Abstract
The transition from embryonic to vegetative growth marks an important developmental stage in the plant life cycle. The turnip (tnp) mutant was identified in a screen for modifiers of POLARIS expression, a gene required for normal root growth. Mapping and molecular characterization of tnp shows that it represents a gain-of-function mutant of LEAFY COTYLEDON1 (LEC1), due to a promoter mutation. This results in the ectopic expression of LEC1, but not of other LEC genes, in vegetative tissues. The LEC class of genes are known regulators of embryogenesis, involved in the control of embryonic cell identity by currently unknown mechanisms. Activation of the LEC-dependent pathway in tnp leads to the loss of hypocotyl epidermal cell marker expression and loss of SCARECROW expression in the endodermis, the ectopic accumulation of starch and lipids, and the up-regulation of early and late embryonic genes. tnp also shows partial deetiolation during dark growth. Penetrance of the mutant phenotype is strongly enhanced in the presence of exogenous auxin and sugars, but not by gibberellin or abscisic acid, and is antagonized by cytokinin. We propose that the role of LEC1 in embryonic cell fate control requires auxin and sucrose to promote cell division and embryonic differentiation.
The early stages of embryogenesis in flowering plants involve the establishment of polarity, radial symmetry, and cellular differentiation as well as the formation of the shoot and root meristems, which determine postembryonic development (Laux et al., 2004). The later stages of embryogenesis see the establishment of the nutrient stores required during germination, as well as desiccation, which prepares the embryo for dormancy (Raz et al., 2001). The transition between the early and later stages of embryogenesis is therefore a key stage in the plant life cycle and has been shown to be under the control of several key genes and plant growth regulators (Ogas et al., 1997, 1999; Parcy et al., 1997; Lotan et al., 1998; Luerßen et al., 1998; Raz et al., 2001; Stone et al., 2001).
The LEAFY COTYLEDON (LEC) class of genes (LEC1 and LEC2 and FUSCA3 and FUS3) have been identified as key regulators of late embryogenesis (Parcy et al., 1997; Lotan et al., 1998; Luerßen et al., 1998; Stone et al., 2001). LEC1 encodes a transcription factor subunit related to the HAP3 subunit of the CCAAT binding factor family (Lotan et al., 1998), while FUS3 and LEC2 encode B3 domain transcription factors (Luerßen et al., 1998; Stone et al., 2001). Loss-of-function mutations in each of these genes result in embryos that are desiccation intolerant and are defective in the production of storage products. The mutant embryos also initiate postgermination processes, including premature activation of the shoot apical meristem (SAM), indicating a role for these genes in inhibiting premature germination (Meinke et al., 1994). The cotyledons of the mutants show leaf-like features such as the presence of trichomes, suggesting that these genes also function in the determination of organ identity.
As well as being key regulators of late embryogenesis, LEC genes have been shown to regulate aspects of early embryogenesis. The suspensors of LEC mutants develop abnormally, and in the case of lec1-2 fus3-3 double mutants the suspensor can continue to proliferate and form secondary embryos, suggesting that LEC genes may act to maintain suspensor cell fate and inhibit the embryonic potential of the suspensor. LEC1 expression is limited to embryogenesis while LEC2 and FUS3 are also expressed at low levels postgermination. Ectopic expression of LEC1 or LEC2 under the control of the cauliflower mosaic virus (CaMV) 35S promoter has been shown to be sufficient to induce embryonic characteristics in vegetative tissue, suggesting that these genes regulate embryogenic competence (Lotan et al., 1998; Luerßen et al., 1998; Stone et al., 2001).
Further evidence that LEC genes are regulators of embryo development has come from studies of the PICKLE (PKL) gene that encodes a CHD3 chromatin-remodelling factor (Ogas et al., 1999). Mutations in PKL result in the vegetative root meristem expressing embryonic traits (Ogas et al., 1997). Analysis of gene expression in pkl mutants reveals that they have high levels of LEC gene expression in vegetative tissue. PKL is therefore required for the repression of LEC genes during and after germination, so preventing activation of embryonic developmental pathways postgermination (Ogas et al., 1999; Dean Rider et al., 2003). An interesting aspect of the pkl mutant is that the mutant phenotype shows low penetrance that can be influenced by growth regulators. The pkl phenotype is suppressed by gibberellins (GAs) while penetrance is increased by growth in the presence of the GA biosynthetic inhibitor, uniconazole-P (Ogas et al., 1997). This, together with the fact that adult pkl plants display shoot phenotypes similar to GA-deficient mutants, suggests that PKL is part of a GA-signaling pathway that promotes the transition from embryonic to vegetative development.
The involvement of growth regulators, particularly auxin, in both zygotic and somatic embryogenesis has been widely reported (Toonen and de Vries, 1996; Fischer-Iglesias et al., 2001; Basu et al., 2002; Ribnicky et al., 2002; Friml et al., 2003). In many species the synthetic auxin is used to induce somatic embryogenesis (Toonen and de Vries, 1996), though the mechanism by which auxin acts is not clear. In zygotic embryogenesis, auxin distribution as determined by the localization and activities of auxin efflux carriers, appears to play a crucial role in the establishment of the axes of polarity (Friml et al., 2003). It is required for the polar expression of genes such as POLARIS (Topping and Lindsey, 1997; Casson et al., 2002). Any possible relationship between auxin and LEC function is currently unknown.
Here we describe the characterization of the turnip (tnp) mutant of Arabidopsis (Arabidopsis thaliana), which we show is a gain-of-function mutant of LEC1. We describe results of experiments to investigate the relationship between LEC1 expression and hormonal and nutritional modifiers of embryonic development, and discuss how LEC1 may act in concert with auxin and sugars to potentiate embryonic pathways.
RESULTS
Identification of the tnp Mutant
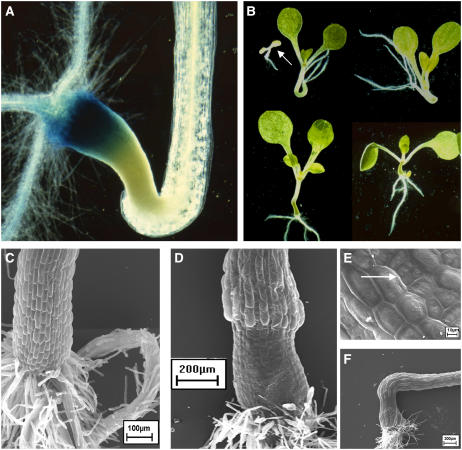
The polaris (pls) mutant is defective in a gene encoding a predicted small polypeptide necessary for correct root growth (Casson et al., 2002). This was initially identified as a promoter-trap line that showed β-glucuronidase (GUS) activity predominantly in the basal region of the embryo and in the seedling root tip (Topping et al., 1994; Topping and Lindsey, 1997). To identify modifiers of PLS expression, the pls promoter-trap line was mutagenized by T-DNA insertion and the transgenic population was screened for mutants in which GUS expression was altered. Line 930 expressed PLS-GUS abnormally at the hypocotyl-root junction, though expression was unaffected elsewhere in the seedling (Fig. 1A). At this position was also formed a structure that was swollen and dense. This phenotype segregated and the mutant was called tnp.
Figure 1.
Phenotype of tnp seedlings. A, Root-hypocotyl junction of a 7-d-old tnp seedling showing GUS activity of the PLS promoter trap in the basal region of the abnormal structure. Magnification ×7. B, Seven-day-old tnp seedlings showing phenotypic variation. Arrow indicates seedling lethal phenotype. Magnification ×2. C, Scanning electron micrograph of the hypocotyl of a 7-d-old pls seedling. Bar = 100 μm. D, Scanning electron micrograph of the hypocotyl of a 7-d-old tnp seedling. Bar = 200 μm. E, Scanning electron micrograph of the tnp structure. Arrow indicates abnormal divisions in a cell file. Bar = 10 μm. F, Scanning electron micrograph of a tnp hypocotyl showing cell elongation above the structure. Bar = 200 μm.
tnp Is Dominant But Shows Incomplete Penetrance
The number of tnp seedlings present in the T2 population was greater than expected for a single, recessive locus, suggesting that the mutation may be dominant (126 wild type/170 tnp). Segregation analysis on T2 seedlings revealed that the tnp mutation was not associated with the presence of a T-DNA (12/55 hygromycin sensitive seedlings were tnp). PCR analysis of the F2 progeny of a tnp/Columbia-0 (Col-0) cross indicated that the mutation was not due to the presence of a partial activation tag nor was it dependent on the pls mutation (data not shown).
While the data indicated that tnp is a dominant mutation, segregation analysis of independent T3 lines showed that the penetrance of the tnp phenotype was highly variable between lines, ranging typically from 0% to 60%. To determine if the incomplete penetrance was due to methylation-dependent gene silencing, individual T3 sibling lines were germinated in the presence of 100 μm 5-azacytidine, a methylation inhibitor (Jones and Taylor, 1980). In each line 5-azacytidine was able to increase penetrance of the tnp phenotype compared to controls, though the effect was highly variable between lines (Table I). However, the results do suggest that methylation-mediated gene silencing may be partially responsible for the incomplete penetrance of tnp.
Table I.
The effect of the methylation inhibitor 5-azacytidine on penetrance of the tnp mutant phenotype
Three independent T3 sibling lines (1, 7, and 21) were grown on 1/2× MS 10 medium or medium supplemented with 100 μm 5-azacytidine (5-AZA-C). The number of seedlings with a wild-type appearance (TNP) or showing a tnp mutant phenotype (tnp) was counted 7 d postgermination.
| T3 Line | Medium | TNP | tnp | % tnp |
|---|---|---|---|---|
| 1 | 1/2× MS 10 | 53 | 21 | 28 |
| 1 | 100 μm 5-AZA-C | 30 | 49 | 62 |
| 7 | 1/2× MS 10 | 120 | 0 | 0 |
| 7 | 100 μm 5-AZA-C | 112 | 9 | 7 |
| 21 | 1/2× MS 10 | 75 | 52 | 41 |
| 21 | 100 μm 5-AZA-C | 71 | 65 | 48 |
The tnp Mutant Shows Altered Cell Identity
tnp seedlings exhibit a high degree of phenotypic variability and on rare occasions prove to be seedling lethal (Fig. 1B, top left, arrow). tnp mutants with the strongest phenotype have the whole hypocotyl replaced by the swollen structure (Fig. 1B, bottom right) while those with a weaker phenotype display extreme curling of the hypocotyl with some evidence of dense, greening cells (Fig. 1B, top middle and right). While the structure was most commonly found at the root-hypocotyl junction, it could be present anywhere along the hypocotyl. Examination of embryos from tnp and control siliques did not reveal any morphological differences, suggesting that the phenotypic defect develops after germination.
