Abstract
The two-component system (TCS), which works on the principle of histidine-aspartate phosphorelay signaling, is known to play an important role in diverse physiological processes in lower organisms and has recently emerged as an important signaling system in plants. Employing the tools of bioinformatics, we have characterized TCS signaling candidate genes in the genome of Oryza sativa L. subsp. japonica. We present a complete overview of TCS gene families in O. sativa, including gene structures, conserved motifs, chromosome locations, and phylogeny. Our analysis indicates a total of 51 genes encoding 73 putative TCS proteins. Fourteen genes encode 22 putative histidine kinases with a conserved histidine and other typical histidine kinase signature sequences, five phosphotransfer genes encoding seven phosphotransfer proteins, and 32 response regulator genes encoding 44 proteins. The variations seen between gene and protein numbers are assumed to result from alternative splicing. These putative proteins have high homology with TCS members that have been shown experimentally to participate in several important physiological phenomena in plants, such as ethylene and cytokinin signaling and phytochrome-mediated responses to light. We conclude that the overall architecture of the TCS machinery in O. sativa and Arabidopsis thaliana is similar, and our analysis provides insights into the conservation and divergence of this important signaling machinery in higher plants.
The two-component His kinases (HK) or two-component system (TCS) is known to play important roles in the regulation of prokaryotic and lower eukaryotic cellular responses to environmental stimuli (Grebe and Stock, 1999; Stock et al., 2000; Urao et al., 2000a, 2000b; Besant et al., 2003). These include antibiotic resistance, chemotaxis, differentiation, and nitrogen metabolism. The existence of a bacterial-type HK in plants was first reported by Chang et al. (1993). Since then, many plants have been documented to possess genes encoding two-component regulators, and their participation in the perception and integration of various extracellular and intracellular signals has been shown (Lohrmann and Harter, 2002; Oka et al., 2002; Grefen and Harter, 2004; Hass et al., 2004; for review, see Stock et al., 2000; Foussard et al., 2001).
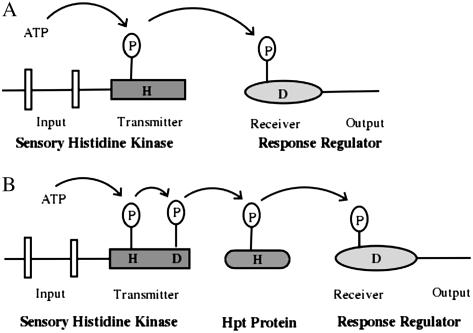
In bacterial systems, the typical machinery, such as that of Escherichia coli Env Z protein (an osmosensor), consists of a membrane-localized HK that senses the input signal and a response regulator (RR) that contains a conserved regulatory domain. In this system, the signaling is initiated when the HK, modulated by the environmental stimulus, autophosphorylates the conserved His residue. The phosphoryl group is then transferred to a conserved Asp residue in the RR, which results in modulation of its activity (Fig. 1A). Autophosphorylation of the HK is a bimolecular reaction between homodimers in which one HK monomer catalyzes the phosphorylation of the His residue in the second monomer (Pan et al., 1993; Surette et al., 1996). Besides directing the forward phosphorylation reaction, many HKs possess a phosphatase activity, enabling them to dephosphorylate their cognate partner (Keener and Kustu, 1988; Lois et al., 1993). The phosphotransfer pathways, which need to be shut down quickly, operate with such bifunctional HKs. More elaborate HKs, also known as hybrid kinases, are typical of eukaryotes and a few prokaryotes and contain multiple phosphodonor and phosphoacceptor sites. Signaling via these pathways uses multiple His phosphotransfers (Hpt) and receiver domains (RDs) or proteins that connect to final RRs or other signaling outputs (Fig. 1B). Examples of such HKs include the SLN1/YPD1/SSK1 phosphorelay in yeast (Saccharomyces cerevisiae), which controls the HOG pathways in response to osmotic stress (Wurgler-Murphy and Saito, 1997). Such a multistep phosphorelay reaction is believed to have the advantage of providing multiple regulatory checkpoints for signal cross talk or negative regulation by certain phosphatases. The existence of multiple phosphorelay reactions in both prokaryotes and eukaryotes suggests that similar mechanisms might be more widely used (Urao et al., 2000a).
Figure 1.
Basic machinery of TCS. A, A simple TCS member. Sensing of an extracellular signal is initiated by the input domain of sensory HK, phosphorylating the conserved His in its transmitter domain. In the next step, the conserved Asp in the RD of the RR is autophosphorylated, resulting in the signal output. B, A hybrid-type TCS in which the conserved His and Asp are found in the same protein, which serves as the sensory HK and is usually membrane bound. The nonmembrane-bound Hpt acts as a mediator for the transfer of the phosphoryl group between the HK and the RR. The TM domains are depicted as vertical bars.
Recently, the TCS signaling machinery has been shown to be present in several plant species, including Arabidopsis (Arabidopsis thaliana; Hwang and Sheen, 2001; Grefen and Harter, 2004). In Arabidopsis, TCS members have been documented to be involved in cytokinin signaling, ethylene signaling, and light perception (Hwang et al., 2002). A member of the family, Arabidopsis HK1 (AtHK1), has been shown to work as a putative osmosensor in the Arabidopsis (Urao et al., 1999). However, the physiological function of most TCS members remains to be elucidated.
With the availability of whole-genome sequences of Oryza sativa and Arabidopsis, we decided to use the tools of bioinformatics to examine the key components of the TCS pathways that are operative in these two model systems. Our analysis is based on the high-quality, finished (The Institute for Genomic Research [TIGR] version 4.0) O. sativa genome sequence (International Rice Genome Sequencing Project, 2005). Genome-wide analyses for TCS signal transduction genes have previously been carried out for fungal pathogens (Catlett et al., 2003) and Arabidopsis (Hwang et al., 2002). In this article, we provide information on the TCS machineries operative in O. sativa and compare them with the similar system of Arabidopsis. Additionally, we have carried out comprehensive analysis of the available massively parallel signature sequencing (MPSS) data as well as the Tos17 database to comment on the expression profile and possible role(s) of the O. sativa TCS members, respectively.
RESULTS AND DISCUSSION
To avoid any ambiguity in nomenclature, we have deliberately opted for naming the various TCS proteins as 1, 2, and 3. In cases where prior information is already available in literature about any of the O. sativa TCS members, the nomenclature has been maintained. However, in case a changed name has been suggested for uniformity purpose, the existing name is also mentioned in parentheses. Further, in some cases, the postscript a, b, or c has been added to represent the products of alternative splicing, as reported by TIGR 4.0 protein pseudo molecule database. Nomenclature of genes has been based on the longest predicted open reading frame (ORF) followed by the number added to the protein. Since the products of alternate spliced genes may also be functionally diverse, we have considered them to represent a separate entity in our multiple sequence alignments; however, for phylogenetic analysis, we have considered only the ones that encode the longest reading frame.
Two-Component Analysis in O. sativa
Our analysis has been carried out with the high-quality, finished O. sativa genome sequence using the HMMER software (for details, see “Materials and Methods”). All the significant hits with positive scores were selected for analysis based on conserved domain search. Initially, we validated the sensitivity and specificity of our methodology using the Arabidopsis genome and compared the output with published reports. We found our results to be in total conformity with those reported by Hwang et al. (2002). Arabidopsis has at least 54 distinct TCS member genes (Hwang et al., 2002). Interrogation of the O. sativa genome identified 51 genes, encoding 73 putative proteins (Fig. 2; Tables I–III), having strong homology with HKs (OsHKs), Hpt proteins (OsHpts), and RRs (OsRRs).
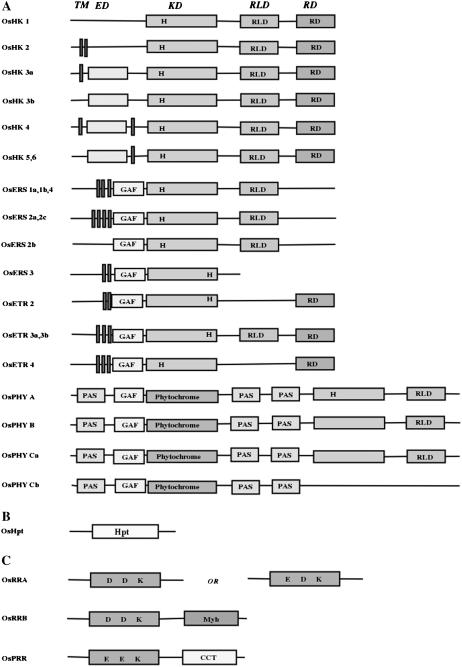
Figure 2.
Representation (unscaled) of the primary domain structures of members of the two-component cell signal system in O. sativa. A, OsHK1 and OsHK2 lack an extracellular domain, while OsHK3, OsHK4, OsHK5, and OsHK6 have similar conserved domains but have TM regions in different positions. OsERS1a, OsERS1b, OsERS4, OsERS2a, and OsERS2c have similar conserved domains, but the latter two have one extra TM region. OsERS2b lacks a TM region. OsERS3 has all conserved domains along with the TM region but lacks the HATPase (RLD) domain. All the OsERSs lack RR domains. OsETR2 and OsETR4 lack HATPase domains, while in OsETR3a and 3b all the domains are present. All the OsETRs have RR domains. OsPHYs, Photosensory proteins in O. sativa show conserved GAF domains, as in ETR/ERS, in addition to the conserved PHY domain (chromophore-binding domain), twin PAS (signal sensor) domains, and a domain showing homology with His protein KD. B, OsHpts, the putative His-containing phosphotransfer proteins. C, RR and PRR proteins. OsRRA, A-type RRs carry only RD and the motifs in RD show variation in two groups of A-type RRs. OsRRB, B-type RR RD and B-motif (DNA-binding motif). OsPRR, PRR protein has atypical RD similar to OsRRB but with E, E, and K motifs. In addition to an RD, it has a CCT domain. Notations: OsHK, putative HK of O. sativa; OsERS, putative ETR without RD of O. sativa; OsETR, putative ETR with RD of O. sativa; OsPHY, PHYs of O. sativa; OsRR, RR of O. sativa; Myb, DNA binding motif.
