al., 2002). The SCFTIR1 complex has been shown to interact with target Aux/IAA proteins in response to 2,4-D and IAA treatment in both in vivo and in vitro systems (Gray et al., 2001; Dharmasiri et al., 2003; Kepinski and Leyser, 2004), thus directly confirming the important biochemical role of this complex in the action of these auxinic compounds. Most significantly, TIR1 has recently been shown to act directly as an auxin receptor because it can bind and functionally respond to IAA and 2,4-D (Dharmasiri et al., 2005a; Kepinski and Leyser, 2005). Three close homologs of TIR1, called auxin signaling F-box protein 1 (AFB1), AFB2, and AFB3, have also been shown to participate in the auxin response (Dharmasiri et al., 2005b). Mutants that are deficient in all four of these proteins are auxin insensitive and exhibit severe developmental and morphological phenotypes. Thus, these four homologs are thought to collectively modulate the plant response to IAA and 2,4-D (Dharmasiri et al., 2005b).
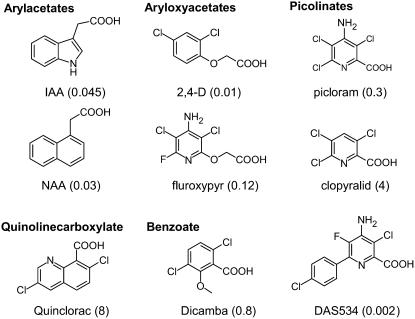
These discoveries have clearly demonstrated the utility of 2,4-D as a chemical surrogate for IAA to dissect the auxin response. 2,4-D has been in commercial use as an herbicide for more than 50 years and is still a staple of many effective herbicide treatments. Further herbicide research has led to the discovery and development of at least 19 commercial herbicides (http://www.plantprotection.org/HRAC/moa2001.htm) and considerably more experimental compounds that act via the auxin mode of action. These comprise at least four structurally distinct classes that include aryloxyacetates, such as 2,4-D, picolinates, benzoates, and quinoline carboxylates, examples of which are shown in Figure 1. These herbicidal auxins exhibit a broad range of potencies and differential selectivities to various plant species (Sterling and Hall, 1997). Despite their structural differences, many studies have shown that these synthetic auxins induce similar morphological, physiological, and molecular events (Chang and Foy, 1983; Boerjan et al., 1992; Grossmann et al., 1996; Dargeviciute et al., 1998; Pufky et al., 2003). However, it is presently unclear whether these structurally different auxinic compounds act via the same mechanisms and signal transduction components that have been characterized for 2,4-D and IAA.
Figure 1.
Chemical structures of IAA, several commercial auxinic herbicides, and the experimental herbicide DAS534. The concentrations of herbicide (μm) required to produce a growth reduction of Arabidopsis roots by 50% relative to untreated controls (GR50) in our assay system are shown in parentheses.
The diversity of synthetic auxins provides a useful suite of compounds to apply a chemical genetic approach to dissecting auxin responses (Blackwell and Zhao, 2003; Zhao et al., 2003; Armstrong et al., 2004). We have screened for Arabidopsis mutations that confer differential resistance to 2,4-D and picolinate auxins using a novel picolinate auxin as an initial probe. We reasoned that if IAA, 2,4-D, and picolinate auxinic compounds all promote common downstream events, as suggested by the similar physiological and morphological responses to the compounds (Chang and Foy, 1983; Boerjan et al., 1992; Grossmann et al., 1996; Dargeviciute et al., 1998; Pufky et al., 2003), then identification of mutants that are selectively resistant to picolinate chemistries could preferentially identify mutations involved in upstream events close to or at the site of chemical interaction. Using this strategy, we have succeeded in isolating and characterizing several mutant alleles at two genetic loci that confer selective resistance to the picolinate class of herbicidal auxins. Our results indicate that picolinate auxins appear to act via the SCF ubiquitination mechanism (like 2,4-D and IAA), but some different upstream components are involved in their action. Significant chemical selectivity therefore resides within these components. These studies also provide chemical genetic tools to differentiate and dissect events in the auxin pathway and to distinguish the action of at least two classes of synthetic auxins.
RESULTS
Structure and Potency of a Novel Picolinate Auxin
One of the symptoms of treatment of Arabidopsis seedlings with natural or synthetic auxins is inhibition of root growth (Estelle and Somerville, 1987). This provides a convenient and quantitative measurement of auxin potency. In our assay system, seedlings were germinated and grown on auxin-containing medium and so were continuously exposed to auxin. The concentrations producing 50% reduction in Arabidopsis root growth (GR50) for a variety of synthetic auxins and for the natural auxin IAA were determined using this method. We found that a novel picolinate auxin, 4-amino-3-chloro-6-(4-chlorophenyl)-5-fluoro-pyridine-2-carboxylic acid (DAS534), had a GR50 of 2 nm, which was 5-fold lower than that of 2,4-D and 22-fold lower than that of IAA (Fig. 1). DAS534 was also more potent on Arabidopsis than several hundred synthetic auxin analogs from the Dow AgroSciences chemical files (data not shown). DAS534 contains a 6-phenyl ring substitution on the picoline ring (Fig. 1) and is a member of a series of 6-aryl-4-aminopicolinate auxinic herbicides (Balko et al., 2004). Because of the significant differences in both structure and potency of DAS534 relative to 2,4-D and IAA, we undertook a chemical genetic screen to assess whether we could recover mutants that were selectively resistant to DAS534 and not resistant to 2,4-D and IAA.
Identification and Genetic Characterization of Mutants Resistant to DAS534
DAS534-resistant plants were isolated by screening ethyl methanesulfonate (EMS)-mutagenized M2 Columbia-0 (Col-0) seedlings growing on agarose media containing 10 nm DAS534. This concentration was sufficient to produce marked auxinic effects on the seedlings, including significant inhibition of root growth. From a total of 780,100 EMS M2 seedlings, 125 putative resistant mutants were identified by visual inspection of root length. Seeds were recovered from 33 of these plants and the phenotype confirmed in secondary screens. We then selected lines that showed low or no cross-resistance to 0.045 μm 2,4-D. Seven lines resistant to DAS534 had negligible resistance to 2,4-D at this concentration and were further evaluated. One additional line (R090) showing robust resistance to both DAS534 and 2,4-D was also retained for comparison.