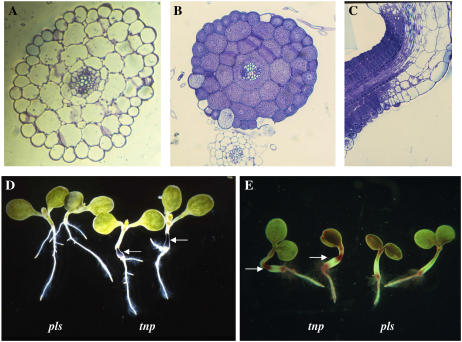
Scanning electron microscopy was used to investigate the surface patterning of the abnormal hypocotyl. The epidermal cells were found to be much smaller and flatter than those of the pls parent (Fig. 1, C and D). While the cells in tnp remained in strict files, occasionally abnormal divisions occurred within a file generating a number of even smaller cells (Fig. 1E). At the boundary of the abnormal hypocotyl, cells were seen to undergo excessive elongation (Fig. 1F). To determine whether this altered morphogenesis was associated with any change in internal cell patterning, transverse and longitudinal sections of the structure were examined. No obvious patterning defects were observed (Fig. 2, A and B). Sectioning revealed, however, that the cells in the abnormal region of the hypocotyl were virtually devoid of a vacuole, and that the transition from abnormal to normal cells did not occur at a strict boundary across the structure (Fig. 2C).
Figure 2.
Sections and accumulation of storage compounds in tnp seedlings. A, Transverse section through the hypocotyl of a 7-d-old pls seedling. Magnification ×20. B, Transverse section of a 7-d-old tnp seedling. Magnification ×20. C, Longitudinal section through the abnormal structure of a 7-d-old tnp seedling. Magnification ×20. D, Seven-day-old pls and tnp seedlings stained with Lugol's solution to visualize starch accumulation. Magnification ×2. E, Seven-day-old pls and tnp seedlings stained with Fat Red 7B solution to visualize triacylglycerol accumulation. Arrows point to the position of the abnormal structure that is in the middle and at the top of the two tnp seedlings. Magnification ×2.
The absence of a large central vacuole and the dense staining of the cells with toluidine blue resembled storage tissue, so the cells were tested for the presence of storage compounds. Staining with Lugol's solution indicated that the cells were packed with starch granules (Fig. 2D), and staining with Fat Red indicated the presence of large amounts of lipids (Fig. 2E).
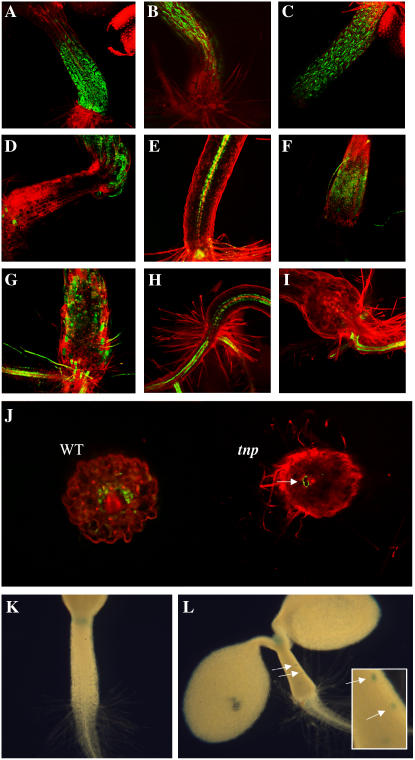
The altered morphogenesis, high levels of starch and lipids, and the altered expression of PLS in the abnormal hypocotyl region suggested an alteration in cell identity. To examine this further, the expression patterns of other markers were investigated. Epidermal cells of the hypocotyl are marked by expression of the Haseloff J2662 and J2601 green fluorescent protein (GFP) marker lines (http://www.plantsci.cam.ac.uk/Haseloff/). In tnp seedlings, expression was absent in the cells of the swollen structure but was present in cells above it (Fig. 3, A–D). The ARR5/IBC6∷GFP (Brandstatter and Kieber, 1998; Casson et al., 2002) marker is normally expressed in pericycle cells of the root and hypocotyl and is also a marker of cytokinin responsiveness. However, in tnp seedlings expression was found to be highly variable both in the abnormal hypocotyl and in morphologically normal hypocotyl cells, most often appearing in the epidermal cell layer (Fig. 3, E–G). Expression of a SCR∷GFP marker (Di Laurenzio et al., 1996; Wysocka-Diller et al., 2000) was used to examine endodermal cell identity. While expression was evident in the root and morphologically normal hypocotyl cells, expression was virtually absent in the abnormal structure. Analysis of transverse sections of tnp hypocotyl did however reveal that rare, vacuolated cells showed SCR∷GFP expression (Fig. 3, H–J).
Figure 3.
Gene expression in the tnp mutant. A and B, J2662 GFP expression in 7-d-old seedling of wild type (A) and tnp (B). C and D, J2601 GFP expression in 7-d-old seedling hypocotyl epidermal cells of wild type (C) and tnp (D). E to G, ARR5/IBC6∷GFP expression in the pericycle of 7-d-old seedlings of wild type (E) and tnp (F and G). H to J, SCR∷GFP expression in the endodermis of hypocotyls of 7-d-old seedlings of wild type (H) and tnp (I). J, Fresh transverse sections of wild-type (WT) and tnp seedlings. Arrow indicates SCR∷GFP expression in a vacuolated cell in the tnp section. K and L, CYCAT1:CDB:GUS activity in the hypocotyl of 7-d-old wild type (K) and tnp (L). Inset and arrows show cells that have undergone cell division.
Ordinarily, growth of the hypocotyl after embryogenesis is purely via cell expansion (Gendreau et al., 1997). To examine if this was the case in tnp seedlings, a CYCAT1:CDB:GUS marker (Hauser and Bauer, 2000) was used to examine cell division events in tnp. As expected, no cell division events were marked in wild-type seedlings, but were evident in tnp (Fig. 3, K and L).
tnp Seedlings Exhibit Defective Dark Growth
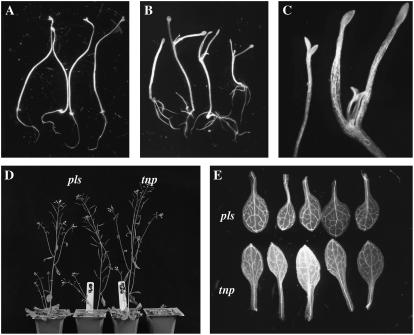
Growing tnp seedlings in the dark and in the presence of 1% Suc resulted in a lower rate of penetrance than light-grown tnp seedlings (11.3% versus 16.7%; n = 200). It was also observed that the dark-grown seedlings underwent partial deetiolation (Fig. 4, A–C). During dark growth, the SAM of the pls control was not activated, whereas in the case of tnp seedlings the petioles of the cotyledons expanded and first leaves developed after 7 d. It has been shown that contact of the SAM with Suc-containing medium gives rise to a similar effect (Roldán et al., 1999). It was found, however, that the SAM was not contacting the growth medium in 60% of tnp seedlings showing partial deetiolation, though the deetiolated phenotype was more pronounced in those seedlings that did show contact. As well as differences in the activity of the SAM, the root system was also different to that of pls, with a greater number of, and more elongated, lateral roots (Fig. 4, A and B). There was no difference in the root architecture of light-grown pls and tnp seedlings (data not shown).
Figure 4.
tnp mutants are defective in other aspects of development. A, Seven-day-old dark-grown pls seedlings showing normal etiolated phenotype. Magnification ×2. B, Seven-day-old dark-grown tnp seedlings showing growth of first true leaves and branched root system. Magnification ×2. C, Shoot apices of pls (left) and tnp (right) 7-d-old dark-grown seedlings. Magnification ×4. D, Forty-day-old pls and tnp plants, showing both normal and late-flowering phenotypes. E, First true leaves of 14-d-old pls and tnp seedlings, showing the more elliptical shape of the tnp leaves. Magnification ×2.
Other aspects of development were also affected in tnp plants. Flowering time was found to be highly variable. The majority of plants flowered at the same time as the pls parental line, though some tnp plants were late flowering (Fig. 4D). Examination of the first true leaves of tnp revealed that they were more elliptical than those of pls (Fig. 4E).
Cloning of TNP
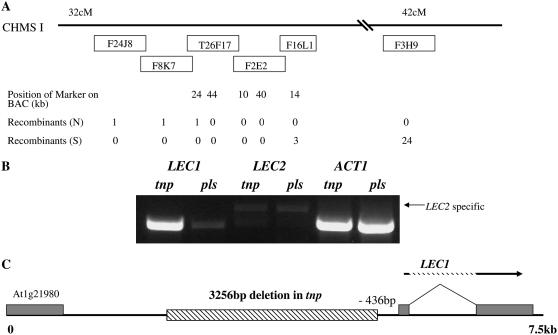
Segregation analysis had shown that the tnp mutation was not associated with a T-DNA. A map-based cloning strategy was therefore used with an F2 mapping population generated by outcrossing tnp (C24 ecotype) to Col-0. TNP was tentatively positioned at approximately 40 cM on chromosome I using the simple sequence length polymorphism (SSLP) marker nga 280 (83 cM). To further map TNP a strategy was developed that would account for the dominant phenotype of tnp linked with the incomplete penetrance. Seed from the F2 mapping population was germinated on medium containing 2% Suc and 25 nm 2,4-dichlorophenoxyacetic acid (2,4-D), which had been found to result in the highest penetrance of tnp without affecting growth (see below), therefore increasing the proportion of TNP/tnp heterozygotes in the population. SSLP analysis was then performed with markers expected to be located on either side of the TNP gene, and plants were identified that had a Col-0 ecotype at one marker and were Col-0/C24 at the second marker, and vice versa. Hence, the dominant tnp heterozygote was used to map TNP. Using this approach, 24/800 plants were found to be Col-0 with the marker nga 248 (42.17 cM, bacterial artificial chromosome [BAC] F3H9) and 1/800 was Col-0 at the marker F24J8 (approximately 32 cM, BAC F24J8). These plants were Col-0/C24 heterozygotes at the alternative marker. Fine mapping was able to place TNP onto either BAC T26F17 or F2E2 (Fig. 5A).