Table I.
Putative HK and related proteins in O. sativa
Genes identified using HMMER search; nomenclature based on domain conservation. Putative alternate spliced protein products have postscript a, b, or c.
| Gene | Protein Notationa | TIGR IDb | BACc | Accession No.d | Gene Locuse | AAf |
|---|---|---|---|---|---|---|
| OsHK1 | OsHK1 | 11976.m08969 | P0709F06 | AP003579 | LOC_Os06g44410 | 968 |
| OsHK2 | OsHK2 | 11976.m05564 | OSJNBb0036B04 | AP007226 | LOC_Os06g08450 | 1,048 |
| OsHK3 | OsHK3a | 11971.m13041 | P0592G05 | AP004672 | LOC_Os01g69920 | 1,023 |
| OsHK3b | 11971.m42798 | P0592G05 | AP004672 | LOC_Os01g69920 | 866 | |
| OsHK4 | OsHK4 | 11973.m10071 | OSJNBa0078A17 | AC091532 | LOC_Os03g50860 | 1,013 |
| OsHK5 | OsHK5 | 11980.m05192 | OSJNBa0073L01 | AC092548 | LOC_Os10g21810 | 1,186 |
| OsHK6 | OsHK6 | 11972.m10082 | P0684F11 | AP005112 | LOC_Os02g50480 | 997 |
| OsETR2 | OsETR2 | 11974.m06161 | OSJNBb0089K06 | AL663013 | LOC_Os04g08740 | 763 |
| OsERS3 | 11974.m35115 | OSJNBb0089K06 | AL663013 | LOC_Os04g08740 | 670 | |
| OsETR3 | OsETR3a | 11972.m10774 | OJ1119_A01 | AP004020 | LOC_Os02g57530 | 770 |
| OsETR3b | 11972.m33481 | OJ1119_A01 | AP004020 | LOC_Os02g57530 | 836 | |
| OsERS4 | 11972.m33988 | P0474F11 | AP004878 | LOC_Os02g57530 | 549 | |
| OsETR4 | OsETR4 | 11977.m05998 | OJ1370_E02 | AP003756 | LOC_Os07g15540 | 777 |
| OsERS1 | OsERS1a | 11973.m34910 | OJ1212_C08 | AC091670 | LOC_Os03g49500 | 636 |
| OsERS1b | 11973.m09949 | OJ1212_C08 | AC091670 | LOC_Os03g49500 | 636 | |
| OsERS2 | OsERS2a | 11975.m05159 | P0431G05 | AC087551 | LOC_Os05g06320 | 635 |
| OsERS2b | 11975.m27546 | P0431G05 | AC087551 | LOC_Os05g06320 | 359 | |
| OsERS2c | 11975.m27547 | P0431G05 | AC087551 | LOC_Os05g06320 | 518 | |
| OsPHYA | OsPHYA | 11973.m10085 | B1377B10 | AC147426 | LOC_Os03g51030 | 1,128 |
| OsPHYB | OsPHYB | 11973.m07375 | OSJNBa0042B10 | AC137071 | LOC_Os03g19590 | 1,171 |
| OsPHYC | OsPHYCa | 11973.m10374 | OJ1112_G08 | AC135225 | LOC_Os03g54084 | 1,137 |
| OsPHYCb | 11973.m78893 | OJ1112_G08 | AC135225 | LOC_Os03g54084 | 957 |
Notation of putative proteins based on conserved domains.
IDs given to pseudomolecules by TIGR (http://www.tigr.org/tdb/e2k1/osa1/) from the O. sativa annotation database.
BAC IDs from the TIGR database.
BAC accession numbers for genomic DNA sequences deposited in the NCBI database (http://www.ncbi.nlm.nih.gov/).
Gene locus.
The number of predicted amino acid.
Table III.
RR and RR-like proteins in O. sativa
Genes identified using HMMER search; nomenclature based on domain conservation. Putative alternate spliced protein products have postscript a, b, or c.
| Gene | Protein Notationa | TIGR IDb | BACc | Accession No.d | Gene Locuse | AAf |
|---|---|---|---|---|---|---|
| A-type RR | ||||||
| OsRRA1 | OsRRA1a | 11981.m04670 | OSJNBa0059J06 | AC123526 | LOC_Os11g04720 | 201 |
| OsRRA1b | 11981.m28648 | OSJNBa0059J06 | AC123526 | LOC_Os11g04720 | 208 | |
| OsRRA2 | OsRRA2 | 11982.m04444 | OSJNBb0034E23 | AL928752 | LOC_Os12g04500 | 201 |
| OsRRA3 | OsRRA3 | 11971.m13268 | P0431G06 | AP003683 | LOC_Os01g72330 | 232 |
| OsRRA4 | OsRRA4 | 11974.m09387 | OSJNBb0065J09 | AL663010 | LOC_Os04g44280 | 134 |
| OsRRA5 | OsRRA5 | 11977.m06949 | OJ1047_A06 | AP003802 | LOC_Os07g26720 | 206 |
| OsRRA6 | OsRRA6 | 11974.m10657 | OSJNBa0018M05 | AL606457 | LOC_Os04g57720 | 170 |
| OsRRA7 | OsRRA7 | 11972.m10856 | OJ1124_D06 | AP004043 | LOC_Os02g58350 | 131 |
| OsRRA8 | OsRRA8 | 11972.m09243 | B1250G12 | AP006452 | LOC_Os02g42060 | 147 |
| OsRRA9 | OsRRA9 | 11974.m08696 | P0076O17 | BX548156 | LOC_Os04g36070 | 231 |
| OsRRA10 | OsRRA10 | 11972.m08606 | OSJNBb0038F20 | AP005808 | LOC_Os02g35180 | 252 |
| OsRRA11 | OsRRA11 | 11978.m06899 | OJ1705_C03 | AP003962 | LOC_Os08g28950 | 167 |
| OsRRA12 | OsRRA12 | 11978.m06712 | P0488B06 | AP005523 | LOC_Os08g26990 | 135 |
| OsRRA13 | OsRRA13 | 11978.m06895 | OJ1705_C03 | AP003962 | LOC_Os08g28900 | 185 |
| OsRRA14 | OsRRA14a | 11973.m10284 | OJ1365_D05 | AC096855 | LOC_Os03g53100 | 127 |
| OsRRA14b | 11973.m35406 | OJ1365_D05 | AC096855 | LOC_Os03g53100 | 127 | |
| OsRRA15 | OsRRA15 | 11974.m06587 | OSJNBa0083 | AL662982 | LOC_Os04g13480 | 132 |
| OsRRA16 | OsRRA16 | 11975.m07520 | OSJNBb0092G21 | AC134932 | LOC_Os05g32880 | 620 |
| OsRRA17 | OsRRA17 | 11974.m07929 | OSJNBa0020I02 | AL662963 | LOC_Os04g28120 | 252 |
| OsRRA18 | OsRRA18 | 11975.m07521 | OSJNBb0092G21 | AC134932 | LOC_Os05g32890 | 368 |
| OsRRA19 | OsRRA19 | 11978.m07556 | P0493A04 | AP004586 | LOC_Os08g35670 | 614 |
| OsPRR3 | OsRRA20a | 11973.m34842 | OSJNBa0044H10 | AC084405 | LOC_Os03g17570 | 533 |
| OsRRA20b | 11973.m34843 | OSJNBa0044H10 | AC084405 | LOC_Os03g17570 | 473 | |
| OsPRR1 | OsRRA21 | 11972.m33775 | OJ1014_H03 | AP003980 | LOC_Os02g40510 | 386 |
| OsRRA22 | OsRRA22 | 11974.m07933 | OSJNBa0038P21 | AL731588 | LOC_Os04g28160 | 380 |
| B-type RR | ||||||
| OsRRB1 | OsRRB1a | 11973.m06685 | OSJNBa0081P02 | AC107226 | LOC_Os03g12350 | 691 |
| OsRRB1b | 11973.m35102 | OSJNBa0081P02 | AC107226 | LOC_Os03g12350 | 691 | |
| OsRRB1c | 11973.m35103 | OSJNBa0081P02 | AC107226 | LOC_Os03g12350 | 691 | |
| OsRRB2 | OsRRB2 | 11972.m06196 | OJ1297_C09 | AP004087 | LOC_Os02g08500 | 626 |
| OsRRB3 | OsRRB3 | 11976.m08921 | OSJNBa0062J02 | AP003517 | LOC_Os06g43910 | 694 |
| OsRRB4 | OsRRB4 | 11976.m05563 | OSJNBb0036B04 | AP007226 | LOC_Os06g08440 | 696 |
| OsRRB5 | OsRRB5 | 11972.m10558 | OJ1004_E04 | AP003975 | LOC_Os02g55320 | 688 |
| OsRRB6 | OsRRB6 | 11971.m12881 | OSJNOa013M08 | AP006838 | LOC_Os01g67770 | 518 |
| OsRRB7 | OsRRB7 | 11974.m07930 | OSJNBa0020I02 | AL662963 | LOC_Os04g28130 | 390 |
| PRR | ||||||
| OsPRR1 | OsPRR1a | 11972.m09137 | OJ1212_C01 | AP004083 | LOC_Os02g40510 | 518 |
| OsPRR1b | 11972.m33776 | OJ1212_C01 | AP004083 | LOC_Os02g40510 | 411 | |
| OsPRR2 | OsPRR2 | 11979.m06571 | P0515E01 | AP005314 | LOC_Os09g36220 | 623 |
| OsPRR3 | OsPRR3a | 11973.m07182 | OSJNBa0044H10 | AC084405 | LOC_Os03g17570 | 767 |
| OsPRR3b | 11973.m34844 | OSJNBa0044H10 | AC084405 | LOC_Os03g17570 | 745 | |
| OsPRR4 | OsPRR4 | 11977.m09154 | P0627E10 | AP005199 | LOC_Os07g49460 | 742 |
| OsPRR5 | OsPRR5a | 11981.m04789 | OSJNBa0052H02 | AC133217 | LOC_Os11g05930 | 699 |
| OsPRR5b | 11981.m28729 | OSJNBa0052H02 | AC133217 | LOC_Os11g05930 | 623 | |
| OsPRR5c | 11981.m28730 | OSJNBa0052H02 | AC133217 | LOC_Os11g05930 | 623 | |
| OsPRR5d | 11981.m28731 | OSJNBa0052H02 | AC133217 | LOC_Os11g05930 | 620 |
Notation of putative proteins based on conserved domains.