To determine the inheritance pattern of the mutations and the number of loci represented, the lines were crossed with each other, with wild-type Col-0 plants, and with the known auxin-resistance mutant axr1-3. Progeny of the crosses were analyzed for resistance to DAS534. All of the mutations were recessively inherited. The mutant R090 showing no auxin selectivity appeared to be allelic with axr1-3 and was not examined further. The other seven selectively resistant mutant lines fell into two complementation groups. Based on our subsequent identification of the genes in which the mutations occurred, the mutations in one complementation group were named afb5-1, afb5-2, afb5-3, and afb5-4. The mutants in the other complementation group were named sgt1b-4, sgt1b-5, and sgt1b-6. All of these mutant lines were phenotypically normal when grown on agarose medium or in soil and had normal fertility. Mutant seedlings exhibited normal gravitropism and had normal morphology under etiolating conditions. The sgt1b mutants typically exhibited slightly longer roots (approximately 15%) than wild-type plants when grown on agarose medium lacking herbicide.
Chemical Selectivity of Resistance
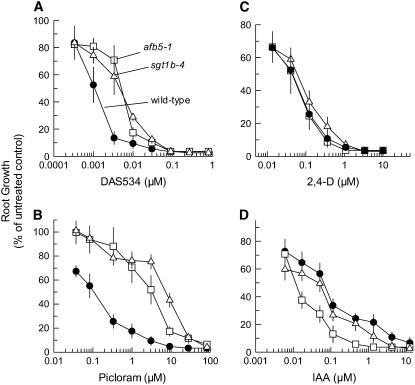
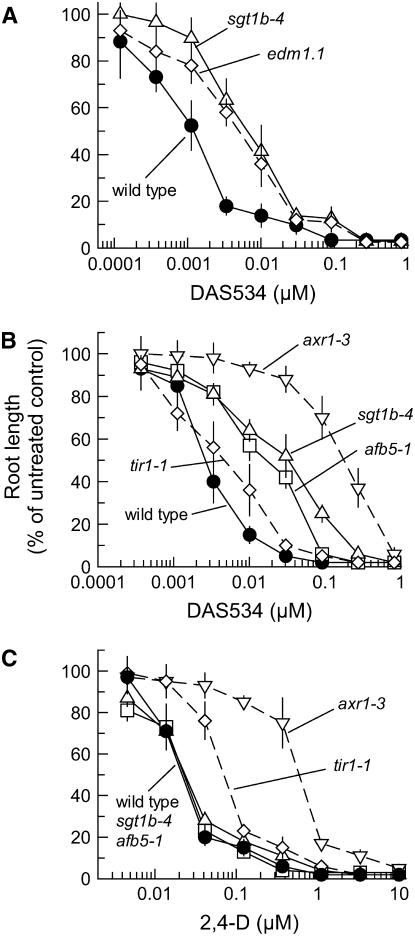
The dose responses of root growth of the mutant lines treated with a variety of auxins were measured to characterize the level and chemical spectrum of resistance in detail. Both afb5 and sgt1b mutants had 6- to 8-fold resistance to DAS534 (Fig. 2A). No significant differences in resistance between lines within each complementation group were noted (data not shown). The mutants were very cross-resistant to the picolinate auxin herbicide picloram (Fig. 2B), showing 26- to 60-fold increases in GR50 over that of wild type. The sgt1b mutants were slightly more resistant than the afb5 mutants. In contrast to the response to DAS534 and picloram, the lines from both complementation groups showed negligible resistance to 2,4-D (Fig. 2C) and a slight increase in sensitivity to IAA (Fig. 2D).
Figure 2.
Differential effects of auxins on root growth of mutants from complementation groups 1 and 2. Root lengths of Arabidopsis seedlings were measured 8 d after seeding on plates containing a range of concentrations of the test compounds. Data are plotted as a percentage of the root length of untreated plants in the same experiment. Root lengths (±sd) for untreated controls were 14.9 (±2.3), 13.9 (±1.3), and 19.4 (±1.7) mm for wild-type, afb5, and sgt1b plants, respectively. The symbols used for wild-type plants, afb5-1, and sgt1b-4 are denoted in A. Treatments are DAS534 (A), picloram (B), 2,4-D (C), and IAA (D). Error bars represent the sd from five individual root measurements.
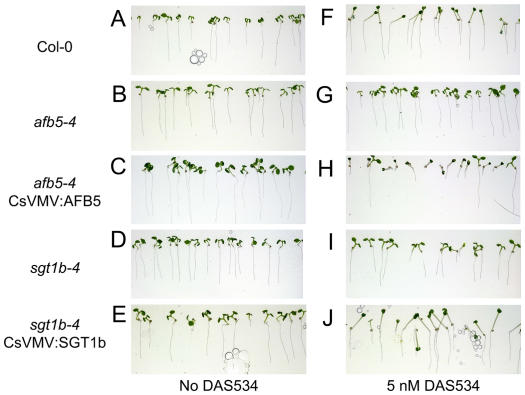
Wild-type, afb5, and sgt1b seedlings growing on 5 nm DAS534 are shown in Figure 3. At this low concentration of DAS534, wild-type plants have elongated hypocotyls and are unable to fully expand their cotyledons (Fig. 3, A and F), whereas hypocotyl elongation and cotyledon expansion of afb5-4 and sgt1b-4 are unaffected by DAS534 (Fig. 3, B, D, G, and I). Four additional herbicidal compounds were also tested on afb5-4. afb5-4 had 50-fold resistance to the picolinate auxinic herbicide clopyralid. However, it showed no resistance to the benzoate auxin dicamba or to a close analog of the aryloxyacetate auxin, fluroxypyr, or to 1-naphthylacetic acid. It also exhibited no difference in response to the auxin transport inhibitor, naphthylphthalamic acid, compared to wild type (data not shown).
Figure 3.
DAS534-resistant mutants and complementation with AFB5 and SGT1b. A to E, Untreated seedlings. F to J, Seedlings grown in the presence of 5 nm DAS534. The primary visible effects at this sublethal concentration of DAS534 are loss of cotyledon decurvature and increased hypocotyl elongation (F). afb5-4 and sgt1b-4 are resistant to this effect (G and I), whereas mutants transformed with the corresponding wild-type gene regain the response (H and J). The afb5-4 CsVMV:AFB5 line (H) appears to have an increased response relative to Col-0 (F).
A foliar spray application of picloram at 200 g/ha on wild-type Col-0 Arabidopsis plants growing in the greenhouse produced profound morphological effects typical of auxinic herbicides and severely inhibited plant growth, whereas the application had minimal effect on afb5-1 (Fig. 4). This is in contrast to the effect of 2,4-D, which induces auxinic symptoms and severe growth reduction to a similar extent on both wild-type and mutant plants at 50 g/ha. Thus, the chemical selectivity of resistance seen in seedling root assays is maintained in adult plants with foliar exposure to the auxin herbicides.
Figure 4.
Selective resistance of afb5-1 to foliar application of picloram. Seventeen-day-old Col-0 and afb5-1 Arabidopsis rosettes growing in the greenhouse were sprayed with picloram at 200 g/ha (top) or 2,4-D at 50 g/ha (bottom). Photographs were taken 12 d after treatment.