Figure 5.
The tnp mutant contains a deletion in the promoter of LEC1, resulting in ectopic expression of LEC1. A, Mapping of the TNP locus. BAC clones and the number of recombinants are shown. B, Semiquantitative RT-PCR analysis of LEC1 and LEC2 transcript levels using RNA extracted from tnp and pls seedlings 1 to 2 d postgermination. Arrow indicates the size of the LEC2 specific product. ACT1 is the amplification control. Control experiments lacking reverse transcriptase showed no amplification products (data not shown). C, Schematic diagram indicating the position of the 3,256 bp deletion of the LEC1 promoter in tnp mutants relative to the transcriptional start site of the LEC1 gene.
BAC T26F17 contains the LEC1 gene (Lotan et al., 1998), which is expressed ectopically after germination in the pkl mutant (Ogas et al., 1999). The pkl root phenotype is reminiscent of the tnp hypocotyl phenotype, suggesting that LEC1 was a potential candidate for TNP. Therefore, the genomic region containing the LEC1 coding sequence was amplified from tnp mutants and sequenced but no nucleotide differences were identified between tnp and the pls parental line.
One possibility we considered was that a nucleotide change in the LEC1 promoter could result in ectopic expression of LEC1, as observed in the pkl mutant. Therefore, semiquantitative reverse transcription (RT)-PCR was performed to determine the levels of LEC1 transcript levels in seedlings at 1 to 2 d postgermination. While low levels of LEC1 were detected in RNA from control germinating seedlings, LEC1 transcript levels were strongly up-regulated in the tnp mutant, whereas LEC2, which is also up-regulated in pkl (Dean Rider et al., 2003), was unaffected (Fig. 5B).
To determine if the up-regulation of LEC1 in tnp was due to a mutation in the promoter region, genomic DNA upstream of the LEC1 transcriptional start site was amplified by thermal asymmetric interlaced PCR (Liu et al., 1995) and sequenced. This revealed that tnp contained a deletion of 3,256 bp, approximately 436 bp upstream of the putative LEC1 transcriptional start site (Fig. 5C). PCR analysis of F2 tnp seedlings from the mapping population were tested for the presence of the deletion and it was found to be present in 40/40 plants. It was concluded that this deletion is responsible for the tnp phenotype.
tnp pkl Double Mutants Show a Combinatorial Phenotype
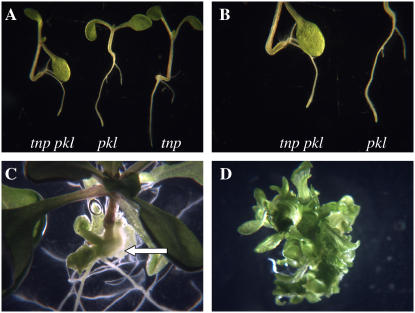
The tnp mutant phenotype is similar to that of the pkl mutant, which is characterized by the development of swollen and greenish roots that accumulate triacylglycerols and protein bodies (Ogas et al., 1997, 1999). To examine the relationship between pkl and tnp, double mutants were generated using the pkl1-1 allele (Ogas et al., 1997) and examined. The tnp pkl double mutants initially showed a simple additive phenotype, with both a tnp hypocotyl and a pkl root (5 d postgermination; Fig. 6, A and B). However, by 14 d postgermination it could be seen that a number of the double mutants were producing adventitious shoots from the tnp hypocotyls (but not the pkl roots), a phenomenon not seen in tnp (Fig. 6C). This occurred in 11/18 (61%) tnp pkl 14 d postgermination seedlings, with some showing numerous leafy structures. The shoots were excised from the hypocotyls and placed on standard 1/2× Murashige and Skoog (MS) 10 medium. Nine days after excision (23 d postgermination) all the shooting explants were still green and proliferating (Fig. 6D), though there was little evidence of rooting, except where root tissue had already been present.
Figure 6.
tnp/pkl double mutants show adventitious shooting. A, Representative tnp/pkl seedlings alongside parental pkl and tnp seedlings 5 d postgermination. Magnification ×2. B, Primary root tips of tnp/pkl and pkl seedlings 5 d postgermination. Magnification ×4. C, Adventitious shooting from a tnp/pkl hypocotyl (arrow) 14 d postgermination. Magnification ×2. D, Shooting tissue 9 d postexcision (23 d postgermination). Magnification ×4.
Auxin and Suc Increase Penetrance of the tnp Phenotype
The penetrance of the pkl mutant phenotype is also variable and is affected by GA and the GA biosynthesis inhibitor, uniconazole-P. To determine if growth factors affect penetrance of the tnp phenotype, tnp seed was germinated and grown in the presence of a number of compounds (Table II).
Table II.
The effect of growth regulators on penetrance of the tnp mutant phenotype
Seedlings were grown on 1/2× MS 10 medium or medium supplemented with the indicated compound, and the number of seedlings with a wild-type (TNP) or tnp mutant phenotype (tnp) was counted 7 d postgermination. Independent experiments are separated by the dashed row. Asterisk (*) indicates that germination frequency was only approximately 25% of controls.
| Medium | TNP | tnp | % tnp |
|---|---|---|---|
| 1/2× MS 10 | 162 | 79 | 32.8 |
| Naphthylacetic acid 10 nm | 83 | 62 | 42.8 |
| IAA 50 nm | 75 | 168 | 69.1 |
| 2,4-D 10 nm | 34 | 121 | 78.1 |
| NPA 10 μm | 86 | 107 | 55.4 |
| NOA 10 μm | 61 | 87 | 58.8 |
| BA 100 nm | 91 | 10 | 9.9 |
| 1-Aminocyclopropane-1-carboxylic acid 10 pm | 104 | 43 | 29.3 |
| GA 10 μm | 96 | 57 | 37.3 |
| 1/2× MS 10 | 145 | 133 | 47.8 |
| GA 10 nm | 129 | 101 | 42.1 |
| GA 100 nm | 123 | 133 | 52.0 |
| GA 1 μm | 117 | 164 | 58.4 |
| GA 10 μm | 107 | 89 | 45.4 |
| Paclobutrazol 10 nm | 32 | 178 | 84.7 |
| Paclobutrazol 100 nm* | 13* | 34* | 72.3* |
| BA 10 nm | 153 | 187 | 55.0 |
| BA 100 nm | 203 | 112 | 35.6 |
| BA 1 μm | 304 | 16 | 5.0 |
| 2,4-D 10 nm | 42 | 192 | 82.1 |
| 2,4-D 100 nm | 9 | 183 | 95.3 |
| 2,4-D 1 μm | 4 | 224 | 98.2 |
As with pkl, penetrance of tnp was increased in the presence of a GA biosynthetic inhibitor, paclobutrazol. Unlike pkl however, GA was not found to suppress penetrance of tnp and indeed had a weak positive effect. Interestingly, when germinated in the presence of both 10 nm paclobutrazol and 10 μm GA, the positive effect of the paclobutrazol was partially suppressed (data not shown).
The natural auxin indole-3-acetic acid (IAA) and synthetic auxins naphthylacetic acid and 2,4-D each had a positive effect on penetrance of the tnp phenotype at low concentration. Of the auxins tested 2,4-D was the most effective, increasing penetrance to nearly 100% at 1 μm. The auxin transport inhibitors napthylphthalamic acid (NPA) and 1-naphthoxyacetic acid (NOA) were also found to have a positive effect on tnp penetrance, whereas the ethylene precursor 1-aminocyclopropane-1-carboxylic acid was found to have little effect. The cytokinin benzyladenine (BA) was the only compound tested that markedly suppressed penetrance of the tnp phenotype, though this was only significant at concentrations above 100 nm. Among other compounds tested, abscisic acid (ABA) was not found to have an effect on penetrance and tnp seedlings showed no difference in sensitivity to ABA in germination studies (data not shown).
Given the large quantities of starch stored in tnp, the effect of sugars on penetrance was examined. Absence of Suc in the medium resulted in a complete loss of penetrance of the tnp phenotype while the highest penetrance was observed with 2% Suc. The addition of 10 nm 2,4-D to the medium always resulted in greater penetrance than with Suc alone, even in the absence of Suc, suggesting that these compounds act via different pathways to increase penetrance (Table III). One-percent Glc or Fru were not as potent as Suc, though the addition of 2,4-D resulted in comparable rates of penetrance, indicating that auxin is more effective at increasing the penetrance than is the carbon source (data not shown).
Table III.
The effect of Suc on penetrance of the tnp mutant phenotype
Seedlings were grown on 1/2× MS medium or medium supplemented with the indicated concentration of Suc and/or 10 nm 2,4-D, and the number of seedlings with a wild-type (TNP) or tnp mutant phenotype (tnp) was counted 7 d postgermination.
| Medium | TNP | tnp | % tnp |
|---|---|---|---|
| 0% Suc | 184 | 0 | 0 |
| 0% Suc + 10 nm 2,4-D | 138 | 3 | 2 |
| 1% Suc | 70 | 107 | 60.5 |
| 1% Suc + 10 nm 2,4-D | 21 | 120 | 85.1 |
| 2% Suc | 53 | 100 | 65.4 |
| 2% Suc + 10 nm 2,4-D | 13 | 155 | 92.3 |
| 3% Suc | 64 | 90 | 58.4 |
| 3% Suc + 10 nm 2,4-D | 23 | 122 | 84.1 |
| 4% Suc | 78 | 86 | 52.4 |
| 4% Suc + 10 nm 2,4-D | 46 | 102 | 68.9 |
| 5% Suc | 79 | 93 | 54.1 |
| 5% Suc + 10 nm 2,4-D | 28 | 131 | 82.4 |
| 6% Suc | 87 | 65 | 42.8 |
| 6% Suc + 10 nm 2,4-D | 50 | 97 | 66.0 |
The ability of auxins and auxin transport inhibitors to increase the penetrance of the tnp phenotype raised the possibility that auxin distribution or levels are affected in tnp mutants. The auxin-inducible reporter IAA2-GUS (Swarup et al., 2001) was introduced into tnp mutants following outcrossing of the PLS promoter trap. In control seedlings, GUS expression was observed at the root-hypocotyl junction, but not significantly elsewhere on the hypocotyl (Fig. 7A). In weak tnp phenotypes, GUS expression was observed in the structure (Fig. 7B), whereas strong tnp phenotypes showed much higher GUS expression throughout the swollen hypocotyl (Fig. 7C). When grown in the presence of auxin, tnp seedlings always displayed a strong phenotype (Fig. 7D).