IDs given to pseudomolecules by TIGR (http://www.tigr.org/tdb/e2k1/osa1/) from the O. sativa annotation database.
BAC IDs from the TIGR database.
BAC accession numbers for genomic DNA sequences deposited in the NCBI database (http://www.ncbi.nlm.nih.gov/).
Gene locus.
The number of predicted amino acid.
Table II.
Hpt proteins in O. sativa
Genes identified using HMMER search; nomenclature based on domain conservation. Putative alternate spliced protein products have postscript a, b, or c.
| Gene | Protein Notationa | TIGR IDb | BACc | Accession No.d | Gene Locuse | AAf |
|---|---|---|---|---|---|---|
| OsHpt1 | OsHpt1 | 11971.m11569 | OJ1150_A11 | AP003928 | LOC_Os01g54050 | 208 |
| OsHpt2 | OsHpt2 | 11978.m08408 | P0439E07 | AP003768 | LOC_Os08g44350 | 147 |
| OsHpt3 | OsHpt3 | 11979.m06878 | OJ1155_H10 | AP005558 | LOC_Os09g39400 | 149 |
| OsHpt4 | OsHpt4 | 11975.m05454 | OJ1725_E07 | AC093920 | LOC_Os05g09410 | 150 |
| OsHpt5 | OsHpt5a | 11975.m64188 | P0483D07 | AC130611 | LOC_Os05g44570 | 152 |
| OsHpt5b | 11975.m08583 | P0483D07 | AC130611 | LOC_Os05g44570 | 104 | |
| OsHpt5c | 11975.m64253 | P0483D07 | AC130611 | LOC_Os05g44570 | 96 |
Notation of putative proteins based on conserved domains.
IDs given to pseudomolecules by TIGR (http://www.tigr.org/tdb/e2k1/osa1/) from the O. sativa annotation database.
BAC IDs from the TIGR database.
BAC accession numbers for genomic DNA sequences deposited in the NCBI database (http://www.ncbi.nlm.nih.gov/).
Gene locus.
The number of predicted amino acid.
The HK Protein Family
O. sativa has at least 14 distinct HKs compared to 16 such genes in Arabidopsis (Hwang et al., 2002). Further, these putative HK homologs in O. sativa belong to three distinct families: the typical OsHK family (that includes cytokinin receptor homologs), an ethylene receptor (ETR) homolog family (that includes OsETRs and OsERSs), and the phytochromes (OsPHYs). Table I summarizes the relevant genomic information for these genes and their products. The predicted proteins range in size from 359 to 1,186 amino acids, thus representing great variations in structure and possible functions. However, the OsHK members share an overall 42% to 77% similarity in amino acid sequence and 30% to 66% identity, excluding the PHY proteins. The identity of PHY proteins with other OsHKs varies from 25% to 27%, while similarity varies between 35% and 41%.
OsHK Family
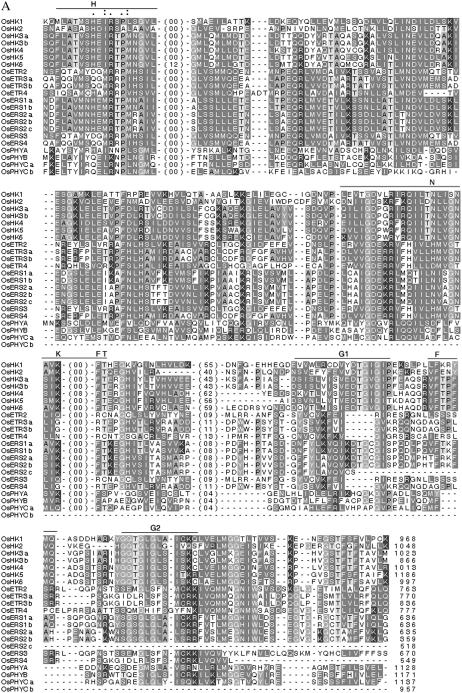
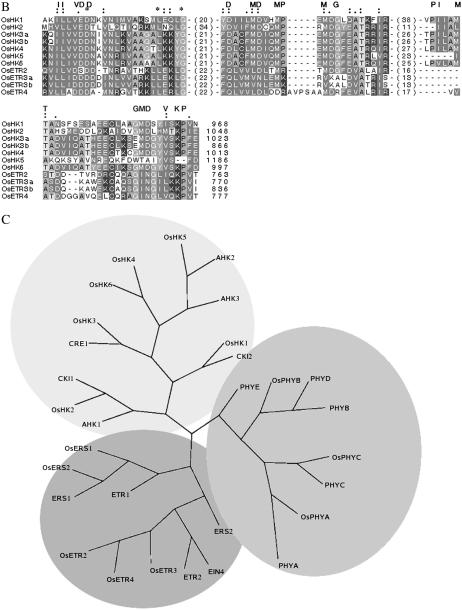
This family contains at least seven typical hybrid type HKs [OsHK1, 2, 3 (a and b), 4, 5, and 6]. To verify the number of HK sequences, an exhaustive TBLASTN search of full-length O. sativa cDNA clones available at Knowledge-based Oryza Molecular biological Encyclopedia (KOME) and a TBLASTX search of the National Center for Biotechnology Information (NCBI) O. sativa genome dataset were also performed, using all seven OsHKs as the query. No additional HK member was found in these searches. The structural details of the OsHKs are presented in Figure 2A, while the sequence details are compared in Figure 3, A (for transmitter domain) and B (for RD). The primary domain analysis of these proteins confirmed them to have a typical hybrid HK-type structure with a conserved His-containing kinase domain (KD) and a complete RD with a highly conserved Asp as the phospho-acceptor, although the number and position of the transmembrane (TM) domain varied (Fig. 2A). All OsHK family members (except OsHK1 and OsHK2) have the extracellular domain (ED), which may be involved in sensing signals. Further, in the case of OsHK1 and OsHK3b, no putative TM domain could be predicted with any significance, while the rest of the members have one or two TM domains, possibly indicating the diverse roles of these members in membrane-associated or nonmembrane-associated sensing operations in plants. A similar variable number of TM domains have been reported in Arabidopsis (Hwang et al., 2002). The five conserved signature motifs, such as H, N, G1, F, and G2, also feature in OsHKs (Fig. 3A). The transmitter domain of HKs has an H box that contains the conserved His residue, which is reversibly phosphorylated in the signaling process. The highly invariable residue Pro, after five residues of His, is conserved across all OsHKs except OsHK2. Further, His has not been found to be conserved in OsETR2, ETR3a, ETR3b, OsERS3, OsERS4, OsPHYB, OsPHYCa, and OsPHYCb. However, this His residue is found to be displaced further down at position 7 on all these members except the OsPHY. Experimental evidence for the participation of this displaced His in phosphorylation is needed. In most cases, a conserved D is missing in the G1 box when the His is missing in the H box.
Figure 3.
Amino acid sequence alignment of HK transmitter (A) and RDs and related proteins (B) in O. sativa. Sequence alignment was done using the T-Coffee program. Conserved motifs are highlighted above the alignment. The numbers indicated in the sequence alignment are the amino acid gaps between the motifs. *, Positions that have a single, fully conserved residue. “:” indicates one of the strong groups is fully conserved, and “.” indicates one of the weaker groups is fully conserved in the sequence alignment. -, Gaps in the alignment. The number at the end of the alignment indicates the total number of amino acids in putative proteins. C, The unrooted relationship tree of HKs and related proteins in O. sativa and Arabidopsis (O. sativa TCS members have been prefixed with Os, while others represent members from Arabidopsis; nomenclature as in Hwang and Sheen, 2001). The tree is branched into three main groups: the ETRs, photoreceptor PHY, and putative HK members (see text for details).
It is now known that members of the TCS (such as AHK2, 3, and CRE1/Ahk4/WOL) are responsible for cytokinin signaling (Kakimoto, 1996; Hwang and Sheen, 2001). Further, the Arabidopsis mutants (e.g. cre1-1 mutant) lacking these members have been reported to lack cytokinin responses (Inoue et al., 2001). Our analysis indicates that the putative cytokinin receptor candidates in O. sativa are OsHK3a, 3b, 4, 5, and 6, which show high identity (50%–62%) with the established cytokinin receptors AHK2, AHK3, and CRE1 from Arabidopsis. However, OsHK3 to 6 show a low level of similarity (28%–30%) with CKI1. This is in agreement with other reports where sequence divergence of CKI1 (only 25% identity with CRE1) has been implicated to be the possible reason for failure of cytokinin binding to CKI1 (Yamada et al., 2001). Except OsHK1 and OsHK2, all OsHK members were found to contain the conserved cyclases/HK-associated sensory extracellular (CHASE) domain. Qiu-min et al. (2004) showed that, in O. sativa, five proteins (OsCRL1a, OsCRL1b, OsCRL2, OsCRL3, and OsCRL4) contained the CHASE domain, but only one of these proteins (OsCRL4) contained, in addition, the putative Ser/Thr protein KD (please note that OsCRL2, OsCRL1b, OsCRL3, and OsCRL1a have been referred to as OsHK3, OsHK4, OsHK5, and OsHK6, respectively, in this study). It has been suggested that OsCRL4 represents a new AtCRE1-like gene family in O. sativa and was shown to be able to functionally complement the phenotype of Atcre-1 mutation, although the structure of the two were quite different (Qiu-min et al., 2004). Similarly, in the case of maize (Zea mays), detailed characterization of the three cytokinin-responsive HKs (ZmHK1, ZmHK2, and ZmHK3a) suggested that each has a capacity to confer cytokinin inducibility to lacZ expression in bacteria having ΔrcsC and cps∷lacZ. This indicates that the cytokinin signaling may be conserved machinery in monocots, though some minor differences do exist with respect to their ligand binding (Yonekura-Sakakibara et al., 2004).