Mapping and Identification of Picolinate Auxin-Resistance Mutations
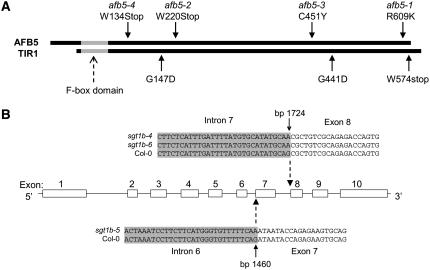
The afb5 and sgt1b mutants displayed clear chemical selectivity in their resistance profiles toward the picolinate class of auxin herbicides. To identify the genes involved in this chemical selectivity, one mutation from each complementation group (afb5-1 and sgt1b-4) was genetically mapped. The mutation in afb5-1 was mapped to a 200-kb interval at around 105 cM on chromosome 5 (see Supplemental Fig. S1). This interval contains 47 genes, including At5g49980, annotated by The Arabidopsis Information Resource (TAIR) as a homolog of TIR1. TIR1 is an F-box protein involved in 2,4-D and IAA-mediated SCF function and has recently been shown to be a receptor for auxin (Dharmasiri et al., 2005a; Kepinski and Leyser, 2005). tir1 mutants are resistant to 2,4-D and IAA (Ruegger et al., 1998). Because of this possible linkage to auxin function, At5g49980 was PCR amplified from genomic DNA from the afb5 mutants and Col-0 plants and compared by DNA sequence analysis. All four afb5 lines contained G-to-A mutations in At5g49980 relative to the Col-0 sequence, resulting in changes in the encoded polypeptide. The mutations in the deduced amino acid sequences from afb5-2 and afb5-4 introduce stop codons at amino acid residues 220 and 134, respectively. The mutations in afb5-1 and afb5-3 introduce amino acid changes in the encoded polypeptide sequence, producing an R-to-K mutation at position 609 in afb5-1 and a C-to-Y mutation at position 451 in afb5-3 (Fig. 5; see multiple sequence alignment in Supplemental Fig. S2).
Figure 5.
Mutation sites in AFB proteins and SGT1b that confer resistance to picolinate auxins. A, Relative positions of the mutations conferring auxin resistance in the polypeptide sequences of AFB5 (this work) and TIR1 (Ruegger et al., 1998; Alonso et al., 2003) were determined by alignment using Clustal within the Vector NTI suite (see Supplemental Fig. S2). The F-box domain is denoted in gray. Note that AFB5 contains a 46-residue N-terminal extension relative to TIR1. A similar length N-terminal extension is also found in AFB4, but not in the four AFBs associated with the IAA and 2,4-D response (Dharmasiri et al., 2005b). B, Exons of SGT1b are shown as boxed regions. Sites of mutations occur at the 3′ splicing site of intron 6 in sgt1b-5 and intron 7 in sgt1b-4 and sgt1b-6 of the gene. The nucleotide sequences of the 3′ intron-exon boundaries are shown. The intron portion is shaded gray and the mutation sites are arrowed.
There are six members of the F-box protein subclass that includes TIR1 in the Arabidopsis genome (Gagne et al., 2002). The three closest homologs of TIR1 have been named AFB1, AFB2, and AFB3, and they have been shown collectively to regulate the response to IAA and 2,4-D (Dharmasiri et al., 2005b). For consistency, we have denoted the two remaining homologs (At4g24390 and At5g49980) as AFB4 and AFB5, respectively. To confirm that the picolinate auxin-resistance phenotype was caused by the identified lesions in AFB5, the auxin-resistance phenotype was complemented by transformation of the afb5-4 mutant line with a construct containing a wild-type copy of AFB5 driven by the cassava vein mosaic virus (CsVMV) promoter (Verdaguer et al., 1998). Ten independent transformants were identified that restored auxin sensitivity to the mutant (Fig. 3H). Some of these transformants appeared to be hypersensitive to DAS534 because their roots were significantly shorter than those of wild-type plants when grown on herbicide-containing media. These data confirm that the mutation in AFB5 is responsible for the resistance phenotype of this complementation group. Because the mutants are resistant to picolinate auxins, and not to 2,4-D, and all contain lesions in the Leu-rich F-box protein AFB5, this protein appears to be specifically involved in mediating the effects of picolinate auxins.
The mutation in sgt1b-4 was mapped to a 100-kb interval on chromosome 4 at around 36 cM (see Supplemental Fig. S1). This interval contains 17 genes, including At4g11260, which encodes the tetratricopeptide repeat-containing protein SGT1b. A mutation in SGT1b has been shown to enhance the level of resistance to 2,4-D in the tir1 mutant background (Gray et al., 2003). The deletion mutant line edm1 lacks seven genes (At4g11220–At4g11280), including SGT1b (Tor et al., 2002). We found that this deletion mutant had a similar level of resistance to DAS534 as sgt1b-4 (Fig. 6A). It was also very cross-resistant to picloram (20-fold; data not shown) similar to the sgt1b mutants. To determine whether the DAS534-resistant plants in this complementation group contained mutations in SGT1b, At4g11260 was PCR amplified from genomic DNA from all three mutants. Sequence analysis showed that all three mutants contained lesions producing alterations of the predicted 3′ intron splice sites of At4g11260. Mutants sgt1b-4 and sgt1b-6 contained G-to-A transitions that disrupt the predicted 3′ splice site of intron 7, whereas mutant sgt1b-5 contained a G-to-A transition that disrupts the 3′ splice site of intron 6 (Fig. 5B).
Figure 6.
Comparison of the effect of DAS534 and 2,4-D on previously characterized mutants. Measurements were performed as described in Figure 2. Root lengths (±sd) for untreated controls were 21.5 (±1.8), 21.2 (±3.2), 24.1 (±3.1), 20.0 (±2.1), 25.0 (±2.1), and 31.5 (±3.1) mm for Col-0, afb5-1, sgt1b-4, edm1, tir1-1, and axr1-3 plants, respectively. A, Effect of DAS534 on edm1 and sgt1b-4. The deletion mutant line edm1 lacks seven genes (At4g11220–At4g11280), including SGT1b (Tor et al., 2002). Effect of DAS534 (B) and 2,4-D (C) on axr1-3 and tir1-1 compared with afb5-1 and sgt1b-4. Symbol labeling is shown in B and is the same for C.
To confirm that the mutations in At4g11260 were responsible for the DAS534-resistant phenotype, the mutant sgt1b-4 was transformed with a construct containing the wild-type At4g11260 gene driven by the constitutive CsVMV promoter (Verdaguer et al., 1998). T2 seeds were tested for DAS534 sensitivity. Using the same process as was employed for identification of AFB5 transformants, 10 independent transformants were identified that were sensitive to DAS534, confirming that the lesions in At4g11260 are responsible for the resistance to DAS534. An example of an sgt1b-4 line containing the transgene that restores sensitivity to DAS534 is shown in Figure 3J.