Figure 7.
IAA2∷GUS expression is up-regulated in tnp mutant structures. All seedlings are 7 d postgermination. A, IAA2∷GUS expression in wild-type seedlings, 7 d postgermination. B, Weak tnp mutants with IAA2∷GUS expression in structures (arrows), 7 d postgermination. C, Strong tnp mutants showing IAA2∷GUS expression, 7 d postgermination. D, A wild-type (left) and a tnp (right) seedling grown on 10 nm 2,4-D, for 7 d postgermination.
High Suc or Glc concentrations in the growth medium are known to inhibit germination and have been used in selection screens for identifying sugar sensitivity mutants (Arenas-Huertero et al., 2000; Laby et al., 2000). To determine if the effect of sugars on tnp penetrance was due to a change in sugar sensitivity, the effect of high Suc or Glc concentrations on germination was examined. No difference was observed between pls and tnp on Suc concentrations ranging between 1% to 10% and on 7% Glc (data not shown). This indicates that tnp is not hypersensitive or insensitive to these sugars.
Given that the penetrance of the tnp phenotype requires the presence of sugars in the growth medium, we monitored starch accumulation to examine whether the continued presence of sugars is required for the maintenance of starch in the tnp hypocotyl. tnp seedlings were germinated in the presence of 1% Suc and at 3 d postgermination were either transferred to the same medium or medium lacking Suc. Eight days after transfer, starch levels in tnp seedlings grown on medium lacking Suc were significantly lower than those maintained on Suc (Supplemental Fig. S1), indicating that a continued supply of Suc is required for starch accumulation.
To eliminate the possibility that the increased penetrance of the tnp phenotype in response to sugars is due to an osmotic stress response, penetrance was determined in the presence of mannitol. It has been shown that under certain conditions osmotic stress can induce somatic embryogenesis in Arabidopsis (Ikeda-Iwai et al., 2003). However, we observed no penetrance of the tnp phenotype when tnp seed was germinated in the presence of 1% mannitol compared to 8.7% penetrance when germinated on 1% Suc (Supplemental Table S1).
Auxin, Paclobutrazol, and Cytokinin Do Not Affect LEC1 Transcript Levels
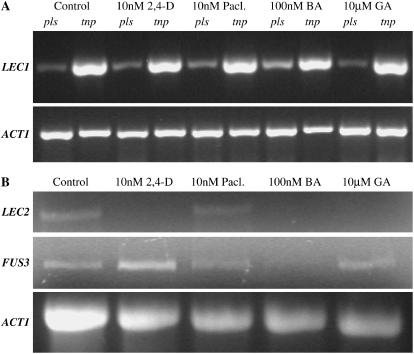
One mechanism by which auxin, paclobutrazol, and cytokinin could affect tnp penetrance would be to alter the levels of LEC1 transcript, with higher transcript levels associated with greater penetrance. Germinating seedlings were therefore treated with these compounds, as well as with GA, which has no effect on penetrance, and RNA was extracted 1 to 2 d postgermination LEC1 transcript levels were determined by semiquantitative RT-PCR and were found to be unaltered in response to these compounds in both tnp and pls controls (Fig. 8A). Therefore, the effect on penetrance by these compounds is not mediated by alterations in LEC1 transcript levels, though posttranscriptional or -translational effects cannot be excluded.
Figure 8.
The effect of plant growth regulators on gene expression in tnp. A, Semiquantitative RT-PCR analysis using RNA extracted from pls and tnp seedlings 1 to 2 d postgermination. Seedlings were grown on 1/2× MS 10 medium (control) or 1/2× MS 10 supplemented with 10 nm 2,4-D, 10 nm paclobutrazol, 100 nm BA, or 10 μm GA. The top section shows LEC1 amplification products and the bottom section shows amplification of the ACT1 control. Control experiments lacking reverse transcriptase showed no amplification products (data not shown). B, Semiquantitative RT-PCR analysis using RNA extracted from tnp seedlings 1 to 2 d postgermination. Treatments were the same as in A. The top section shows LEC2 amplification products, the middle section shows FUS3 amplification products, and the bottom section shows amplification of the ACT1 control. Control experiments lacking reverse transcriptase showed no amplification products (data not shown).
An alternative possibility is that these compounds act by altering the expression of other key embryonic regulators to alter penetrance. FUS3 and LEC2 play key roles in embryogenesis and the transition to germination (Luerßen et al., 1998; Stone et al., 2001; Kroj et al., 2003; Gazzarrini et al., 2004). Semiquantitative RT-PCR was used to determine if the transcript levels for these genes are affected in tnp seedlings 1 to 2 d postgermination in response to these compounds (Fig. 8B). Despite the slightly uneven loading of RNA, it can be seen that 2,4-D, BA, and GA treatments significantly reduce the levels of LEC2 transcripts in tnp seedlings (no detectable signal; Fig. 8B), whereas paclobutrazol had no effect. In the case of FUS3, 2,4-D treatment was found to increase transcript levels whereas BA resulted in a reduction. Paclobutrazol and GA did not significantly alter FUS3 transcript levels in the tnp mutant.
Genes for Embryonic Competence and Late Embryogenesis Are Up-Regulated in tnp
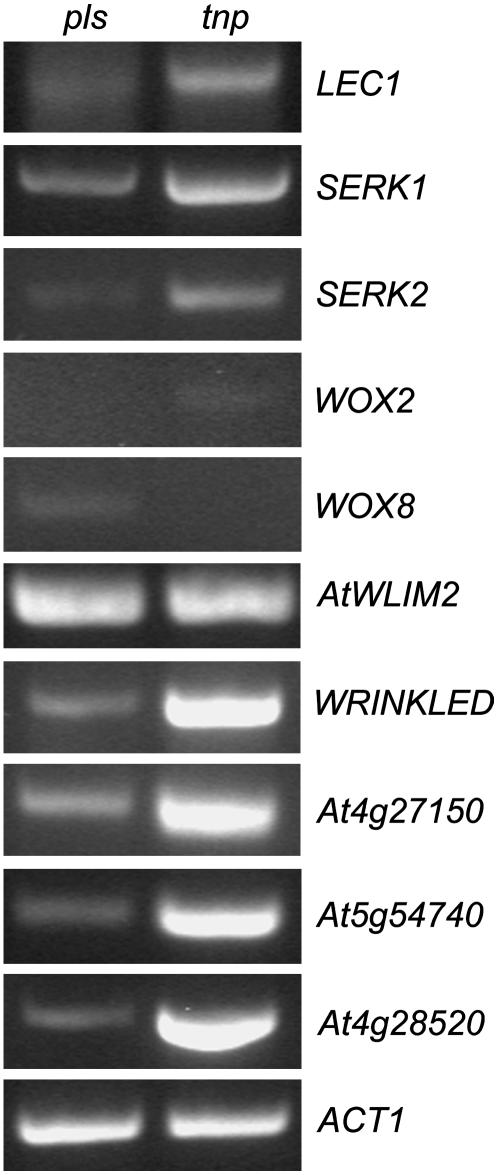
The hypocotyl of the tnp mutant appears to have acquired embryo-like characteristics. While LEC1 expression is up-regulated in tnp seedlings, the expression of other LEC-family members LEC2 and FUS3 is not affected. To further investigate the nature of the gene expression changes occurring in tnp mutants we examined the expression of several genes associated with both early and late embryogenesis using semiquantitative RT-PCR (Fig. 9). The somatic embryogenesis receptor kinase (SERK1) gene has been found to enhance the embryonic competence of cultured cells, while SERK2 is its most closely related family member (Hecht et al., 2001). The WUSCHEL-RELATED HOMEOBOX genes WOX2 and WOX8 are only expressed in the early stages of embryogenesis and determine cell fate (Haecker et al., 2004). At4g27150 and At5g54740 encode 2S storage protein-like genes, At4g28520 (CRU3; Pang et al., 1988) encodes a 12S cruciferin seed storage protein, and WRINKLED1 is an APETALA2/ethylene-responsive element-binding protein transcription factor required for the control of storage compound biosynthesis (Cernac and Benning, 2004). AtWLIM2 is predicted to encode a LIM domain transcription factor (Eliasson et al., 2000) that was shown to be up-regulated in pkl mutants and expressed strongly in developing siliques (Dean Rider et al., 2003).
Figure 9.
Expression analysis of early and late embryo genes in tnp mutants Semiquantitative RT-PCR analysis using RNA extracted from pls (left) and tnp (right) seedlings 2 d postgermination. ACT1 is used as a loading control. See text for details of specific genes.
Both SERK1 and SERK2 were found to be up-regulated in 2-d-old tnp seedlings (Fig. 9). The expression of WOX2 and WOX8 was extremely low and variable. In contrast, all the genes associated with storage compound production were up-regulated in tnp seedlings, indicating that seed maturation pathways are activated in tnp vegetative tissues. Finally, AtWLIM2 expression, like LEC2, was not altered in tnp, indicating that only LEC1, and most probably not other PKL targets, are affected in tnp mutants.