ETR Family
In Arabidopsis, ethylene is perceived by a family of five receptors (ETR1, ETR2, ERS1, ERS2, and Ein4) that display significant homology to a prokaryotic family of signal transducers (Chang et al., 1993; Hua et al., 1995, 1998; Hua and Meyerowitz, 1998; Sakai et al., 1998). In O. sativa, OsETR and OsERS are members of the putative ETR family of HKs with or without a RD (Fig. 2). There are five genes that encode three OsETRs and four OsERS. Yau et al. (2004) showed, in O. sativa L. subsp. Indica var IR36, the existence of at least five ETR genes: OS-ERS1, OS-ERS2, OS-ETR2, OS-ETR3, and OS-ETR4. In our analysis, OsERS1a represents the alternate splice product for OsERS1b, while OsERS2a represents the same for OsERS2b and OsERS2c (Table I). Similarly, OsETR3a and OsETR3b are also reported in databases as alternative spliced products (http://www.tigr.org/tdb/e2k1/osa1/expression/alt_spliced.info.shtml). The primary domain analysis of these proteins indicated that ETRs OsETR2, 3 (a and b), and OsETR4 have a hybrid HK-type structure, with their KDs having the conserved His and their RD having a highly conserved Asp as a phospho-acceptor (Fig. 2). All the ERS members of O. sativa lacked the RD domain; OsERS3 lacked both the RD and receiver-like domain (RLD). All the O. sativa ETR members were predicted to possess two or more TM domains (except the OsERS2b) along with the GAF (cyclic GMP adenylyl cyclase FhlA) domain. GAF domains bind molecules such as cAMP and cGMP. ETRs and phytochromes (PHYs) are the only plant proteins known to contain GAF and HK-related domains. Various ETRs of O. sativa, along with their alternate splice products, have been aligned and compared in Figure 3A and show that the G2 and F domains are missing in OsERS2c. The autophosphorylating His residue is absent at the usual conserved position in OsERS3, OsERS4, OsETR2, OsETR3a, and OsETR3b, but is shifted six amino acids toward the C terminus. It is not known if this affects activity of HKs. Arabidopsis has a similar situation where H is replaced by E (in ETR2) and D (in ERS2), and none of the PHYs except PHYC has conserved H (Hwang et al., 2002). OsETR4 presents a unique situation where H is present at both positions; which of these participates in the phosphorelay needs to be established. An analysis of the RD domain of the ETR family is presented in Figure 3B. The conserved D, which is essential for the phosphorelay, is replaced by Q in all the ETRs. Further, the second D (five positions down to the first one) is also replaced by N in two of these ETR members (OsETR3a and b). Also, the PIMT motif in the RD in all the OsETRs as well as in the OsHK5 shows deletion of all six residues. The functional significance of these structural changes is not known.
PHY Family
The PHY photoreceptors regulate growth and development in response to red and far-red light signals (Smith, 2000). In Arabidopsis, this family consists of PHYA, B, C, D, and E (Sharrock and Quail, 1989; Clack et al., 1994). All dicots seem to be similar, but monocots seem to have only PHYA, B, and C (Mathews and Sharrock, 1996, 1997). In Arabidopsis, phya, b, d, or e mutants have been instrumental in deciphering the specific roles of the individual proteins in photomorphogenesis (Whitelam and Devlin, 1997; Devlin et al., 1998). PHYs have been identified in tobacco (Nicotiana tabacum), tomato (Solanum lycopersicum), oat (Avena sativa), maize, and potato (Solanum tuberosum), but relatively very few PHY genes have been fully characterized from monocots (Basu et al., 2000). Our analysis identified three PHY genes (OsPHYA, OsPHYB, and OsPHYC) in O. sativa. Among these, OsPHYC encodes alternatively spliced products for OsPHYCa and b. The proteins encoded by the OsPHY members vary in length between 957 and 1,171 amino acids (Table I). The putative domain analysis of the OsPHY family members show a structure similar to that in Arabidopsis, i.e. having a GAF domain, light-sensing domain, duplicated PAS [Per (period circadian protein), Arnt (ah receptor nuclear translocator protein), Sim (single-minded protein)] domains, and RLD (Fig. 2). PAS domains are present in many signaling proteins in archaea, bacteria, and eukaryotes, and act as signal sensor domains. Both RLD and KD seem to be missing from OsPHYCb; the functional significance of this is unknown (Fig. 2A). Analysis of OsPHYs using pfam (version 20.0) has clearly indicated toward presence of an additional PAS domain before GAF domain that has not been found in Arabidopsis. These minor structural and functional variations may have arisen through the gene duplication events and modification of gene functions as needed in evolution. However, experiments where the PHYs have been expressed in O. sativa from other systems, such as oat, have indicated toward the functional conservation of the transduction pathway components and the promoter sequences involved in regulation of PHY expression between the two systems (Bruce et al., 1989; Clough et al., 1995).
The unrooted relationship tree of the HK and the related proteins in O. sativa and Arabidopsis indicates the presence of subgroups within the protein subfamilies (Fig. 3C). This analysis indicates that OsHK3 to 6, AtHK2 and 3, as well as CRE1 have a close relationship with established cytokinin receptors, while the putative ETR proteins of O. sativa and Arabidopsis form a distinct cluster. In Arabidopsis, Hwang et al. (2002) have reported that CRE is close to the HK genes. Within the ETR family, two separate branches have evolved, one comprising ERS proteins and the other ETRs. However, ETR1 and ERS2 seem to be clear exceptions in this regard. It seems, from this tree, that the ERSs and ETRs of O. sativa and Arabidopsis have remained more heterogeneous within the ETR subfamily unlike the PHY and the cytokinin receptor families. Hence, the “in group” heterogeneity is higher in the ETR subgroup when compared to the other two. In the tree, all the PHYs from both O. sativa and Arabidopsis form a distinct group. The Arabidopsis PHYE remains in a distinct branch because O. sativa has no reported PHYE. OsPHYB appears to be more distant in evolution from Arabidopsis PHYB and is closer to the Arabidopsis PHYD. Perhaps the OsPHYB evolved before the evolution of PHYD in dicots as in Arabidopsis.
In Arabidopsis, a specific member of the HK group has been documented to play an important role as an osmosensor. The gene AtHK1 (or AHK1), showing high structural similarity with the yeast osmosensing HK SLN1 (synthetic lethal of N-end rule), is able to functionally complement the yeast double mutant lacking its two osmosensors, i.e. sln1 and sho (Urao et al., 1999), indicating the functional conservation of the osmosensing machinery between the two systems. Further, employing yeast two-hybrid analysis, AtHK1 has also been shown to interact physically with AHP2 (Urao et al., 2000b), suggesting that the transduction of the stress signal could occur via a multistep phosphorelay. Moreover, the AtHK1 transcript has been found to be most abundant in roots and is up-regulated by external osmolarity changes. Nonetheless, the precise mechanism of the functioning of the osmosensor in Arabidopsis is yet to be deciphered. Our analysis of the O. sativa genome has indicated that the overall percent identity of OsHKs with AtHK1 ranges from 25% to 29% with OsHK2 being the closest relative of AtHK1 (Fig. 3C). It remains to be proven experimentally which of the O. sativa HKs functions as an osmosensor.
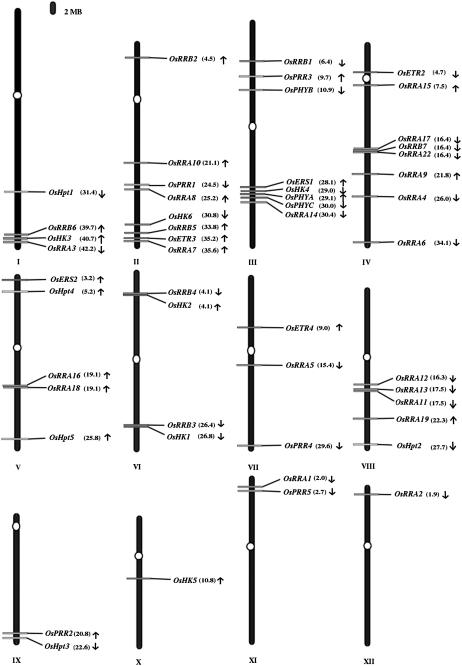
Genes encoding TCS members were distributed over the 12 different chromosomes of O. sativa (Fig. 6). The largest number (nine) of the TCS members is located on chromosome II, followed by eight each on chromosomes III and IV, while there is only one on chromosomes X and XII. Genes for OsHK family members show the following distribution: chromosome I (OsHK3), II (OsHK6, OsETR3), III (OsPHYB, OsERS1, OsHK4, OsPHYA, OsPHYC), IV (OsETR2 ), V (OsERS2), VI (OsHK1 and 2), VII (OsETR4), and X (OsHK5). These distributions highlight some interesting aspects. All the PHY genes are clustered on chromosome III; two of them, OsPHYA and OsPHYC, are placed within 1 Mb of each other. In a previous observation, PHYA and PHYC were located on long arm, while PHYB had been located on the short arm of chromosome III of O. sativa var IR36 (Basu et al., 2000). This is in synteny with the three PHY genes from sorghum (Sorghum bicolor) that are located on the same linkage group (referred to as a linkage group C by Paterson et al. [1995] and as a linkage group A by Childs et al. [1997]). In contrast, PHY genes are distributed on chromosomes I, V, VI, and IX in maize and on chromosomes I, II, and V in Arabidopsis (Hwang et al., 2002).
Figure 6.
Graphical (scaled) representation of location of putative genes for TCS on O. sativa chromosomes. The centromeres on chromosomes have been marked in ovals. The position of first exon of genes (in Mb) has been marked in the parentheses along with their names at same location on chromosomes. Arrow marks the direction of the ORF specific to the TCS gene. Where there is a case of alternative splicing, the genes located on chromosomes represent the longest ORF encoding loci.