Comparison with Other Auxin-Resistant Mutants
The mutations we identified in AFB5 and SGT1b conferring picolinate-selective resistance implicate the SCF ubiquitin ligase complex in the molecular mode of action of these herbicides. Several other mutants associated with this complex (e.g. tir1, axr1) or its ubiquitination targets (e.g. axr2) in the auxin response have been characterized and possess varying phenotypes and levels of resistance to 2,4-D or IAA (Lincoln et al., 1990; Ruegger et al., 1998; Nagpal et al., 2000). However, their chemical resistance profile to picolinate auxins has not been previously established so their relative resistance to 2,4-D, DAS534, and picloram was evaluated.
There was no significant chemical selectivity in axr1-3 because it had a high level of resistance to DAS534 as well as to 2,4-D (Fig. 6, B and C). It was also resistant to picloram (Table I). The relative degree of resistance of axr1-3 to the picolinate auxins was somewhat higher than that of afb5 and sgt1b mutants, indicating that axr1 has a greater overall effect on auxinic responses. This is consistent with the general role of AXR1 in the activation of SCF complexes by rubinylation (Schwechheimer et al., 2002). tir1-1 mutants have significantly lower levels of resistance to 2,4-D than axr1 (Ruegger et al., 1998; Fig. 6C), consistent with the findings that additional SCF complex components (AFB1–AFB3) are involved in response to this synthetic auxin (Dharmasiri et al., 2005b). We found that the modest level of resistance of tir1-1 to DAS534 was similar to that of 2,4-D (Fig. 6, B and C; Table I). Thus, the marked chemical selectivity we observed with the afb5 and sgt1b mutants was not apparent with this mutant. The tir1-1 mutant was slightly more resistant to picloram than DAS534, but this resistance was considerably less than the strong resistance of afb5 or sgt1b mutants to this compound (Table I). The resistance levels of tir1-1 and axr1-3 to 2,4-D in our study were similar to those previously described (Estelle and Somerville, 1987; Ruegger et al., 1998), indicating that the results we obtained in our assay system are comparable to previous work. AXR2 is one of the short-lived Aux/IAA transcriptional regulators targeted for SCF-mediated ubiquitination and mutations in this locus have been found to confer a dominant 2,4-D resistance phenotype (Wilson et al., 1990; Nagpal et al., 2000). The level of resistance of axr2-1 to DAS534, picloram, and 2,4-D was similar (Table I), so there was also no pronounced chemical specificity in the synthetic auxin resistance of this mutant.
Table I.
Resistance of afb5 and sgt1b and three other auxin-resistant mutants to various herbicidal auxins
Root lengths were measured as described in Figure 2. The fold increase in resistance was calculated by dividing the mutant root GR50 by the wild-type GR50. ND, Not determined.
| Mutants | Fold Increase in Resistance over Wild Type
|
|||
|---|---|---|---|---|
| DAS534 | Picloram | 2,4-D | IAA | |
| afb5 | 5.6 | 26 | 1.5 | 0.4 |
| sgt1b | 8.4 | 60 | 1.8 | 0.7 |
| axr1-3 | 80 | 160 | 28 | ND |
| tir1-1 | 2.0 | 7.7 | 2.8 | ND |
| axr2-1 | 6.7 | 5.0 | 7.5 | ND |
Resistance Phenotype of afb5 sgt1b Double Mutants
Mutations in either AFB5 or SGT1b cause similar levels of resistance to DAS534. Plants containing homozygous mutations in both AFB5 and SGT1b were generated to determine whether the level of resistance to DAS534 was similar or increased over that of the single-mutant lines. Three double-mutant lines were compared with the single-mutant parental lines for resistance to DAS534, picloram, and 2,4-D. The single and double mutants exhibited similar levels of resistance (5- to 10-fold; data not shown); thus, the resistance mechanisms in afb5 and sgt1b are not additive. The double mutants also exhibited no obvious deleterious phenotype in growth or fertility, similar to the single mutants. This result suggests that AFB5 and SGT1b are involved in the same response pathway to DAS534.
DISCUSSION
Chemically Selective Auxin Resistance
We have used a novel picolinate auxin to uncover mutants in two loci that exhibit chemically selective auxin resistance. Both loci encode components associated with the SCF ubiquitination pathway, one of which is a homolog of the IAA and 2,4-D receptor protein. This pathway has been shown to be the signal transduction mechanism for plant responses to 2,4-D and IAA (Leyser, 2002; Dharmasiri and Estelle, 2004). Although other auxin-resistant mutants such as rib1, aux1, and eir1 have been observed to have some chemical specificity, their differential resistance is due to the specificity of pumps and cellular uptake mechanisms (Yamamoto and Yamamoto, 1998; Marchant et al., 1999; Poupart and Waddell, 2000) rather than in components of auxin signal transduction. None of the three examples of mutants within the auxin signal transduction pathway that we tested, tir1, axr1, and axr2, displayed the marked chemical selectivity in responses that we observed with afb5 and sgt1b. Thus, the mutants we have identified appear to be unique in showing differential resistance in the signal transduction pathway. This indicates that there are significant differences in chemical perception of synthetic auxins within upstream components of the pathway. This may contribute to subtle variations in the effects of different synthetic auxins and suggests that they do not necessarily have equivalent downstream effects. For example, some evidence for differential gene expression in etiolated Arabidopsis seedlings treated with 2,4-D, IAA, and picloram has been found (Pufky et al., 2003).
AFB Redundancy and Interaction with Auxins
Several studies have indicated that the F-box protein TIR1 acts as the recognition component of the auxin-responsive SCF complex and interacts with certain Aux/IAA proteins to target them for ubiquitination and proteasomal degradation (Gray et al., 2001; Dharmasiri et al., 2003; Kepinski and Leyser, 2004). The five homologs of TIR1 in the Arabidopsis genome share 45% to 87% amino acid identity (see phylogenetic tree in Supplemental Fig. S2). The three proteins that are most closely related to TIR1 (61%–72% amino acid identity) have been named AFB1, AFB2, and AFB3 and appear to have similar and overlapping roles to TIR1 (Dharmasiri et al., 2005b). Combinations of mutations in these four genes show increasingly severe 2,4-D resistance phenotypes such that the quadruple mutant is insensitive to 2,4-D (Dharmasiri et al., 2005b). No characterization of the roles of the more distantly related AFB4 and AFB5 (45% and 46% amino acid identity to TIR1, respectively) has been described, to our knowledge. Our data show that mutations in AFB5 confer selective resistance to picolinate auxins, especially picloram, while conferring very low or no cross-resistance to 2,4-D and IAA. This suggests that a significant proportion of plant responses to picolinate auxins is mediated via an SCFAFB5 ubiquitin ligase. This is exemplified by the observation that adult afb5 plants have normal sensitivity to treatment with herbicidal levels of 2,4-D, but are quite resistant to picloram (Fig. 4). The only remaining AFB without phenotypic characterization is AFB4, a close homolog of AFB5 (80% amino acid sequence identity). Both AFB4 and AFB5 contain an N-terminal extension of 46 residues relative to TIR1 and other AFBs (Fig. 4; alignment in Supplemental Fig. S2). The similarity of AFB4 and AFB5 may indicate that AFB4 could also interact with picolinate auxins.