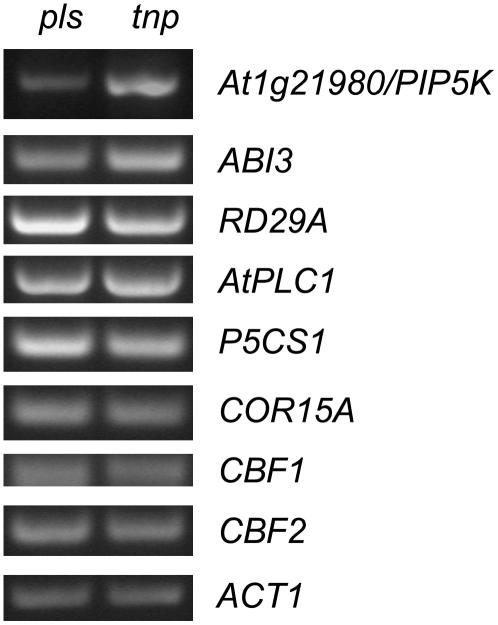
A Putative 1-Phosphatidylinositol-4-P 5-Kinase Is Up-Regulated in tnp Mutants But Downstream Stress Pathways Are Not Activated
The deletion in tnp mutants lies in the region between the LEC1 gene and another gene, At1g21980, a putative 1-phosphatidylinositol-4-P 5-kinase (PIP5K; Mikami et al., 1998; Elge et al., 2001; Westergren et al., 2001) with the start codon of At1g21980 approximately 1.9 kb from the start of the deletion (Fig. 5C). Since LEC1 transcript levels are up-regulated in tnp mutants, we determined whether those of PIP5K are similarly affected. Semiquantitative RT-PCR analysis revealed that transcript levels of PIP5K are up-regulated in tnp seedlings (Fig. 10), indicating that PIP5K may contribute to the tnp phenotype. PIP5K has been previously reported as being expressed in procambial cells (Elge et al., 2001), is induced by water stress and ABA (Mikami et al., 1998), and catalyzes the synthesis of the phosphoinositide signaling intermediates phosphatidylinositol-3,4-bisphosphate and phosphatidylinositol-4,5-bisphosphate (Westergren et al., 2001). Phosphoinositide signaling has been implicated as playing a role in stress responses such as salt, cold, and osmotic stress (Hirayama et al., 1995; Zhu, 2002; Williams et al., 2005). We therefore examined the expression of a number of stress-induced genes such as the cold stress-induced genes CBF1, CBF2, and COR15a (Gilmour et al., 1998); RD29a and AtPLC1 that are induced in response to cold, salt, and osmotic stress (Hirayama et al., 1995; Narusaka et al., 2003; Nakashima et al., 2006) in tnp mutant seedlings. No significant differences in expression were observed when compared to the pls parental line (Fig. 10).
Figure 10.
Expression analysis of stress-induced genes in tnp mutants. Semiquantitative RT-PCR analysis using RNA extracted from pls (left) and tnp (right) seedlings 2 d postgermination. ACT1 is used as a loading control. See text for details of specific genes.
DISCUSSION
LEC1 is an important regulator of both early and late embryogenesis and is also required for somatic embryogenesis (Gaj et al., 2005). LEC1 expression is restricted to embryogenesis and is repressed in vegetative tissue postgermination in part by PKL, a putative chromatin-remodelling factor (Lotan et al., 1998; Ogas et al., 1999). Therefore, the repression of LEC1 expression is a feature of the transition from embryonic to vegetative growth. Here we report the isolation and characterization of the tnp mutant, which shows ectopic expression of LEC1 due to a deletion of part of the gene promoter; therefore, we can designate the mutant lec1-dtnp.
The tnp Mutation Derepresses LEC1 Expression Postembryonically
The tnp mutation mapped to a deletion of part of the regulatory region of the LEC1 gene, leading to an increased transcript abundance in seedlings. Since lec1-dtnp embryos did not display any defects in morphology or desiccation tolerance, we conclude that the levels of LEC1 mRNA are not altered enough to disrupt normal embryo development. In contrast to previous reports (Lotan et al., 1998; Ogas et al., 1999), low levels of LEC1 expression were found in germinating wild-type seedlings, though this may be due to ecotype-specific differences (C24 compared to Wassilewskija for LEC1 and Col-0 for PKL). The high levels of LEC1 expression in lec1-dtnp suggest the deleted promoter sequence contains elements that are required for the repression of LEC1 in vegetative tissue.
PKL is a member of the CHD3 family of chromatin-remodeling factors, which form part of a NuRD histone deacetylase complex that has been shown to be involved in transcriptional repression in animal systems (Ahringer, 2000; Vignali et al., 2000). Since LEC1 is also derepressed in pkl mutants, it is possible that histones bound to the deleted promoter region may be targeted for deacetylation by the NuRD complex in vegetative tissue, resulting in repression of LEC1. An alternative explanation is that this region contains the binding site(s) for other as yet unidentified transcriptional repressor(s). A further possibility is that the deleted region brings other regulatory sequences, associated with upstream genes, in closer association with LEC1 5′ flanking sequences to activate transcription. We cannot yet distinguish between these three possibilities. The fact that At1g21980, adjacent to LEC1, is also up-regulated in tnp suggests the existence of a mechanism that coordinately represses both genes.
At1g21980 encodes a putative 1-phosphatidylinositol-4-P 5-kinase (PIP5K) that is involved in the synthesis of the intermediates phosphatidylinositol-3,4-bisphosphate and phosphatidylinositol-4,5-bisphosphate. Phosphoinositide signaling is believed to be involved in stress responses (Zhu, 2002). Altering the pool of phosphatidylinositol-4,5-bisphosphate, such as in the sac9 mutant (Williams et al., 2005), results in constitutive activation of the stress pathway. However, while the deletion in tnp mutants results in increased expression of PIP5K, which potentially can mimic water stress (Mikami et al., 1998), we did not observe any significant changes in the expression of several stress-induced genes. Also, the penetrance of the tnp phenotype was not affected by the presence of mannitol in the growth medium, which mimics osmotic stress. Furthermore, tnp does not show an altered sensitivity to ABA in germination experiments (data not shown). We therefore conclude that while the altered expression of PIP5K in tnp mutants may contribute to some other aspects of the mutant phenotype, it does not directly contribute to the embryonic phenotype. This is further supported by our own data that show that PIP5K is expressed at only very low levels during embryogenesis (Casson et al., 2005).
Ectopic LEC1 Expression Promotes Embryonic Identity in Seedlings
Previous analysis of lec1 mutants has shown that LEC1 is required to specify embryonic organ identity (lec1 mutants develop cotyledons with leaf-like features), and is also involved in activating pathways involved in storage product accumulation (Meinke et al., 1994; West et al., 1994). Overexpressing LEC1 under the control of the CaMV 35S promoter was shown to result in a high degree of seedling lethality, with those seedlings showing embryo-like morphology (Lotan et al., 1998). The rare 35S∷LEC1 seedlings that did survive were found to produce embryo-like structures from vegetative tissues, indicating that LEC1 is sufficient to induce embryonic developmental pathways in vegetative tissue. In the case of lec1-dtnp seedlings, lethality was rare and no ectopic embryos were ever observed to develop on vegetative tissue. A possible explanation for this difference may be that lower levels, or different patterns, of LEC1 transcription occur in lec1-dtnp compared to the 35S∷LEC1 seedlings.
Nevertheless, the ectopic expression of LEC1 in lec1-dtnp did result in some or all of the hypocotyls acquiring embryonic traits, most notably the activation of embryonic genes and altered patterns of hypocotyl markers (Figs. 3 and 9). The most evident phenotypic effect was the accumulation of large amounts of starch and lipids. Starch is not normally a major storage product in mature Arabidopsis embryos, and lec1 mutants themselves accumulate more starch than wild-type embryos. There are suggestions that starch is a default storage deposition pathway (Lin et al., 1999). Some of our semiquantitative RT analysis was performed on populations of tnp plants that lacked penetrance of the tnp phenotype and yet still had a molecular phenotype. It may be the case that the concentration of Suc (or other sugars) in the medium overloads the lipid storage pathways, so that excess carbon is stored as starch rather than lipid. It was observed that the levels of starch in the structure fell dramatically when plants were transferred to medium lacking Suc (Supplemental Fig. S1).
The analysis of gene expression in tnp mutants reveals that along with LEC1, the transcript levels of SERK1 and SERK2 are up-regulated in comparison to control seedlings. This supports the view that at least some cells in tnp mutants have acquired embryonic identity. That LEC1 is required for embryonic competence is supported by the observation that the ability of cultured cells to undergo somatic embryogenesis is severely impaired in lec1 mutants (Gaj et al., 2005). A number of genes associated with the seed maturation in which proteins and lipids are synthesized and stored are also up-regulated in tnp mutants, a process that LEC1 is implicated as having a key role in controlling (West et al., 1994; Kagaya et al., 2005). WOX2 and WOX8 are only expressed in early stage embryos and are implicated in cell fate and pattern formation (Haecker et al., 2004). The transcript levels for these genes are extremely low and do not correlate with the tnp phenotype. This suggests that while vegetative cells of tnp mutants can acquire embryonic competence and certain embryo-like aspects such as the synthesis of storage compounds, the early events of embryogenesis in which pattern formation and cell fate are determined are probably not activated in tnp mutants.
As seen in the tnp/pkl cross, other embryonic pathways must be activated to activate organogenesis. A key factor here may be the level LEC1 expression. For example, Kagaya et al. (2005) report that inducible LEC1 expression results in the induction of LEC2 as well as genes for seed storage proteins, yet in this study we could not detect a significant increase in LEC2 expression. Furthermore, constitutive expression of LEC1 in 35S:LEC1 plants generally results in seedling lethality (Lotan et al., 1998). The 35S:LEC1 seedlings that do survive resemble the seedling lethal tnp mutant seedlings (compare figure 6 in Lotan et al., 1998 to Fig. 1B). In 35S:LEC1 plants that survive, and also in dexamethasone-inducible LEC1 plants, genes for seed storage production were induced, which is also observed in tnp. The fact that the expression levels of both LEC2 and AtWLIM2 are not affected in tnp mutants, whereas both of these genes, along with LEC1, are strongly derepressed in pkl mutants (Dean Rider et al., 2003), indicates that at least in tnp, LEC1 cannot overcome PKL-dependent repression of these other embryonic genes.