The Hpt Protein Family
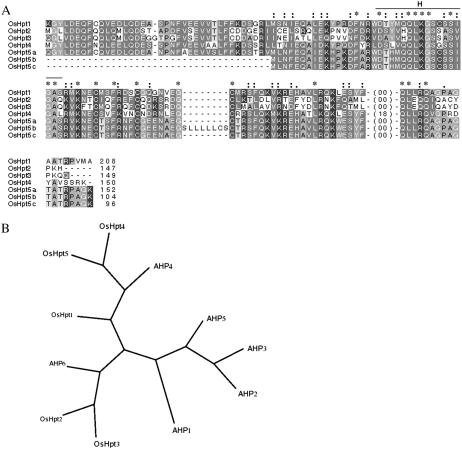
In Arabidopsis, five putative Hpt proteins (AHP1–5) might mediate the transfer of phosphoryl groups from membrane-localized HKs to nuclear-localized RRs (Hwang et al., 2002). Hwang and Sheen (2001) showed the movement of an Hpt into the nucleus by green fluorescent protein tag fusion. The analysis of the OsHpts in O. sativa indicated five genes (OsHpt1–5) coding for seven putative Hpts (OsHpt5 has three alternate splice products: OsHpt5a, b, and c) ranging in length from 96 to 208 amino acids (Table II). Different members of the OsHpt family lack overall sequence similarity but share a common four-helix bundle motif without any TM domain indicating their cytoplasmic location (Fig. 2B). All Hpt proteins of Arabidopsis contain a consensus sequence of XHQXKGSSXS with the position of H conserved. However, in O. sativa, only OsHpt2 and OsHpt3 show this consensus; all others lack it (Fig. 4A). The OsHpt5a, 5b, and 5c have a conserved H one position before the conserved H in OsHpts 2 and 3. These putative OsHpts lacking H but showing conserved Hpt domain may have some alternative degenerated phosphorelay role in cell signaling. The unrooted relationship tree for these different OsHpts is shown in Figure 4B, indicating the absence of any clustering among them. However, it can be seen that all AHP members (except AHP4 and 6) make a group that is separate from OsHpts, possibly indicating their diverse evolutionary pattern. The overall percentage identity within OsHpt family ranges from 37% to 71% and similarity varies from 64% to 84%. These putative OsHpts (1–5) are located on chromosomes I, V, VIII, and IX (Fig. 6).
Figure 4.
Putative Hpt proteins in O. sativa. A, Depiction of the deduced aligned amino acid sequence of OsHpts. *, Positions that have a single, fully conserved residue; “:” indicates one of the strong groups is fully conserved, and “.” indicates one of the weaker groups is fully conserved in the sequence alignment. -, Gaps in the alignment. The number at the end of the alignment indicates the total number of amino acids in putative proteins. B, Unrooted relationship tree of OsHpts and Arabidopsis Hpts (AHPs). T-Coffee program was used for alignment, and PHYLIP was used for graphical presentation of the relationship among the OsHpts and AHPs.
The RR Family
In most systems, RRs represent the terminal component of the TCS pathway, functioning as phosphorylation-activated switches that catalyze the phosphoryl transfer from the phospho-His of the HK to a conserved His in its own regulatory domain. RRs can catalyze phosphoryl transfer independent of assistance from an HK because small molecules such as acetyl phosphate, carbamoyl phosphate, imidazole phosphate, and phosphoramidate can serve as phosphodonors to them (Lukat et al., 1992). The autodephosphorylation of RRs is also known, thus limiting the lifetime of their activated state.
Genome-wide analysis in Arabidopsis has indicated the existence of 32 genes encoding putative RRs and related proteins that are not fused to the His protein KD (Hwang et al., 2002; Schaller et al., 2002). The majority of these RRs have two main domains: a conserved N-terminal regulatory domain and a variable C-terminal effector domain. O. sativa has a total of 32 genes that code for 44 putative OsRR proteins (Table III). Based on the predicted structural features shown in Figure 2C, the OsRR proteins in O. sativa can be divided into three distinct categories similar to those in Arabidopsis: OsRRAs, which possess only a RD; OsRRBs, containing RD fused to a DNA-binding domain; and OsPRRs, which are RR-like proteins or pseudo-RRs (PRRs; sometimes also referred to as RRLs).
O. sativa possesses 20 genes coding for the A-type RRs (Table III, OsRR1 through OsRRA19 and OsRRA22; note that OsRRA20 and 21 are encoded by OsPRR3 and OsPRR1, respectively) in contrast to 11 such genes in Arabidopsis. The identity among the A-type RRs is between 26% to 90% and similarity 48% to 93%. All OsRRA-type RRs have conserved DDK or EDK motifs as in Arabidopsis. A-type RR are involved in cytokinin signaling. The expression of some RRs has been reported to be sensitive to environmental stresses such as drought, salinity, and low temperature (Urao et al., 1998), indicating a possible link between stresses and cytokinin signaling.
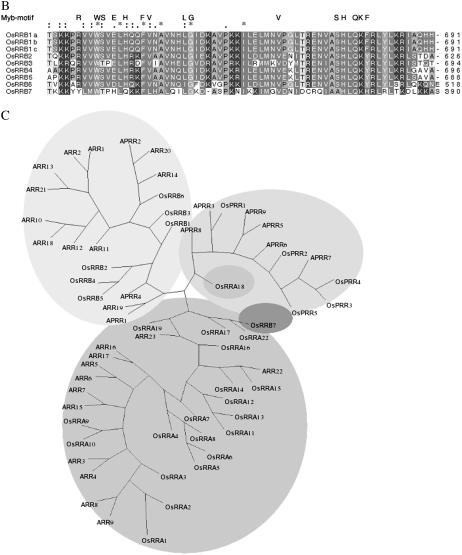
In O. sativa, B-type RRs (OsRRBs) share 27% to 94% identity and all are more than 48% similar. They show weak relationships with A-type RRs. The B-type RRs have a highly conserved C-terminal stretch of around 70 amino acids. This stretch encodes for protein with high structural similarity to the DNA-binding domain of the c-MYB (Figs. 2C and 5B). There are seven B-type RR genes in O. sativa, compared to 12 in Arabidopsis. OsRRB1 encodes three proteins, OsRRB1a, b, and c. The alignment of the putative RD and DNA-binding motifs of the B-type RRs is shown in Figure 5, A and B, respectively. Detailed investigations on a B-type RR, Ehd1 (Early heading date 1) from O. sativa, have shown that it can function as a floral inducer under short-day conditions (Doi et al., 2004). Further, Ehd1 activates downstream genes such as FT-like genes and MADS-box genes, which are closely related to floral transition (Doi et al., 2004). The acute expression of Ehd1 in response to “light-on” signals suggests that its expression is regulated by PHYs. B-type RRs are also involved in cytokinin and ethylene signaling in Arabidopsis (Hwang et al., 2002), while the O. sativa Ehd1 is possibly also involved in developmental and environmental signals mediated by light, cytokinin, and ethylene (Doi et al., 2004).
Figure 5.
Deduced multiple sequence alignment of putative RR and PRR proteins in O. sativa. A, Amino acid sequences of transmitter domains of putative RR and PRR proteins. The highly conserved amino acids are highlighted, and the conserved motifs are marked above the consensus sequences. The gaps between the motifs have been marked by the numbers in each sequence. B, Alignment of putative DNA-binding motifs of B-type RRs. Multiple sequence alignment was done using T-Coffee software. C, Unrooted relationship tree of putative RR and PRR protein in O. sativa and Arabidopsis (O. sativa TCS members have been prefixed with Os, while others represent members from Arabidopsis; nomenclature as in Hwang and Sheen, 2001).
In Arabidopsis, nine genes encoding for PRRs have been identified (Hwang et al., 2002). There are five such genes in the O. sativa genome that encode 10 OsPRRs lacking a typical RD (Table III; Fig. 2C). These five PRRs have previously been reported (Murakami et al., 2003). In this analysis, we have followed the nomenclature as OsPRR1, OsPRR2, OsPRR3, and OsPRR4 representing reported OSPRR1, OSPRR95, OSPRR73, and OSPRR37, respectively. However, the sequence for reported OSPRR59 is not available for analysis. The RD of the OsPRRs shows about 40% similarity with the RDs of the OsRRBs. The identity within OsPRRs ranges from 40% to 70%. Further, the three amino acid residues (D-D-K) in typical RRs are not conserved in PRRs (Fig. 2C), and the phospho-accepting D of the RD is replaced by the E, N, or Q.
Arabidopsis has five members of the circadian-associated PRRs, including APRR1/TOC1. Each has a common structural design containing the pseudo-RD of about 120 amino acids at its N terminus and a short CCT motif of about 50 amino acids at the C-terminal end. Based on these criteria, O. sativa has at least five genes homologous to APRRs, both in their amino acid sequence structural motifs. Recently, the analysis of the PRR in japonica O. sativa (var Nipponbare) has given a new insight in the control of circadian rhythm (Murakami et al., 2003). The OsPRRs are nuclear localized and all have a pseudo-RD, which is similar to the phosphoaccepting RD of the authentic RRs (Mizuno, 1998; Imamura et al., 1999). The expression of OSPRR genes is under circadian control. They are expressed in diurnal and sequential manner in O. sativa, which is reminiscent of the circadian oscillations of the APRR1/TOC1 in Arabidopsis. The extension of this analysis in an indica variety (Kasalath) of O. sativa by Murakami et al. (2005) showed that the indica genome also has homologous members of the OSPRR family. Interestingly, among them, OSPRR37 (OsPRR4 in this article) has been mapped very close to the photoperiod sensitivity Hd2 (Heading date2) quantitative trait locus, which is the major locus that enhances the photoperiod sensitivity of flowering in Nipponbare. Further, Kasalath has a severe mutational lesion in this OSPRR37 coding sequence (i.e. there is premature termination codon in its CCT motif) as compared to that of Nipponbare (Murakami et al., 2005).