Recent studies have indicated that IAA and 2,4-D can directly bind to the TIR1 F-box protein of the SCF complex to increase its affinity for the target Aux/IAA proteins (Dharmasiri et al., 2005a; Kepinski and Leyser, 2005). AFB1, AFB2, and AFB3 are also implicated in the response to IAA and 2,4-D (Dharmasiri et al., 2005a, 2005b). Our data are consistent with a similar mechanism for picolinate auxins that involves AFB5. The SCFAFB5 ubiquitin ligase complex may be readily modulated by picolinate auxins and not by 2,4-D. Because the tir1 mutant we tested showed slight resistance to the picolinate auxins, as well as to 2,4-D (Fig. 5, B and C), SCFTIR1 may mediate some portion of the response to all of these compounds. This could account for the 7-fold resistance of tir1-1 to picloram (Table I). The high nonselective level of resistance of axr1 to all of the auxins we tested indicates that multiple SCFs are involved in both 2,4-D and picolinate auxin responses, consistent with AXR1 activating a wide range of SCF complexes via rubinylation (Schwechheimer et al., 2002). However, the relative contribution of each SCF complex to the response may vary depending on the auxin chemistry applied.
The afb5 mutant alleles exhibited a high level of resistance (approximately 30-fold) to picloram, whereas they had relatively less resistance (approximately 6-fold) to the more potent auxin DAS534. One rationale for the increased auxinic potency of DAS534 may be that it interacts strongly with all AFBs, whereas picloram may interact primarily via AFB5. Similarly, IAA and 2,4-D appear to act primarily via TIR1 and the homologous AFB1, AFB2, and AFB3 (Dharmasiri et al., 2005a). Plant responses to IAA may not be significantly mediated by SCFAFB5 because none of the afb5 alleles gave resistance to IAA and the mutants had no obvious endogenous auxin-related phenotypes as seedling or adult plants as do many IAA/2,4-D-resistant mutants (Woodward and Bartel, 2005). Indeed, the slight hypersensitivity of afb5 alleles to IAA may indicate that some indirect negative interaction could occur. Differences in response could arise from a lower affinity of AFB5 for IAA and 2,4-D (relative to TIR1 and AFB1, AFB2, or AFB3), thus leading to a significantly higher threshold for response. Additional biochemical assays of SCFAFB function using the appropriate components (Dharmasiri et al., 2003) will be required to determine the relative affinities of the AFBs for various auxins. The functional diversity and variations in response of AFBs to various auxins could also be affected by differences in the time and location of expression of the AFB proteins. Detailed analysis of the relative expression of the AFB gene family may help elucidate this. An alternative and intriguing explanation of the lack of cross-resistance of afb5 plants to IAA is that AFB5 interacts with another undetermined endogenous small molecule. Herbicidal picolinate auxins may therefore mimic the activity of this compound rather than IAA.
The LRR F-box proteins are composed of an N-terminal F-box that interacts with other components of the SCF complex and a C-terminal LRR domain that interacts with proteins targeted for ubiquitination (Gray et al., 1999; Gagne et al., 2002). The four resistance mutations in AFB5 are all in the LRR domain of the protein, two of which introduce stop codons into the N-terminal portion of the domain and two of which introduce amino acid changes into the C-terminal region. We did not detect any phenotypic difference between the four alleles, indicating that a highly conservative amino acid substitution at the extreme C terminus of the protein (R609K) has the same effect as truncations at the N terminus (W134stop, W220stop). Both sites of amino acid substitutions, Arg-609 and Cys-451, are completely conserved in all six TIR1-related AFBs, suggesting that they are essential for auxin-related F-box protein functionality (Supplemental Fig. S2). The three characterized tir1 alleles are also point mutations in the C-terminal LRR domain in amino acids that are conserved in the six TIR1-related AFBs (Fig. 5A), rather than in the N-terminal F-box domain (Ruegger et al., 1998; Alonso et al., 2003). As TIR1 has been shown to directly bind auxins (Dharmasiri et al., 2005a; Kepinski and Leyser, 2005), the residues conferring auxin resistance in TIR1 and AFB5 may be close to the site of chemical interaction.
Role of SGT1b in Auxin Action
The second locus that we identified conferring picolinate-specific auxin resistance encodes SGT1b, a tetratricopeptide-repeat protein that is highly conserved throughout eukaryotes (Lingelbach and Kaplan, 2004). In yeast (Saccharomyces cerevisiae), it is an essential gene that physically associates with a variety of multiprotein complexes, including SCF ubiquitin ligase. It has been shown to interact in substoichiometric amounts with Skp1p (the cullin scaffold protein) of the SCF complex, but is not required for in vitro ubiquitin ligase enzymatic activity (Kitagawa et al., 1999). Genetic evidence has indicated that it may be involved in promoting association and turnover of SCF and other complexes with the appropriate target (Lingelbach and Kaplan, 2004). In plants, SGT1b has recently been shown to be a component of R gene-mediated disease resistance, and mutant plants lacking SGT1b have increased susceptibility to certain plant pathogens (Muskett and Parker, 2003). In addition, a mutation that increases the rather weak auxin resistance of tir1-1 (eta3) was recently discovered to be an allele of SGT1b (Gray et al., 2003). The eta3 mutants showed decreased degradation of Aux/IAA proteins in response to IAA, suggesting that SGT1b is required for some elements of auxin-mediated SCF function.
Our data indicate that the effect of mutations in SGT1b has a significantly greater effect on resistance to picolinate auxins than on resistance to 2,4-D or IAA. The level of resistance of the three sgt1b alleles (and the deletion mutant edm1) to DAS534 and picloram was approximately 8-fold and 60-fold, respectively, whereas there was negligible resistance to 2,4-D (approximately 1.7-fold). Gray et al. (2003) observed a modest level of resistance to 2,4-D (approximately 2-fold) in a sgt1b mutant that increased to 7-fold resistance in the sgt1b tir1-1 double mutant. In contrast, we observed no difference in picolinate-resistance of sgt1b afb5 double mutants relative to that of the single mutants. This may indicate that SCFAFB5 function has a greater dependency on SGT1b than SCFTIR1 such that loss of function of either SGT1b or AFB5 is equivalent in terms of picolinate resistance, whereas both SGT1b and TIR1 functions need to be attenuated to manifest significant resistance to 2,4-D. SGT1b may alter the affinity of the various SCF complexes for their auxin effector (Gray et al., 2003) It will be of interest to evaluate whether differences in synergy by mutations in SGT1b also occur in combination with mutations in other AFBs. It is also intriguing to speculate whether there is any direct chemical interaction of picolinate auxins with SGT1b, possibly in complex with AFB5 and its targets.