It has been reported recently that LEC2 activates the maturation phase genes IAA30 and AGL15 and that these genes may also have a role in early embryogenesis (Braybrook et al., 2006). This raises the possibility that LEC1 similarly activates early genes, a possibility also suggested by observed early defects in the lec1 mutant embryo (West et al., 1994).
Ectopic LEC1 Expression Is Associated with Altered Cell Division and Cytokinin Signaling
Examination of CYCAT1:CDB:GUS activity in lec1-dtnp revealed that cells in the swollen hypocotyl continue to divide, consistent with the altered cell shape (Figs. 1 and 3). Since growth of the wild-type hypocotyl epidermal and cortical cells is normally by cell expansion, with all the cells in place by the end of embryogenesis (Gendreau et al., 1997), our results indicate that suppression of LEC1 is necessary to restrict cell division postgermination. Interestingly, while hypocotyl cells in lec1-dtnp undergo division, they do not undergo subsequent cell expansion, and remain small. It is possible that this may be due to physical constraints imposed by the surrounding cells, which are densely packed with storage products.
Altered cytokinin distribution or sensitivity, marked by ectopic IBC6/ARR5∷GFP activity in lec1-dtnp seedlings, may also mechanistically contribute to the ectopic cell division activity. The IBC6/ARR5 gene is a member of the Arabidopsis response regulator gene family, and is specifically up-regulated in response to cytokinins (Brandstatter and Kieber, 1998; D'Agostino et al., 2000). In lec1-dtnp seedlings, IBC6/ARR5∷GFP expression is found in patches throughout the outer cell layer of the abnormal hypocotyl instead of the pericycle. This suggests a change in either the localization of cytokinin or its perception, and this may result in the observed activation of cell division directly (Riou-Khamlichi et al., 1999) and/or a cytokinin-mediated blocking of cell expansion (Davies, 1995).
A potential change in cytokinin localization or sensing may also explain the partially deetiolated phenotype of lec1-dtnp seedlings when grown in the dark. Growth in the presence of cytokinin is known to cause deetiolation (Chory et al., 1994). Suc can also cause deetiolation and root branching (Roldán et al., 1999), and possible altered carbon utilization in the lec1-dtnp mutant (visualized as ectopic lipid and starch accumulation) may contribute to the dark-grown phenotype.
Furthermore, the presence of cytokinin in the epidermis may explain in part why shoots developed from the embryo-like hypocotyl tissue in lec1-dtnp pkl double mutants, but not from single lec1-dtnp. PKL is required for the repression of key embryonic genes other than LEC1, for example LEC2 (Dean Rider et al., 2003), which is not up-regulated in lec1-dtnp. The additional expression of these genes in the double mutant may result in the swollen hypocotyl tissue being more embryogenically competent than in single lec1-dtnp mutants. A high cytokinin-to-auxin ratio has long been known to cause shooting of embryonic callus in tissue culture (e.g. Smigocki and Owens, 1988). This may explain the differentiation of the embryo-like tissue and shoot development.
Auxin and Sugars Affect Embryonic Cell Identity in lec1-dtnp
The modulation of penetrance of the lec1-dtnp phenotype by exogenous signaling molecules provides new information on their possible interaction with the LEC1-mediated pathway of embryonic development.
The penetrance of the lec1-dtnp phenotype is almost entirely dependent on the presence of a carbon source in the growth medium, with Suc found to be the most effective. Additionally, treatment with the auxin 2,4-D causes low levels of penetrance in the absence of Suc. The dramatic accumulation of starch and lipids in the lec1-dtnp hypocotyl is a major feature of the lec1-dtnp phenotype. It has been shown in the cotyledons of Vicia faba embryos that relatively high Suc concentrations promote storage cell differentiation and starch production (Borisjuk et al., 2002). In this example, the high Suc concentrations are established by uptake by epidermal cells, which leads to the induction of starch biosynthesis genes and starch accumulation. Therefore, in Vicia Suc can act as an initiation signal for starch accumulation. We suggest that in lec1-dtnp, Suc may similarly act as a signal to induce starch production, potentiated by the ectopic expression of LEC1.
While Suc increases penetrance of the lec1-dtnp phenotype, plant hormones were also found to be influential. This observation is similar to that for the pkl mutant, where GA and GA inhibitors significantly affect penetrance (Ogas et al., 1997, 1999; Henderson et al., 2004). In the case of lec1-dtnp the GA inhibitor, paclobutrazol, was found to increase penetrance. Interestingly, and unlike pkl, GA was not found to directly repress penetrance of lec1-dtnp. However, in double treatment experiments, GA was able to reduce but not abolish the effectiveness of paclobutrazol (data not shown), an observation that appears somewhat contradictory. Furthermore, while paclobutrazol increases penetrance, adult lec1-dtnp plants do not resemble GA-deficient or GA-signaling mutants, as is the case with pkl. The region of the LEC1 promoter that is deleted in tnp is unlikely to account directly for the paclobutrazol and GA effects since neither appears to alter LEC1 transcript levels in either lec1-dtnp or wild-type controls. GA and paclobutrazol have an effect on the transcription of LEC2 but not FUS3 in a lec1-dtnp background, and it may be these differential effects on LEC2 and other embryonic genes that explain the GA response differences between lec1-dtnp and pkl.
The two hormones that most dramatically and differentially affected penetrance of lec1-dtnp were auxin and cytokinin, both of which have been widely used to induce embryogenic competence in somatic cells (e.g. Toonen and de Vries, 1996). In the case of auxin, we found this effect was independent of the requirement for Suc. The importance of auxin in early embryogenesis has been reported in a number of studies. A number of seedling pattern mutants are defective in auxin-responsive genes, including MONOPTEROS and BODENLOS (Hardtke and Berleth, 1998; Hamann et al., 2002). Furthermore, the distribution of auxin plays a vital role in embryonic axis formation and the organization of the meristems (Sabatini et al., 1999; Friml et al., 2003).
PLS expression is auxin regulated (Topping and Lindsey, 1997; Casson et al., 2002) and is mislocalized to the tnp hypocotyl, and both auxin and auxin transport inhibitors strongly enhance the penetrance of the tnp phenotype. Expression of the IAA2:GUS reporter was much higher in the abnormal hypocotyl of tnp mutants with strong phenotypes compared to those with weaker phenotypes (compare Fig. 7, B and C). This indicates that the swollen hypocotyl either has increased auxin responsiveness or higher levels of auxin. The fact that tnp mutants grown in the presence of auxin always display a strong phenotype may indicate that it is the auxin concentration in the hypocotyl that determines the strength of the phenotype.
The auxin transport inhibitors NPA and NOA were also able to increase the penetrance of lec1-dtnp. One possible explanation is that, by blocking auxin transport, there is an accumulation of auxin in some cells. Alternatively the transport inhibitors might disrupt the balance of auxin and cytokinin activities. The observation that the concentrations of 2,4-D required to increase lec1-dtnp penetrance are much lower than those required to initiate somatic embryogenesis (typically 4–20 μm) indicate that the cells of the lec1-dtnp hypocotyl are already primed by the ectopic expression of LEC1.
The results of the semiquantitative RT-PCR analysis indicate that the actions of auxin and cytokinin may be partly due to their effect on LEC2 and FUS3 expression. Cytokinin treatment resulted in repression of these two genes while 2,4-D repressed LEC2 while increasing the levels of FUS3, supporting other studies that have shown that auxin induces FUS3 expression (Gazzarrini et al., 2004). Therefore the ability of cytokinin to repress penetrance of lec1-dtnp may be linked in part to its effect on these other regulators of embryogenesis. In the case of auxin, the increase in FUS3 levels may, along with other unknown factors, counteract the reduction in LEC2 levels. These data strongly support the role of auxin as an important regulator of embryonic identity through its effect on LEC and FUS genes. It should be noted that in a number of our experiments, the penetrance of the visible lec1-dtnp phenotype was zero, yet gene expression differences were always observed, indicating a molecular phenotype in the absence of a structural one.
Treatment with the methylation inhibitor 5-azacytidine also increased the penetrance of the lec1-dtnp phenotype. Methylation is one mechanism by which genes can be transcriptionally repressed (Fransz and de Jong, 2002). Some developmental processes, such as the role of vernalization in flowering time, may be determined by gene methylation status (Sheldon et al., 2000). This result therefore suggests that at least some of the downstream targets of LEC1 may be transcriptionally repressed by methylation, most probably during the later stages of embryogenesis. There is also the possibility that other LEC1-independent regulators of embryogenesis are repressed by methylation and that derepression of these may contribute to the increase in penetrance.
Concluding Remarks
In summary, the results demonstrate that the role of LEC1 as a key regulator of embryogenesis is promoted by auxin and Suc. Repression of the LEC1 pathway in postembryonic tissues, which may in part be controlled by DNA methylation, is necessary not only to suppress storage product accumulation, but also cell division and cell fate. Further analysis of lec1-dtnp should help elucidate the role of these signals in determining the embryogenic competence of plant cells.
MATERIALS AND METHODS
Materials and Growth Conditions
The tnp mutant was isolated in an activation-tagging screen of the pls mutant. The pls line (Arabidopsis [Arabidopsis thaliana] ecotype C24) contains the promoter trap pΔgusBin 19 (Topping et al., 1991, 1994; Casson et al., 2002). pls plants were transformed with the activation tag construct consisting of a tandem repeat of 4× CaMV 35S enhancer in the binary vector pMOG1006 (a gift from Mogen, Leiden, The Netherlands). Plant transformation was performed by the floral dip method (Clough and Bent, 1998) using the Agrobacterium tumefaciens C58C1 (Dale et al., 1989).
For in vitro growth studies, seeds were vernalized and surface sterilized (Clarke et al., 1992) and plated on growth medium (half-strength MS medium, Sigma), 1% Suc, and 2.5% phytagel (Sigma) at 22°C ± 2°C at a photon flux density of approximately 150 μmol m−2 s−1. For hormone application experiments, seeds were germinated aseptically on growth medium containing various concentrations of hormones.