The unrooted tree of all members of the PRR family from O. sativa and Arabidopsis shows three distinct groups, each comprised of RRA-type, RRB-type, and PRR members; this suggests independent coevolution of these distinct subspecies in these two plant genomes (Fig. 5C). The OsRRs are distributed over all chromosomes except X (Fig. 6). The maximum numbers of OsRRs are present on contigs of chromosome IV. Further, overlapping ORFs also occurs in the O. sativa TCS genes. For example, OsPRR1 is predicted to encode for OsRRA21 in addition to OsPRR1, and the cluster is located on the longer arm of chromosome II. This association or coupling-in of two different types of RRs (i.e. OsPRR1 and OsRRA21) may throw some light on their close interlinks or cross talks at the signaling level. Likewise, the gene encoding the OsPHYC is predicted to encode for a Type II DNA topoisomerase, indicating the possible coregulation of some light-induced process at the nuclear level. There are several other cases where TCS members are in close proximity. These include the TCS clustering of OsRRB6, OsRRA3, and OsHK3 at the distal arm of chromosome I, which may allow correlated expression and regulation of these members. A similar situation occurs for the lower arm of chromosome II. On chromosome V, OsRRA16 and OsRRA18 are very close to each other. On chromosome IV, the OsRRA17, OsRRB7, and OsRRA22 are predicted to be arranged back-to-back and there is low identity among them. On chromosome VI, OsRRB4 and OsRRB3 are close to OsHK2 and OsHK1, respectively (but on the opposite coding strands in the case of OsHK2 and OsRRB4). Another interesting feature of this genome distribution is the strong clustering or condensing of the TCS member genes within a region of chromosomes (for example, see the short and long arms of chromosome III). In contrast, in Arabidopsis there is a more uniform distribution of the HK genes between chromosomes (Hwang et al., 2002).
Analysis of Expression Profiles for TCS Members in O. sativa Employing MPSS Database
MPSS is a valuable tool to have an insight into gene expression (Brenner et al., 2000). MPSS involves the cloning of a cDNA library on beads and the acquisition of 17- to 20-nt signatures (tags) from these cDNAs using an unconventional sequencing method. Signatures of this length have a high specificity and may match to just one position in a complex genome. MPSS has been used previously for genome-level expression analysis in several systems including Arabidopsis (Meyers et al., 2004). To extract information about the relative abundance of transcripts of O. sativa TCS members, we have carried out the analysis in the available MPSS database (http://mpss.udel.edu/rice/), and the results are presented as Supplemental Table S2. This database is derived from the TIGR O. sativa genome sequence version 3.0, and our search has been performed employing the 20-nt-long signatures. On an overall basis, we found high variability in abundance of TCS-specific mRNA tags (measured as transcript per million; TPM) in different libraries. For example, tags corresponding to OsHK4, 5, 6, OsETR2, OsPHYA, and B are relatively more abundant, while the rest of the OsHK family members showed very insignificant presence. Similarly, among the various Hpt proteins, OsHpt2 has been found to be most abundant, being represented in almost all libraries analyzed here. We found that OsRRA1 and OsRRA2 (located on chromosomes 11 and 12, respectively), though regulated differentially, show a hit score >1 in MPSS analysis, reconfirming their gene duplication status. In addition, we found OsRRB3, 4, 5, and 7 are relatively insignificant in expression, and OsRRB1, 2, and 6 are more abundant. Further, all PRR (except OsPRR2) are abundant in these expression libraries. These observations indicate that the TCS-related transcriptome of O. sativa is very complex, showing a tissue-specific differential regulation for most of the members.
Search into Possible Role(s) of O. sativa TCS Members Employing the Tos17 Mutant Database
One of the important goals of functional genomics is to construct saturation mutant libraries for O. sativa. Random mutant population has been generated in O. sativa employing endogenous transposon Tos17 tag (http://tos.nias.affrc.go.jp/). Studies have clearly established that Tos17 prefer to integrate in genic regions and hotspots for integration are distributed throughout the genome, making them more suitable for functional genomic studies as compared to other tagged lines (Miyao et al., 2003). Thus, these insertion-tagged lines serve as a useful tool to get an insight into possible functions of novel genes. Because the multiple copy nature of Tos17 as well as background mutations that accompany prolonged tissue culture are known, it is often difficult to establish the relationship between the disrupted gene and the corresponding mutant phenotype. However, it has also been suggested that if two or more lines have independent insertion in the same gene and the same phenotype is obtained, it is likely that the observed phenotype results from disruption of that gene (Miyao et al., 2003). We have carried out in silico screening for O. sativa TCS members by BLAST search against flanking sequences from the mutant lines (with cutoff value e−02). The results are presented as Supplemental Table S3. We found that phenotypes such as dwarfism, tillering, and heading (late or no heading) are the most common responses as generated by mutation in OsPHYs and OsRRs, while specific phenotypes are associated with some of the other members. Examples of the later category include phenotypes such as clum morphology (mutation in OsETR4 and OsERS1), stenophyllous and rolled leaves (mutations in OsHK1, OsHpt3, OsRRA16, and OsRRA18), vivipary (mutations in OsRRA1 and OsRRA2), and chlorina (mutations in OsHK1, OsHK4, OsPHYA, and OsRRB3). OsPHYs appears to have a role in yield, grain size, kernel, and leaf morphology, while mutations in OsHpts, OsHK1, and OsHK4 exhibit lethality. Mutant lines of OsPHYs, OsPRRs, OsRRB4, OsRRB6, OsRRA1, OsRRA2, and OsHpt5 show low fertility and sterility. Similarly, all OsRRBs and OsPHYs, OsRRA1, OsRRA2, and OsPRR5 show lesion mimic. Though Tos17 has been reported to be biased for insertion in kinase genes (Miyao et al., 2003), no mutants could be found for TCS members such as OsHK6, OsRTR2, OsETR3, OsRRA3, OsRRA10, and OsRRA15. However, approaches such as specific gene silencing employing RNAi may prove to be helpful in assigning the precise role(s) to these TCS members.
CONCLUSION
In view of the involvement of TCS-based signaling in diverse life processes, several genomic studies have been attempted. For example, in E. coli, 30 HKs and 32 RRs have been shown to be present (Mizuno, 1997). This is in contrast to yeast, where only one HK system (SLN1/YPD1/SSK1) has been shown to play a definite role in osmosensing and adaptation to the environment (Maeda et al., 1994). In this work, screening of the high-quality, finished whole-genome sequence of O. sativa has been carried out for the presence of genes that code for putative TCS members. This analysis has revealed that the O. sativa genome has 51 genes that code for 73 putative TCS family members (encoding typical OsHKs, OsHpts, and OsRRs). In contrast, the Arabidopsis genome reportedly has 54 genes coding for 54 putative TCS members (Hwang et al., 2002). However, our analysis of Arabidopsis genome sequence (TIGR version 5.0) has suggested a total of 63 TCS proteins encoded by 54 genes (Table IV). Based on structural similarities of TCS members in Arabidopsis and O. sativa, we propose that O. sativa TCS members participate in diverse physiological phenomenon ranging from light perception to osmotic adjustment as well as regulation of flowering and hormone signaling. Our analysis of the O. sativa genome shows the presence of at least 22 putative OsHKs and related proteins, seven putative OsHpts, and 44 putative RRs.
Table IV.
Comparison of the TCS architecture in O. sativa and Arabidopsis genomes
| Gene Family | No. of Genes | Arabidopsis No. of Proteins
|
O. sativa (japonica)
|
||
|---|---|---|---|---|---|
| TIGR Version 5.0 | Hwang et al. (2002) | No. of Genes (TIGR Version 4.0) | No. of Putative Proteins (TIGR Version 4.0) | ||
| TCS proteins | 54 | 63 | 54 | 51 | 73 |
| HKs and related protein | 16 | 19 | 16 | 14 | 22 |
| Hpts | 6 | 7 | 6 | 5 | 7 |
| RR | 32 | 37 | 32 | 32 | 44 |
Our analysis of the TCS machinery in the O. sativa genome and its comparison with Arabidopsis has revealed that the number of putative TCS genes in the two evolutionarily diverse model systems is very similar in both species (54 versus 51). The overall structure of the TCS machinery is very similar in both species, i.e. maximum number of components belonging to the RR family and minimum number of components belonging to the Hpt family, indicating the architectural conservation of the TCS machinery (Table IV). Apart from the numbers, the individual components from O. sativa also show significant sequence conservation with related members of Arabidopsis (Figs. 3–5). However, with respect to TCS members, the genomes of these two model plants possess some unique features, which are of great importance from the evolutionary point of view. Although the total number of putative genes in O. sativa is much greater in Arabidopsis, the portion of the genome that encodes TCS components has not expanded in O. sativa. This economy is presumably compensated for by more prevalent cases of alternative splicing of TCS members in O. sativa, where several protein variants are translated by single ORFs with differential editing of pre-mRNA. In prokaryotes, the number of TCS members has been associated with the varied environments they live in; pathogens that live in very narrow environment fluctuations possess few HKs. Does it mean that the same observation is true for eukaryotes as well? In contrast to this observation, Jain et al. (2006) found that the auxin-responsive GH3 gene family is enlarged in Arabidopsis as compared to O. sativa. Zhang et al. (2005) have reported lineage-specific expansion of certain gene families involved in pathogen resistance. However, the genome-wide analysis of these gene families in other plant genomes will help to test this hypothesis. Nonetheless, the O. sativa and Arabidopsis genomes offer interesting research materials that can be exploited to answer some basic questions related to molecular evolution of genomes.
Barakat et al. (1998) have reported that related Arabidopsis genes are fairly evenly distributed on all the chromosomes, but the O. sativa genes are clustered, indicating that organization of O. sativa and Arabidopsis genomes are substantially different. Our results are in agreement with these observations because we found distinct clustering (in terms of physical proximity) of TCS members on chromosomes (see Fig. 6; for example, chromosomes I, II, III, and VIII). However, Jain et al. (2006), who analyzed the auxin-responsive GH3 gene family in O. sativa, found no evident clusters on the chromosomes. In the O. sativa genome, chromosomal segment duplication had occurred several million years ago and there has been a massive ongoing duplication of individual genes, which provides a continuous source of raw material for the gene genesis (Guyot and Keller, 2004; Yu et al., 2005). It has been established that almost 65% of the O. sativa genome is segmentally duplicated as 18 distinct pairs spread across the genome. Most of these TCS members analyzed in this study are part of the duplicated set of segments and show 55% or more identity as paralogous genes. The OsRRA1 and OsRRA2 that are part of recent segmental duplication of chromosome 11 and 12, respectively, show 96% identity in their encoded proteins. Similarly, OsHpt1 and OsHpt5 show 82% identity, and OsHpt2 and OsHpt3 show 76% identity. OsPRR3 and OsPRR4 show 63% identity in sequence. We have observed that genes encoding OsRRB2 and OsRRB3, OsHK6 and OsHK2, though mapped within segmentally duplicated regions, show low identity of 40% and 25%, respectively. Such variation in sequence identity among duplicated genes may be due to gene evolution and accumulation of insertion and deletion events (Taylor and Raes, 2004). Besides segmental duplications, there are clear indications for the occurrence of tandem duplications within TCS members in O. sativa. For example, the lower arm of chromosome VIII shows the presence of three members of OsRRA family having a minimum 84% identity with each other (Fig. 6).