Relevance to Herbicidal Auxin Resistance and Selectivity in the Field
There are now many well-characterized auxin-resistant Arabidopsis mutants (Woodward and Bartel, 2005). However, there are relatively few cases of field resistance developing to herbicidal auxins, especially in comparison to other herbicidal modes of action in widespread use. This lack of resistance development in the field may be due to the redundancy in auxin receptors (AFBs) and other components of the auxin signal response and the fitness penalties that may occur because of mutations in the pathway. However, an isolated occurrence of field resistance to picolinate auxins has been documented in yellow starthistle (Centaurea solstitialis) and appears to have arisen from heavy localized use of picloram. This resistant biotype carries a recessive mutation (Sabba et al., 2003) that confers selective resistance to picloram and clopyralid, but not to 2,4-D, and it also has normal growth and morphology (Fuerst et al., 1996). Thus, the picloram-resistant field biotype appears to have some of the characteristics associated with the mutations that we have identified in Arabidopsis. In contrast, another well-characterized auxin-resistant biotype of wild mustard (Brassica kaber syn. Sinapis arvensis) contains a dominant mutation that confers nonselective resistance to 2,4-D and picloram (Jugulam et al., 2005).
Herbicidal auxins can have varying potencies and selectivities toward various plant species that can be dependent on foliar uptake, translocation, and metabolism of the compound, as well as plant morphology. However, in several cases, this does not completely account for their weed spectrum or potency (Hall and Vanden Born, 1988; Sterling and Hall, 1997; Grossmann, 2000). This suggests that some degree of selectivity may occur at or close to the target site. There is evidence for differential cellular uptake of synthetic auxins by auxin permeases (Delbarre et al., 1996; Imhoff et al., 2000; Hoessel et al., 2005) that could account for some differences in herbicidal efficacy between different auxins. Our present data suggest that some degree of synthetic auxin selectivity and potency may also lie within components of the SCF ubiquitin ligase machinery, including the F-box protein receptors.
Utility of Auxin Diversity in Chemical Genetics
Our work demonstrates the utility of using a differential screen with structurally diverse chemistries to uncover and characterize novel phenotypes and mutations. For example, in this case, the mutations we describe would not emerge from resistance screens using 2,4-D or IAA. The structural diversity within synthetic auxins may have additional utility in carefully designed chemical genetic studies to differentiate and dissect auxin-mediated biological processes. Conversely, better understanding of the molecular interactions of these compounds with the upstream signaling components may lead to new and improved herbicidal molecules.
MATERIALS AND METHODS
Materials
Seeds of tir1-1, axr1-3, and axr2-1 were obtained from the Arabidopsis Biological Resource Center (ABRC). Seeds of edm1 were provided by Dr. Eric Holub (Horticulture Research International). DAS534 was synthesized as described in Supplemental Methods S1. Clopyralid was obtained from the Dow AgroSciences compound files. All other chemical reagents were obtained from Sigma Chemical Company.
Mutant Screens
Sterilized EMS-mutagenized M2 Col-0 seeds (Lehle Seeds) were sterilized by washing four times in 15% commercial bleach, 0.5% SDS solution, then rinsed with sterile water. Seeds were stratified at 4°C for 3 to 4 d. Petri plates (15-cm diameter) were prepared containing 50 mL of modified Murashige and Skoog basal medium (Sigma) with 1 mm MES buffer, 0.8% Suc, 0.42% agarose (SeaKem), and 10 nm DAS534. Sterilized seeds were distributed on top of the medium and 30 mL of medium were then poured on top and allowed to solidify. The plates were moved to a growth chamber at 22°C, illuminated at 75 μE m−2 s−1 on a 16-h-light/8-h-dark cycle for 10 d. Resistant seedlings were identified as plants with long roots and fully expanded cotyledons. Herbicide-resistant seedlings were transferred to petri plates containing Murashige and Skoog medium lacking DAS534 and allowed to recover in the growth chamber for 7 to 10 d. After recovery, seedlings were transferred to soil (HP Promix) and allowed to grow to maturity in a greenhouse at 22°C with supplemented light on a 16-h-light/8-h-dark cycle. Selection of lines for detailed study was based on the strength of resistance, elimination of potential siblings, and health and fertility of adult plants.
Root Growth Assays
Arabidopsis (Arabidopsis thaliana) seedling auxin response assays were performed using growth medium supplemented with 0.8% Suc and 0.4% agarose. Compounds were added to the medium as concentrated solutions dissolved in dimethyl sulfoxide. Controls had the appropriate amount of dimethyl sulfoxide added without inhibitor. Plates were incubated for 8 d at 23°C under continuous fluorescent lighting (50 to 100 μE m−2 s−1). Root measurements were made by carefully extracting individual plants from the medium and measuring the length of the tap root. Five roots were measured at each concentration of compound.
Greenhouse Tests
Arabidopsis seedlings were grown for 2 weeks in a growth chamber (23°C; continuous light at 120–150 μE m−2 s−1), then taken to the greenhouse for 3 d after which plants were sprayed with picloram or 2,4-D using a track sprayer to deliver the appropriate rate. Plants were then grown in the greenhouse for 12 d (22°C under supplemented light with a 14-h-light/10-h-dark cycle) prior to photography and evaluation.
Genetic Analyses and Mapping
F1 seeds from crosses between DAS534-resistant and wild-type lines were tested for resistance to DAS534 by plating the seeds on medium containing 10 nm DAS534. The roots of DAS534-resistant seedlings were as long as seedlings plated on medium lacking the herbicide, whereas DAS534-sensitive roots were approximately 50% shorter.