Arabidopsis seeds transgenic for the IBC6/ARR5∷GFP gene fusion were kindly provided by Joe Kieber (University of North Carolina, Chapel Hill). SCR∷GFP seeds were kindly donated by Philip Benfey (Duke University, North Carolina). CYCAT1:CDB:GUS seeds were kindly provided by Marie-Theres Hauser (University of Agricultural Sciences, Vienna). IAA2∷GUS seeds were kindly provided by Malcolm Bennett (University of Nottingham, UK). J2662, J2601, and pkl1-1 seeds were obtained from the Nottingham Arabidopsis Stock Centre (Nottingham, UK). J2662 and J2601 are part of the Jim Haseloff enhancer-trap GFP lines. Marker lines were crossed with tnp and F2 seedlings examined.
Microscopy and Histology
For scanning electron microscopy, tissues were fixed overnight in 2% paraformaldehyde, 2.5% glutaraldehyde in 0.1 m phosphate buffer (pH 7.0), dehydrated in an acetone series of 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 95%, and 100%. Tissue was sputter coated with gold palladium (5–50 Å) following critical point drying. Samples were viewed using a JEOL JSM IC848 microscope (JEOL). Tissues were cleared and mounted for light microscopy in chloral hydrate (Topping and Lindsey, 1997) or 20% glycerol. Leaves were cleared by incubating at 70°C for 30 min in 90% ethanol, followed by incubation at 70°C for 30 min in lactic acid:phenol:glycerol:water (1:1:1:1) and viewed under dark-field illumination (Telfer and Poethig, 1994). For the preparation of histological sections, plant tissues were fixed and embedded in Historesin (Jung/Leica) essentially according to the manufacturer's instructions. Tissue was vacuum infiltrated with the fixative, 1.5% glutaraldehyde/0.3% paraformaldehyde in 25 mm PIPES, dehydrated in an ethanol series, and embedded in Historesin. Ten-micrometer sections were cut on a Reichert omU3 microtome and stained with 0.005% (w/v) toluidine blue at 60°C for 30 s. Staining of seedlings for the presence of starch was done by placing seedlings in Lugol's solution (Sigma) for 5 min. Staining with Fat Red 7B was carried out as described by Ogas et al. (1997).
Light micrographs were taken using a CoolSNAP and compared with digital camera (Photometrics, Roper Scientific) with Openlab 3.1.1 software (Improvision) on Leica MZ125 (Leica Microsystems), Olympus SZH10 (Olympus), or Zeiss Axioskop (Carl Zeiss) microscopes. Confocal images were taken with a Bio-Rad Radiance 2000 microscope after counterstaining of tissues with 10 μg/mL propidium iodide. Images were processed in Adobe Photoshop 5.0.
Cloning of TNP
tnp plants of the C24 ecotype were crossed to plants of the Col-0 ecotype to generate a mapping population and 800 F2 progeny showing the tnp phenotype were isolated. DNA was extracted from these progeny according to the method of Edwards et al. (1991). SSLP markers nga280 and nga248, described at www.Arabidopsis.org, placed TNP at approximately 35 cM on chromosome I. Further SSLP and single nucleotide polymorphisms were detected by sequencing PCR-amplified regions from both C24 and Col-0 DNA. Fine mapping placed TNP on either BAC T26F17 or F2E2 that contain the LEC1 gene. Sequence upstream of the LEC1 transcriptional start site was amplified from tnp mutants by thermal asymmetric interlaced PCR using the degenerate AD2 and AD2 oligonucleotides as described by Liu et al. (1995) and the LEC1 promoter-specific oligonucleotides P1: GGTCAGTGGTTAGTTACCACG, P2: CGTGGGCGTACGTAATCTGAG, and P3: CGTGTTGGAGCTATTCGACAC. To confirm that the deletion was present in tnp mutants, PCR was performed using the oligonucleotide pair CCATTCCATATTCAAGGCATC and CGATTATCGAACGGCTGAG, which generates a 144 bp product if the deletion is present and a 3,400 bp product in wild-type plants. The deletion was present in 40/40 plants from the F2 mapping population.
Gene Expression Analysis
Tissue localization of GUS enzyme activity was performed as described (Topping and Lindsey, 1997). For transcript analysis, RNA was extracted using the RNeasy Plant RNA Extraction kit (Qiagen). RT-PCR was performed using the OneStep RT-PCR kit (Qiagen) as detailed in the manufacturer's instructions. Oligonucleotide pairs used for amplification were: LEC1: 5′-GCAACCACCATGTGTGGCTCG-3′ and 5′-GAAGAGCCACCACCAACACTGG-3′, which generate a 515 bp product from mRNA. LEC2: 5′-GACGAAGATGGCAAGGATCAACAGG-3′ and 5′-CTTCCACCACCATATCACCACCACTC-3′, which generate a 794 bp product from mRNA. For FUS3: 5′-TCTCCCGGGCTGAAACCCAAAGAGATCCACC-3′ and 5′-TATGGTACCGCTGATCACCATCAAGAAACC-3′, which generate a 1,113 bp product from mRNA. SERK1: 5′-GTGGACCTGTTACAAGTCACC-3′ and 5′-CGTTAGCTAGCCGAGCGAGATCG-3′, which generate a 935 bp product from mRNA. SERK2: 5′-GGTATAGTGCTACTGGAGCC-3′ and 5′-GTCTTATTGACCAGGCTAGAGG-3′, which generate a 515 bp product from mRNA. Wox2: 5′-CCCAACGAAAGATCAGATCACG-3′ and 5′-CGAGTAGAAGTAGAACCACCAG-3′, which generate a 710 bp product from mRNA. WOX8: 5′-CGATACTCCATCTTACATGCAC-3′ and 5′-CCGTTATTAACGGTAGAGAATGC-3′, which generate a 533 bp product from mRNA. At5g54740: 5′-CCTCTTCATCCTCCTAGCCAACG-3′ and 5′-CACACATCTTGTCCACTTGCC-3′, which generate a 307 bp product from mRNA. At4g27150: 5′-CTTCCATCTACCGCACTGTTGTCG-3′ and 5′-GGGCATTCACCAACTTGCTGG-3′, which generate a 417 bp product from mRNA. At4g28520: 5′-CATCATCGCTCTTCTCGACATCG-3′ and 5′-GGAAACCATTTGATATGACCTCC-3′, which generate a 793 bp product from mRNA. AtWLIM2: 5′-GTGGAGCTTCTCTCAGCTGATG-3′ and 5′-GCTGAGCGAAATGGTGCTTGCAG-3′, which generate a 435 bp product from mRNA. At1g21980: 5′-GACAAGGTTTCCACCAGAAGGGAC-3′ and 5′-CCTTTGAGGTCAAACCGTCTCTGG-3′, which generate a 431 bp product from mRNA. Wri1: 5′-GATGGACTGGGAGATTCGAG-3′ and 5′-GATGGTTAGCTTGGTTCACAGG-3′, which generate a 530 bp product from mRNA. AtPLC1: 5′-GAACCGCAAAGGAGGGTTGAAG-3′ and 5′-AGCCGAACGGCACGAATACC-3′, which generate a 711 bp product from mRNA. ABI3: 5′-GGAAGACATCGGAACCTCTCG-3′ and 5′-GTAAAAACCCGGACCCCGAC-3′, which generate a 504 bp product from mRNA. RD29A: 5′-GATGATGACGAGCTAGAACCTG-3′ and 5′-GCCCACCGGGAAAACAACTCCTG-3′, which generate a 723 bp product from mRNA. ACT1: 5′-GATCCTAACCGAGCGTGGTTAC-3′ and 5′-GACCTGACTCGTCATACTCTGC-3′, which generate a 529 bp product from mRNA. Oligonucleotide pairs for COR15a, P5CS1, CBF1, and CBF2 are as described previously (Williams et al., 2005). Total RNA was treated with DNase according to the method of Sanyal et al. (1997) and 500 ng of RNA was used per reaction. Typical reaction conditions were 50°C for 30 min, 95°C for 15 min followed by two cycles of 94°C denaturation for 30 s, 65°C primer annealing for 30 s, and 72°C extension for 60 s. This was followed by 20 to 40 cycles of 94°C denaturation for 30 s, 55°C primer annealing for 30 s, and 72°C extension for 60 s, and a final extension at 72°C for 7 min. Minus RT control experiments were performed by adding enzyme after the 50°C incubation.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Starch accumulation is dependent on the continuous presence of sugars in the growth medium.
Supplemental Table S1. Osmotic stress does not induce penetrance of tnp.
Supplementary Material
This work was supported by funding from the Biotechnology and Biological Sciences Research Council (to K.L.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Keith Lindsey (keith.lindsey@durham.ac.uk).
The online version of this article contains Web-only data.