The possibility of close interaction of the TCS pathway with other signaling pathways in plants is being explored. In one such study, comparative microarray analysis carried out on loss-of-function mutant arr2 in Arabidopsis revealed more than 600 genes having altered expression, including genes related to auxin, ethylene, biotic and abiotic signal transduction, photomorphogenesis, protein folding, degradation, and development (Hass et al., 2004). In another study, transgenic Arabidopsis plants ectopically expressing the constitutively active B-type ARR showed alterations in the activity of several plant signaling pathways (Tajima et al., 2004). Similarly, combinatorial microarray analysis performed with wild-type Arabidopsis and those having altered cytokinin-mediated His-Asp phosphorelay signaling circuitry revealed the involvement of 214 cytokinin-up-regulated genes (including those belonging to the AP2/EREBP, MYB, and GATA families), indicating the elaborate and interconnected nature of this TCS machinery in cytokinin signaling (Kiba et al., 2005). These observations support a model where a TCS-dependent cross talk integrates light (phyB) input and hormone (cytokinin)-regulated homeostasis, enabling the plant to react most efficiently to environmental cues (Grefen and Harter, 2004).
In the cyanobacterium Synechocystis, at least five HKs have been shown to be involved in the perception of osmotic stress, which in turn regulate a distinct set of downstream genes (Paithoonrangsarid et al., 2004). This allows the system to have a complex network of Hik-Rre components where salt and hyperosmotic stress signals could be differentially perceived (Shoumskaya et al., 2005). The O. sativa genome contains at least seven members of the OsHK family that show close relationship (25%–29% overall identity) with reported osmosensor AtHK1 from Arabidopsis. However, it remains to be established whether there is a functional homolog of AtHK1 in O. sativa, or if O. sativa is similar to Synechocystis with each OsHK member sensing a different signal. Future investigations employing novel molecular and genetic tools and techniques (such as RNAi) will shed light on the functioning of this important signaling machinery and will pave the way to engineer O. sativa plants for better adaptation to environmental cues.
MATERIALS AND METHODS
Search of Proteins Involved in the TCS in Oryza sativa
The domain structure of the TCS, and the related proteins, was used to identify and classify the TCS proteins using the TIGR O. sativa genome sequence version 4.0. Profiles unique to HKs were used to screen all predicted proteins using the HMMER software (version 3.3.2; http://hmmer.wustl.edu/). These unique profiles are for Pfam HMM of HK A (phosphoacceptor) domain (accession no. PF00512), Hpt (accession no. PF01627), RR RD (accession no. PF0072), CHASE domain (accession no. PF03924), and HATPase (accession no. PF02518). We have used these profiles as default parameters in the hmmsearch program of the HMMER package. All the significant hits with positive scores were selected for classification and were then examined individually for accessory domains that are usually present. This was accomplished by searching the sequences against the Pfam database (version 20.0) to map the known domains, such as HK A, Hpt, and the RR myb and CCT (see Fig. 2 for a schematic representation). This step, besides mapping accessory domains to the TCS and providing the basis of classifying the proteins as members of the TCS, also helped to identify false positives, such as retrotransposon or transposable elements containing fragments of TCS sequences. TM regions were mapped using the PSORT program (Nakai and Horton, 1999). To validate the specificity and sensitivity of the methodology, similar analyses were performed on the datasets having all known and well-characterized HKs and related proteins of the TCS in Arabidopsis (Arabidopsis thaliana; http://mips.gsf.de/proj/thal/db/index.html). The percentage values of the sequence similarity and identity within the groups were obtained using BLAST (Altschul et al., 1990).
The protein sequences of all the HKs, RRs, and Hpts from O. sativa were used to search in KOME (28,469 full-length cDNA clones) using the TBLASTN program (Altschul et al., 1997). Details of these analyses are presented in Supplemental Table S1. The protein sequences of the TCS in Arabidopsis were retrieved from the Munich Information Center for Protein Sequences (http://mips.gsf.de/proj/thal/) database. TIGR has used FGENESH, an HMM-based tool for ab initio gene prediction using known properties of coding and noncoding sequence, to predict genes and gene-related information, such as the reading frame and the coding statistics (codon usage and GC contents). FGENESH has proved to be the most sensitive and specific method when compared with other ab initio gene prediction methods (Mathé et al., 2002), especially for the monocots (Yuan et al., 2005). We have also used an HMM-based tool in an unsupervised manner (i.e. no prior selection criterion) to find out the TCS in pseudomolecule protein dataset. We have used an extrinsic approach, based on similarity searches using BLAST and TBLASTN in other data sets, such as KOME and NCBI databases, to exclude the possibility of any additional member of TCS that might have been left unreported in TIGR.
Multiple Sequence Alignments and Phylogenetic Analysis
Multiple sequence alignments of related proteins belonging to each class from both Arabidopsis and Oryza were performed using T-Coffee (Notredame et al., 2000) with editing of the subsequent alignments using Seaview (Galtier et al., 1996). Alignments of the protein domain sequences were used as input for phylogenetic analysis using protpars, a maximum parsimony routine from the PHYLIP package (version 3.6; Felsenstein, 2004). Unrooted trees were plotted using “draw tree” from the same package.
Analysis of MPSS Database for Expression Profiles
To gain insight into expression profiles of TCS members in O. sativa, the MPSS database (http://mpss.udel.edu/rice/) was searched (opting 20-nt signature sequences) using the locus ID given in the TIGR database. The data thus obtained have been analyzed and grouped based on tissue specificity (Supplemental Table S2).
Analysis of Tos17 Mutant Population
Protein sequences of individual TCS members were used as a query to search the Tos17-tagged lines (http://tos.nias.affrc.go.jp/). For better precision in picking up the phenotypes, only a high negative exponential E score value for FST hits has been the selection criteria. The information thus obtained has been presented in Supplemental Table S3.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. ESTs for TCS members in O. sativa available at KOME database.
Supplemental Table S2. Analysis of expression profiles of O. sativa TCS members using MPSS database.
Supplemental Table S3. Tos17-tagged lines available for TCS members in O. sativa.
Supplementary Material
Acknowledgments
We thank Professor Roger A. Leigh and Professor Sudhir Sopory for a critical reading of this manuscript; we are indebted to Professor Govindjee for his suggestions to improve the language of this manuscript.
This work was supported by the Government of India (Department of Science and Technology and Department of Biotechnology), by the International Foundation for Science (Sweden), by the International Atomic Energy Agency (Austria), and by the Council of Scientific and Industrial Research, India (Junior Research Fellowship to A.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ashwani Pareek (ashwanip@mail.jnu.ac.in).