To generate mapping populations, homozygous afb5-1 and sgt1b-4 M3 plants (in the Col-0 background) were crossed with wild-type Landsberg erecta (Ler). F2 seeds were germinated on medium containing 10 nm DAS534. Plants resistant to the herbicide were identified and allowed to recover on medium without herbicide for 7 d, then transplanted to soil. A single leaf was removed at the rosette stage for genomic DNA isolation. Mapping was performed using single-nucleotide polymorphism (SNP) markers from Cho et al. (1999) or identified by comparing sequences from the Ler Random Sequence Database (The Institute for Genomic Research [TIGR]) with the complete sequence of Arabidopsis Col-0 (TAIR). Verified SNPs were used as molecular markers to map the mutation using a fluorescence polarization SNP-mapping technique (Hsu et al., 2001). All primers were custom synthesized by Operon. All other reagents were supplied in the AcycloPrime-FP SNP detection kit from Perkin-Elmer. Fluorescence polarization was read in a LJL BioSystems Analyst 96-384 meter (Molecular Devices). Information on the markers used in the mapping experiments is listed in Supplemental Table S1. The genes At5g49980 and At4g11260 were PCR amplified from afb5-1 by Pfu Turbo DNA Hotstart DNA polymerase (Stratagene) and sequenced using Applied BioSystems BigDye terminator, version 3.1, cycle-sequencing kit and run on an ABI 3100.
Complementation of Mutant Phenotypes
The gene At5g49980 was PCR amplified from genomic DNA of Col-0 using the primers 5′-CACCAAAAATGACACAAGATCGCTCAGAAATGTC-3′ and 5′-TCCTACACTTACCCATTTCATTCCCG-3′ and the gene At4g11260 similarly amplified using the primers 5′-CACCAAAAATGGCCAAGGAATTAGCAGAGAAA-3′ and 5′-TCGAGCAGAAACCAAAAAAGAAAAACA-3′. The PCR products were cloned into pENTR 1A (Invitrogen) and independently recombined into the binary plant transformation vector pNT4274 using Gateway (Invitrogen) with the strong constitutive CsVMV (Verdaguer et al., 1998) and the nos terminator. The transformation vector also contained a nptII gene as a selectable marker. Vectors were transformed into Agrobacterium strain GV3101 containing the helper plasmid pMP90RK and used to transform either afb5-4 (for the strain containing the At5g49980 vector) or sgt1b-4 (for the strain containing the At4g11260 vector) via the floral-dip procedure (Clough and Bent, 1998). Kanamycin-resistant T1 plants were transplanted to soil and allowed to self-fertilize. T2 seed was tested for susceptibility to DAS534 as described above.
Identification of Double Mutants
Genomic DNA was isolated from 95 F2 plants generated from a cross of afb5-1 and sgt1b-4. The At5g49980 (AFB5) and At4g11260 (SGT1b) genes were amplified by PCR and evaluated by DNA sequence analysis for mutations. Eight plants containing mutations in both genes were identified and progeny from three of them were compared with the single-mutant parental lines for resistance to DAS534, picloram, and 2,4-D.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Map-based cloning of DAS534-resistance mutations.
Supplemental Figure S2. Sequence comparisons of AFBs.
Supplemental Table S1. Synthesis of DAS534.
Supplemental Methods S1. Description of SNP markers used in mapping.
Supplementary Material
Acknowledgments
We thank Cathy Hironaka, Darcie Otter, Michelle Leal, Karin Conners, and Alan Lammers for technical assistance at Exelixis Plant Sciences, Dr. Eric Holub (Horticulture Research International, Wellesbourne, UK) for kindly providing seeds of edm1, and Dr. Max Ruegger (Dow AgroSciences) for helpful comments. We also thank Paul Schmitzer, Andrew Toft, and Cliff Gerwick (Dow AgroSciences) for greenhouse evaluation of mutants, and Terry Balko (Dow AgroSciences) for the description of the synthetic preparation of DAS534.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Terence A. Walsh (tawalsh@dow.com).
The online version of this article contains Web-only data.
References
- Alonso JM, Stepanova AN, Solano R, Wisman E, Ferrari S, Ausubel FM, Ecker JR (2003) Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc Natl Acad Sci USA 100: 2992–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong JI, Yuan S, Dale JM, Tanner VN, Theologis A (2004) Identification of inhibitors of auxin transcriptional activation by means of chemical genetics in Arabidopsis. Proc Natl Acad Sci USA 101: 14978–14983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balko TW, Buysse AM, Epp JB, Fields SC, Lowe CT, Keese RJ, Richburg JS III, Ruiz JM, Weimer MR, Green RA, et al, inventors. August 31, 2004. 6-Aryl-4-aminopicolinates and their use as herbicides. U.S. Patent No. 6784137
- Blackwell HE, Zhao Y (2003) Chemical genetic approaches to plant biology. Plant Physiol 133: 448–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Genetello C, Van Montagu M, Inze D (1992) A new bioassay for auxins and cytokinins. Plant Physiol 99: 1090–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang IK, Foy CL (1983) Rapid growth responses of dwarf corn coleoptile sections to picloram. Pestic Biochem Physiol 19: 203–209 [Google Scholar]
- Cho RJ, Mindrinos M, Richards DR, Sapolsky RJ, Anderson M, Drenkard E, Dewdney J, Reuber TL, Stammers M, Federspiel N, et al (1999) Genome-wide mapping with biallelic markers in Arabidopsis thaliana. Nat Genet 23: 203–207 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dargeviciute A, Roux C, Decreux A, Sitbon F, Perrot-Rechenmann C (1998) Molecular cloning and expression of the early auxin-responsive Aux/IAA gene family in Nicotiana tabacum. Plant Cell Physiol 39: 993–1002 [DOI] [PubMed] [Google Scholar]
- Davies PJ (1995) Plant Hormones: Physiology, Biochemistry and Molecular Biology. Kluwer Academic Publishers, Dordrecht, The Netherlands
- del Pozo JC, Dharmasiri S, Hellmann H, Walker L, Gray WM, Estelle M (2002) AXR1-ECR1-dependent conjugation of RUB1 to the Arabidopsis cullin AtCUL1 is required for auxin response. Plant Cell 14: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Estelle M (1999) The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc Natl Acad Sci USA 96: 15342–15347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbarre A, Muller P, Imhoff V, Guern J (1996) Comparison of mechanisms controlling uptake and accumulation of 2,4-dichlorophenoxy acetic acid, naphthalene-1-acetic acid, and indole-3-acetic acid in suspension-cultured tobacco cells. Planta 198: 532–541 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005. a) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Jones AM, Estelle M (2003) Auxin action in a cell-free system. Curr Biol 13: 1418–1422 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Juergens G, Estelle M (2005. b) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Estelle M (2004) Auxin signaling and regulated protein degradation. Trends Plant Sci 9: 302–308 [DOI] [PubMed] [Google Scholar]
- Estelle MA, Somerville C (1987) Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol Gen Genet 206: 200–206 [Google Scholar]
- Fuerst EP, Sterling TM, Norman MA, Prather TS, Irzyk GP, Wu Y, Lownds NK, Callihan RH (1996) Physiological characterization of picloram resistance in yellow starthistle. Pestic Biochem Physiol 56: 149–161 [Google Scholar]
- Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99: 11519–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M (1999) Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev 13: 1678–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Hellmann H, Dharmasiri S, Estelle M (2002) Role of the Arabidopsis RING-H2 protein RBX1 in RUB modification and SCF function. Plant Cell 14: 2137–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCFTIR1-dependent degradation of Aux/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Gray WM, Muskett PR, Chuang H, Parker JE (2003) Arabidopsis SGT1b is required for SCF(TIR1)-mediated auxin response. Plant Cell 15: 1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann K (2000) The mode of action of quinclorac: a case study of a new auxin-type herbicide. In AH Cobb, RC Kirkwood, eds, Herbicides and Their Mechanisms of Action. Academic Press, Sheffield, UK, pp 181–214
- Grossmann K, Scheltrup F, Kwiatkowski J, Caspar G (1996) Induction of abscisic acid is a common effect of auxin herbicides in susceptible plants. J Plant Physiol 149: 475–478 [Google Scholar]
- Guilfoyle TJ, Hagen G (2001) Auxin response factors. J Plant Growth Regul 20: 281–291 [Google Scholar]
- Hall JC, Vanden Born WH (1988) The absence of a role of absorption, translocation, or metabolism in the selectivity of picloram and clopyralid in two plant species. Weed Sci 36: 9–14 [Google Scholar]
- Hellmann H, Hobbie L, Chapman A, Dharmasiri S, Dharmasiri N, del Pozo C, Reinhardt D, Estelle M (2003) Arabidopsis AXR6 encodes CUL1 implicating SCF E3 ligases in auxin regulation of embryogenesis. EMBO J 22: 3314–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoessel D, Schmeiser C, Hertel R (2005) Specificity patterns indicate that auxin exporters and receptors are the same proteins. Plant Biol 7: 41–48 [DOI] [PubMed] [Google Scholar]
- Hsu TM, Chen X, Duan S, Miller RD, Kwok PY (2001) Universal SNP genotyping assay with fluorescence polarization detection. Biotechniques 31: 560–564 [DOI] [PubMed] [Google Scholar]
- Imhoff V, Muller P, Guern J, Delbarre A (2000) Inhibitors of the carrier-mediated influx of auxin in suspension-cultured tobacco cells. Planta 210: 580–588 [DOI] [PubMed] [Google Scholar]
- Jugulam M, McLean MD, Hall JC (2005) Inheritance of picloram and 2,4-D resistance in wild mustard (Brassica kaber). Weed Sci 53: 417–423 [Google Scholar]
- Kepinski S, Leyser O (2004) Auxin-induced SCFTIR1-Aux/IAA interaction involves stable modification of the SCFTIR1 complex. Proc Natl Acad Sci USA 101: 12381–12386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Kitagawa K, Skowyra D, Elledge SJ, Harper JW, Hieter P (1999) SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol Cell 4: 21–33 [DOI] [PubMed] [Google Scholar]
- Leyser HMO, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M (1993) Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364: 161–164 [DOI] [PubMed] [Google Scholar]
- Leyser HMO, Pickett FB, Dharmasiri S, Estelle M (1996) Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J 10: 403–413 [DOI] [PubMed] [Google Scholar]
- Leyser O (2002) Molecular genetics of auxin signaling. Annu Rev Plant Biol 53: 377–398 [DOI] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M (1990) Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2: 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingelbach LB, Kaplan KB (2004) The interaction between Sgt1p and Skp1p is regulated by HSP90 chaperones and is required for proper CBF3 assembly. Mol Cell Biol 24: 8938–8950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Reed JW (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49: 387–400 [PubMed] [Google Scholar]
- Ljung K, Hull AK, Kowalczyk M, Marchant A, Celenza J, Cohen JD, Sandberg G (2002) Biosynthesis, conjugation, catabolism, and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol Biol 49: 249–272 [PubMed] [Google Scholar]
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ (1999) AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J 18: 2066–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muskett P, Parker J (2003) Role of SGT1 in the regulation of plant R gene signalling. Microbes Infect 5: 969–976 [DOI] [PubMed] [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW (2000) AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol 123: 563–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poupart J, Waddell CS (2000) The rib1 mutant is resistant to indole-3-butyric acid, an endogenous auxin in Arabidopsis. Plant Physiol 124: 1739–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pufky J, Qiu Y, Rao MV, Hurban P, Jones AM (2003) The auxin-induced transcriptome for etiolated Arabidopsis seedlings using a structure/function approach. Funct Integr Genomics 3: 135–143 [DOI] [PubMed] [Google Scholar]
- Risseeuw EP, Daskalchuk TE, Banks TW, Liu E, Cotelesage J, Hellmann H, Estelle M, Somers DE, Crosby WL (2003) Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J 34: 753–767 [DOI] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M (1998) The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev 12: 198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabba RP, Ray IM, Lownds N, Sterling TM (2003) Inheritance of resistance to clopyralid and picloram in yellow starthistle (Centaurea solstitialis L.) is controlled by a single nuclear recessive gene. J Hered 94: 523–527 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Serino G, Deng X (2002) Multiple ubiquitin ligase-mediated processes require COP9 signalosome and AXR1 function. Plant Cell 14: 2553–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling TM, Hall JC (1997) Mechanism of action of natural auxins and the auxinic herbicides. In MR Roe, RJ Kuhr, JD Burton, eds, Herbicide Activity: Toxicology, Biochemistry and Molecular Biology. IOS Press, Amsterdam, pp 111–141
- Tor M, Gordon P, Cuzick A, Eulgem T, Sinapidou E, Mert-Turk F, Can C, Dangl JL, Holub EB (2002) Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell 14: 993–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaguer B, de Kochko A, Fux CI, Beachy RN, Fauquet C (1998) Functional organization of the cassava vein mosaic virus (CsVMV) promoter. Plant Mol Biol 37: 1055–1067 [DOI] [PubMed] [Google Scholar]
- Wilson AK, Pickett FB, Turner JC, Estelle M (1990) A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol Gen Genet 222: 377–383 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Yamamoto KT (1998) Differential effects of 1-naphthaleneacetic acid, indole-3-acetic acid and 2,4-dichlorophenoxyacetic acid on the gravitropic response of roots in an auxin-resistant mutant of Arabidopsis, aux1. Plant Cell Physiol 39: 660–664 [DOI] [PubMed] [Google Scholar]
- Yang X, Lee S, So JH, Dharmasiri S, Dharmasiri N, Ge L, Jensen C, Hangarter R, Hobbie L, Estelle M (2004) The IAA1 protein is encoded by AXR5 and is a substrate of SCFTIR1. Plant J 40: 772–782 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Dai X, Blackwell HE, Schreiber SL, Chory J (2003) SIR1, an upstream component in auxin signaling identified by chemical genetics. Science 301: 1107–1110 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.