References
- Ahringer J (2000) NuRD and SIN3 histone deacetylase complexes in development. Trends Genet 16: 351–356 [DOI] [PubMed] [Google Scholar]
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, Leon P (2000) Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev 14: 2085–2096 [PMC free article] [PubMed] [Google Scholar]
- Basu S, Sun H, Brian L, Quatrano RL, Muday GK (2002) Early embryo development in Fucus distichus is auxin sensitive. Plant Physiol 130: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisjuk L, Walenta S, Rolletschek H, Mueller-Klieser W, Wobus U, Weber H (2002) Spatial analysis of plant metabolism: sucrose imaging within Vicia faba cotyledons reveals specific developmental patterns. Plant J 29: 521–530 [DOI] [PubMed] [Google Scholar]
- Brandstatter I, Kieber JJ (1998) Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell 10: 1009–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braybrook SA, Stone SL, Park S, Bui AQ, Le BH, Fischer RL, Goldberg RB, Harada JJ (2006) Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc Natl Acad Sci USA 103: 3468–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson S, Spencer M, Walker K, Lindsey K (2005) Laser capture microdissection for the analysis of gene expression during embryogenesis of Arabidopsis. Plant J 42: 111–123 [DOI] [PubMed] [Google Scholar]
- Casson SA, Chilley PM, Topping JF, Evans IM, Souter MA, Lindsey K (2002) The POLARIS gene of Arabidopsis encodes a predicted peptide required for correct root growth and leaf vascular patterning. Plant Cell 14: 1705–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernac A, Benning C (2004) WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J 40: 575–585 [DOI] [PubMed] [Google Scholar]
- Chory J, Reinecke D, Sim S, Washburn T, Brenner M (1994) A role for cytokinins in de-etiolation in Arabidopsis: det mutants have an altered response to cytokinins. Plant Physiol 104: 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke MC, Wei W, Lindsey K (1992) High frequency transformation of Arabidopsis thaliana by Agrobacterium tumefaciens. Plant Mol Biol Report 10: 178–189 [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 6: 735–743 [DOI] [PubMed] [Google Scholar]
- D'Agostino IB, Deruere J, Kieber JJ (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale PJ, Marks MS, Brown MM, Woolston CJ, Gunn HV, Mullineaux PM, Lewis DM, Kemp J, Chen DF, Gilmour DM, (1989) Agroinfection of wheat-inoculation of in vitro grown seedlings and embryos. Plant Sci 63: 237–245 [Google Scholar]
- Davies PJ, editor (1995) Plant Hormones: Physiology, Biochemistry and Molecular Biology, Ed 2. Kluwer Academic Publishers, Dordrecht, The Netherlands
- Dean Rider S Jr, Henderson JT, Jerome RE, Edenberg HJ, Romero-Severson J, Ogas J (2003) Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J 35: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elge S, Brearley C, Xia HJ, Kehr J, Xue HW, Mueller-Roeber B (2001) An Arabidopsis inositol phospholipid kinase strongly expressed in procambial cells: synthesis of PtdIns(4,5)P2 and PtdIns(3,4,5)P3 in insect cells by 5-phosphorylation of precursors. Plant J 26: 561–571 [DOI] [PubMed] [Google Scholar]
- Eliasson A, Gass N, Mundel C, Baltz R, Krauter R, Evrard JL, Steinmetz A (2000) Molecular and expression analysis of a LIM protein gene family from flowering plants. Mol Gen Genet 264: 257–267 [DOI] [PubMed] [Google Scholar]
- Fischer-Iglesias C, Sundberg B, Neuhaus G, Jones A (2001) Auxin distribution and transport during embryonic pattern formation in wheat. Plant J 26: 115–129 [DOI] [PubMed] [Google Scholar]
- Fransz PF, de Jong JH (2002) Chromatin dynamics in plants. Curr Opin Plant Biol 5: 560–567 [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Gaj MD, Zhang S, Harada JJ, Lemaux PG (2005) Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta 222: 977–988 [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P (2004) The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev Cell 7: 373–378 [DOI] [PubMed] [Google Scholar]
- Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Höfte H (1997) Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol 114: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J 16: 433–442 [DOI] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T (2004) Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131: 657–668 [DOI] [PubMed] [Google Scholar]
- Hamann T, Benkova E, Bäurle I, Kientz M, Jürgens G (2002) The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev 16: 1610–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J 17: 1405–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser M-T, Bauer E (2000) Histochemical analysis of root meristem activity in Arabidopsis thaliana using a cyclin:GUS (beta-glucuronidase) marker line. Plant Soil 226: 1–10 [Google Scholar]
- Hecht V, Vielle-Calzada JP, von Recklinghausen I, Hartog M, Zwartjes C, Schmidt E, Boutilier K, Grossniklaus U, de Vries S (2001) The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol 127: 803–816 [PMC free article] [PubMed] [Google Scholar]
- Henderson JT, Li H-C, Dean Rider S, Mordhorst AP, Romero-Severson J, Cheng J-C, Robey J, Sung ZR, de Vries SC, Ogas J (2004) PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiol 134: 995–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Ohto C, Mizoguchi T, Shinozaki K (1995) A gene encoding a phosphatidylinositol-specific phospholipase-C is induced by dehydration and salt stress in Arabidopsis thaliana. Proc Natl Acad Sci USA 92: 3903–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda-Iwai M, Umehara M, Satoh S, Kamada H (2003) Stress-induced somatic embryogenesis in vegetative tissues of Arabidopsis thaliana. Plant J 34: 107–114 [DOI] [PubMed] [Google Scholar]
- Jones P, Taylor SM (1980) Cellular differentiation, cytidine analogs and DNA methylation. Cell 20: 85–93 [DOI] [PubMed] [Google Scholar]
- Kagaya Y, Toyoshima R, Okuda R, Usui H, Yamamoto A, Hattori T (2005) LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol 46: 399–406 [DOI] [PubMed] [Google Scholar]
- Kroj T, Savino G, Valon C, Giraudat J, Parcy F (2003) Regulation of storage protein gene expression in Arabidopsis. Development 130: 6065–6073 [DOI] [PubMed] [Google Scholar]
- Laby RJ, Kincaid MS, Kim D, Gibson SI (2000) The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J 23: 587–596 [DOI] [PubMed] [Google Scholar]
- Laux T, Würschum T, Breuninger H (2004) Genetic regulation of embryonic pattern formation. Plant Cell 16: S190–S202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Sun L, Nguyen LV, Rachubinski RA, Goodman HM (1999) The Pex16p homolog SSE1 and storage organelle formation in Arabidopsis seeds. Science 284: 328–330 [DOI] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Matsudaira Yee K, West MAL, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Luerßen H, Kirik V, Herrmann P, Miséra S (1998) FUSCA3 encodes a protein with a conserved VP1/ABI3-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J 15: 755–764 [DOI] [PubMed] [Google Scholar]
- Meinke DW, Franzmann LH, Nickle TC, Yeung EC (1994) Leafy Cotyledon mutants of Arabidopsis. Plant Cell 6: 1049–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami K, Katagiri T, Iuchi S, Yamaguchi-Shinozaki K, Shinozaki K (1998) A gene encoding phosphatidylinositol-4-phosphate 5-kinase is induced by water stress and abscisic acid in Arabidopsis thaliana. Plant J 15: 563–568 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29$ in seeds, germinating embryos, and seedlings in Arabidopsis. Plant Mol Biol 60: 51–68 [DOI] [PubMed] [Google Scholar]
- Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Interaction between two cis acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J 34: 137–148 [DOI] [PubMed] [Google Scholar]
- Ogas J, Cheng J-C, Sung ZR, Somerville C (1997) Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277: 91–94 [DOI] [PubMed] [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C (1999) PICKLE is a CHD3 chromatin-remodelling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci USA 96: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang PP-Y, Pruitt RE, Meyerowitz EM (1988) Molecular cloning, genomic organization, expression and evolution of 12S seed storage protein genes of Arabidopsis thaliana. Plant Mol Biol 11: 805–820 [DOI] [PubMed] [Google Scholar]
- Parcy F, Valon C, Kohara A, Miséra S, Giraudat J (1997) The ABSCISIC ACID-lNSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell 9: 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz V, Bergervoet JHW, Koornneef M (2001) Sequential steps for developmental arrest in Arabidopsis seeds. Development 128: 243–252 [DOI] [PubMed] [Google Scholar]
- Ribnicky D, Cohen J, Hu W, Cooke T (2002) An auxin surge following fertilization in carrots: a mechanism for regulating plant totipotency. Planta 214: 505–509 [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JAH (1999) Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283: 1541–1544 [DOI] [PubMed] [Google Scholar]
- Roldán M, Gómez-Mena C, Ruiz-García L, Salinas J, Martínez-Zapater JM (1999) Sucrose availability on the aerial part of the plant promotes morphogenesis and flowering of Arabidopsis in the dark. Plant J 20: 581–590 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al (1999) An auxin-dependent organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472 [DOI] [PubMed] [Google Scholar]
- Sanyal A, O'Driscoll SW, Bolander MA, Sarkar G (1997) An effective method of completely removing contaminating genomic DNA from an RNA sample to be used for PCR. Mol Biotechnol 8: 135–137 [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Finnegan EJ, Rouse DT, Tadege M, Bagnall DJ, Helliwell CA, Peacock WJ, Dennis ES (2000) The control of flowering by vernalization. Curr Opin Plant Biol 3: 418–422 [DOI] [PubMed] [Google Scholar]
- Smigocki AC, Owens LD (1988) Cytokinin gene fused with a strong promoter enhances shoot organogenesis and zeatin levels in transformed plant cells. Proc Natl Acad Sci USA 85: 5131–5135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Friml J, Marchant A, Ljung K, Sandberg G, Palme K, Bennett M (2001) Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev 15: 2648–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A, Poethig RS (1994) Leaf development in Arabidopsis. In EM Meyerowitz, CR Somerville, eds, Arabidopsis. Cold Spring Harbor Press, Cold Spring Harbor, NY, pp 379–401
- Toonen MAJ, de Vries SC (1996) Initiation of somatic embryos from single cells. In TL Wang, A Cuming, eds, Embryogenesis: The Generation of a Plant. Bios Scientific Publishers, Oxford, pp 173–189
- Topping JF, Agyeman F, Henricot B, Lindsey K (1994) Identification of molecular markers of embryogenesis in Arabidopsis thaliana by promoter trapping. Plant J 5: 895–903 [DOI] [PubMed] [Google Scholar]
- Topping JF, Lindsey K (1997) Promoter trap markers differentiate structural and positional components of polar development in Arabidopsis. Plant Cell 9: 1713–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topping JF, Wei W, Lindsey K (1991) Functional tagging of regulatory elements in the plant genome. Development 112: 1009–1019 [DOI] [PubMed] [Google Scholar]
- Vignali M, Hassan AH, Neely KE, Workman JL (2000) ATP-dependent chromatin-remodeling complexes. Mol Cell Biol 20: 1899–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M, Yee KM, Danao J, Zimmerman JL, Fischer RL, Goldberg RB, Harada JJ (1994) LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 12: 1731–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergren T, Dove SK, Sommarin M, Pical C (2001) AtPIP5K1, an Arabidopsis thaliana phosphatidylinositol phosphate kinase, synthesizes PtdIns(3,4)P(2) and PtdIns(4,5)P(2) in vitro and is inhibited by phosphorylation. Biochem J 359: 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ME, Torabinejad J, Cohick E, Parker K, Drake EJ, Thompson JE, Hortter M, Dewald DB (2005) Mutations in the Arabidopsis phosphoinositide phosphatase gene SAC9 lead to overaccumulation of PtdIns(4,5)P2 and constitutive expression of the stress-response pathway. Plant Physiol 138: 686–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka-Diller JW, Helariutta Y, Fukaki H, Malamy JE, Benfey PN (2000) Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127: 595–603 [DOI] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.