The online version of this article contains Web-only data.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat A, Matassi G, Bernardi G (1998) Distribution of genes in the genome of Arabidopsis thaliana and its implications for the genome organization of plants. Proc Natl Acad Sci USA 95: 10044–10049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu D, Dehesh K, Schneider-Poetsch HJ, Harrington SE, McCouch SR, Quail PH (2000) Rice PHYC gene: structure, expression, map position and evolution. Plant Mol Biol 44: 27–42 [DOI] [PubMed] [Google Scholar]
- Besant PG, Tan E, Attwood PV (2003) Mammalian protein histidine kinases. Int J Biochem Cell Biol 35: 297–309 [DOI] [PubMed] [Google Scholar]
- Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, et al (2000) Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol 18: 630–634 [DOI] [PubMed] [Google Scholar]
- Bruce WB, Christesen AH, Klein T, Fromm N, Quail PH (1989) Photoregulation of a phytochrome gene promoter from oat transferred into rice by particle bombardment. Proc Natl Acad Sci USA 86: 9692–9696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catlett NL, Yoder OC, Turgeon BG (2003) Whole-genome analysis of two-component signal transduction genes in fungal pathogens. Eukaryot Cell 2: 1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM (1993) Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262: 539–544 [DOI] [PubMed] [Google Scholar]
- Childs KL, Miller FR, Cordonnier-Pratt MM, Pratt LH, Morgan PW, Mullet JE (1997) The sorghum photoperiod sensitivity gene, Ma3, encodes a phytochrome B. Plant Physiol 113: 611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack T, Mathews S, Sharrock RA (1994) The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol 25: 413–427 [DOI] [PubMed] [Google Scholar]
- Clough RC, Casal JJ, Jordan ET, Christou P, Vierstra RD (1995) Expression of functional oat phytochrome A in transgenic rice. Plant Physiol 109: 1039–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Patel SR, Whitelam GC (1998) Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell 10: 1479–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A (2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev 18: 926–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J (2004) PHYLIP (Phylogeny Inference Package) Version 36. Distributed by the author, Department of Genetics, University of Washington, Seattle
- Foussard M, Cabantous S, Pedelacq J, Guillet V, Tranier S, Mourey L, Birck C, Samama J (2001) The molecular puzzle of two-component signaling cascades. Microbes Infect 3: 417–424 [DOI] [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C (1996) SeaView and Phylo_win, two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci 12: 543–548 [DOI] [PubMed] [Google Scholar]
- Grebe TW, Stock JB (1999) The histidine protein kinase superfamily. Adv Microb Physiol 41: 139–227 [DOI] [PubMed] [Google Scholar]
- Grefen C, Harter K (2004) Plant two-component systems: principles, functions, complexity and cross talk. Planta 219: 733–742 [DOI] [PubMed] [Google Scholar]
- Guyot R, Keller B (2004) Ancestral genome duplication in rice. Genome 47: 610–614 [DOI] [PubMed] [Google Scholar]
- Hass C, Lohrmann J, Albrecht V, Sweere U, Hummel F, Yoo SD, Hwang I, Zhu T, Schäfer E, Kudla J, et al (2004) The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis. EMBO J 23: 3290–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Chang C, Sun Q, Meyerowitz EM (1995) Ethylene insensitivity conferred by Arabidopsis ERS gene. Science 269: 1712–1714 [DOI] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM (1998) Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94: 261–271 [DOI] [PubMed] [Google Scholar]
- Hua J, Sakai H, Nourizadeh S, Chen QG, Bleecker AB, Ecker JR, Meyerowitz EM (1998) EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell 10: 1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Chen HC, Sheen J (2002) Two-component signal transduction pathways in Arabidopsis. Plant Physiol 129: 500–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389 [DOI] [PubMed] [Google Scholar]
- Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Kiba T, Ueguchi C, Sugiyama T, Mizuno T (1999) Compilation and characterization of Arabidopsis thaliana response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol 40: 733–742 [DOI] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T (2001) Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Jain M, Kaur N, Tyagi AK, Khurana JP (2006) The auxin-responsive GH3 gene family in rice (Oryza sativa). Funct Integr Genomics 6: 36–46 [DOI] [PubMed] [Google Scholar]
- Kakimoto T (1996) CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science 274: 982–985 [DOI] [PubMed] [Google Scholar]
- Keener J, Kustu S (1988) Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NTRB and NTRC of enteric bacteria: roles of the conserved amino-terminal domain of NTRC. Proc Natl Acad Sci USA 85: 4976–4980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Naitou T, Koizumi N, Yamashino T, Sakakibara H, Mizuno T (2005) Combinatorial microarray analysis revealing Arabidopsis genes implicated in cytokinin responses through the His→Asp phosphorelay circuitry. Plant Cell Physiol 46: 339–355 [DOI] [PubMed] [Google Scholar]
- Lohrmann J, Harter K (2002) Plant two-component signaling systems and the role of response regulators. Plant Physiol 128: 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois AF, Weinstein M, Ditta GS, Helinski DR (1993) Autophosphorylation and phosphatase activities of the oxygen-sensing protein FixL of Rhizobium meliloti are coordinately regulated by oxygen. J Biol Chem 268: 4370–4375 [PubMed] [Google Scholar]
- Lukat GS, McCleary WR, Stock AM, Stock JB (1992) Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci USA 27: 718–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Wurgler-Murphy SM, Saito H (1994) A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369: 242–245 [DOI] [PubMed] [Google Scholar]
- Mathé C, Sagot M-F, Schiex T, Rouzé P (2002) Current methods of gene prediction, their strengths and weaknesses. Nucleic Acids Res 30: 4103–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews S, Sharrock RA (1996) The phytochrome gene family in grasses (Poaceae): a phylogeny and evidence that grasses have a subset of the loci found in dicot angiosperms. Mol Biol Evol 13: 1141–1150 [DOI] [PubMed] [Google Scholar]
- Mathews S, Sharrock RA (1997) Phytochrome gene diversity. Plant Cell Environ 20: 666–671 [Google Scholar]
- Meyers BC, Vu TH, Tej SS, Ghazal H, Matvienko M, Agrawal V, Ning J, Haudenschild CD (2004) Analysis of the transcriptional complexity of Arabidopsis thaliana by massively parallel signature sequencing. Nat Biotechnol 22: 1006–1011 [DOI] [PubMed] [Google Scholar]
- Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, Abe K, Shinozuka Y, Onosato K, Hirochika H (2003) Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15: 1771–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T (1997) Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res 4: 161–168 [DOI] [PubMed] [Google Scholar]
- Mizuno T (1998) His-Asp phosphotransfer signal transduction. J Biochem (Tokyo) 123: 555–563 [DOI] [PubMed] [Google Scholar]
- Murakami M, Ashikari M, Miura K, Yamashino T, Mizuno T (2003) The evolutionarily conserved OsPRR quintet: rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol 44: 1229–1236 [DOI] [PubMed] [Google Scholar]
- Murakami M, Matsushika A, Ashikari M, Yamashino T, Mizuno T (2005) Circadian-associated rice pseudo response regulators (OsPRRs): insight into the control of flowering time. Biosci Biotechnol Biochem 69: 410–414 [DOI] [PubMed] [Google Scholar]
- Nakai K, Horton P (1999) PSORT: a program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem Sci 24: 34–35 [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J (2000) T-coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302: 205–207 [DOI] [PubMed] [Google Scholar]
- Oka A, Sakai H, Iwakoshi S (2002) His-Asp phosphorelay signal transduction in higher plants: receptors and response regulators for cytokinin signaling in Arabidopsis thaliana. Genes Genet Syst 77: 383–391 [DOI] [PubMed] [Google Scholar]
- Paithoonrangsarid K, Shoumskaya MA, Kanesaki Y, Satoh S, Tabata S, Los DA, Zinchenko VV, Hayashi H, Tanticharoen M, Suzuki I, et al (2004) Five histidine kinase perceive osmotic stress and regulate distinct sets of genes in Synechocystis. J Biol Chem 279: 21531–21538 [DOI] [PubMed] [Google Scholar]
- Pan CX, Fukunaga R, Yonehara S, Nagata S (1993) Unidirectional cross-phosphorylation between the granulocyte colony-stimulating factor and interleukin 3 receptors. J Biol Chem 268: 25818–25823 [PubMed] [Google Scholar]
- Paterson AH, Lin Y-R, Li Z, Schertz KF, Doebley JF, Pinson SRM, Liu S-C, Stansel JW, Irvine JE (1995) Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science 269: 1714–1717 [DOI] [PubMed] [Google Scholar]
- Qiu-min H, Hua-wu J, Xiao-peng QI, Jie YU, Ping WU (2004) A CHASE domain containing protein kinase OsCRL4 represents a new AtCRE1-like gene family in rice. J Zhejiang Univ Sci 5: 629–633 [DOI] [PubMed] [Google Scholar]
- Sakai H, Hua J, Chen QG, Chang C, Medrano LJ, Bleecker AB, Meyerowitz EM (1998) ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA 95: 5812–5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Mathews D, Gribskov M, Walker JC (2002) Two-component signaling elements and histidyl-to-aspartyl phosphorelays. In C Somerville, E Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD
- Sharrock RA, Quail PH (1989) Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev 3: 1745–1757 [DOI] [PubMed] [Google Scholar]
- Shoumskaya MA, Paithoonrangsarid K, Kanesaki Y, Los DA, Zinchenko VV, Tanticharoen M, Suzuki I, Murata N (2005) Identical Hik-Rre systems are involved in perception and transduction of salt signals and hyperosmotic signals but regulate the expression of individual genes to different extents in Synechocystis. J Biol Chem 280: 21531–21538 [DOI] [PubMed] [Google Scholar]
- Smith H (2000) Phytochromes and light signal perception by plants: an emerging synthesis. Nature 407: 585–591 [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN (2000) Two-component signal transduction. Annu Rev Biochem 69: 183–215 [DOI] [PubMed] [Google Scholar]
- Surette MG, Levit M, Liu Y, Lukat G, Ninfa EG, Ninfa A, Stock JB (1996) Dimerization is required for the activity of the protein histidine kinase CheA that mediates signal transduction in bacterial chemotaxis. J Biol Chem 271: 939–945 [DOI] [PubMed] [Google Scholar]
- Tajima Y, Imamura A, Kiba T, Amano Y, Yamashino T, Mizuno T (2004) Comparative studies on the type-B response regulators revealing their distinctive properties in the His-to-Asp phosphorelay signal transduction of Arabidopsis thaliana. Plant Cell Physiol 45: 28–39 [DOI] [PubMed] [Google Scholar]
- Taylor JS, Raes J (2004) Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet 38: 615–643 [DOI] [PubMed] [Google Scholar]
- Urao T, Miyata S, Yamaguchi-Shinozaki K, Shinozaki K (2000. b) Possible His to Asp phosphorelay signaling in an Arabidopsis two-component system. FEBS Lett 478: 227–232 [DOI] [PubMed] [Google Scholar]
- Urao T, Yakubov B, Satoh R, Yamaguchi-Shinozaki K, Seki M, Hirayama T, Shinozaki K (1999) A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 11: 1743–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Yakubov B, Yamaguchi-Shinozaki K, Shinozaki K (1998) Stress-responsive expression of genes for two-component response regulator-like proteins in Arabidopsis thaliana. FEBS Lett 427: 175–178 [DOI] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki K, Shinozaki K (2000. a) Two-component systems in plant signal transduction. Trends Plant Sci 5: 67–74 [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Devlin PF (1997) Roles of different phytochromes in Arabidopsis photomorphogenesis. Plant Cell Environ 20: 752–758 [Google Scholar]
- Wurgler-Murphy SM, Saito H (1997) Two-component signal transducers and MAPK cascades. Trends Biochem Sci 22: 172–176 [DOI] [PubMed] [Google Scholar]
- Yamada H, Suzuki T, Terada K, Takei K, Ishikawa K, Miwa K, Mizuno T (2001) The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol 42: 1017–1023 [DOI] [PubMed] [Google Scholar]
- Yau CP, Wang L, Yu M, Zee SY, Yip WK (2004) Differential expression of three genes encoding an ethylene receptor in rice during development, and in response to indole-3-acetic acid and silver ions. J Exp Bot 55: 547–556 [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Kojima M, Yamaya T, Sakakibara H (2004) Molecular characterization of cytokinin-responsive histidine kinases in maize differential ligand preferences and response to cis-zeatin. Plant Physiol 134: 1654–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, Li S, Li H, Zhou J, Ni P, Dong W, Hu S, Zeng C, et al (2005) The genomes of Oryza sativa: a history of duplications. PLoS Biol 3: 266–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Ouyang S, Wang A, Zhu W, Maiti R, Lin H, Hamilton J, Haas B, Sultana R, Cheung F, et al (2005) The institute for genomic research Osa1 rice genome annotation database. Plant Physiol 138: 18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JY, Broekling CD, Blancaflor EB, Sledge MK, Summer LW, Wang ZY (2005) Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J 42: 689–707 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.