Abstract
CesA1 and CesA3 are thought to occupy noninterchangeable sites in the cellulose synthase making primary wall cellulose in Arabidopsis (Arabidopsis thaliana L. Heynh). With domain swaps and deletions, we show that sites C terminal to transmembrane domain 2 give CesAs access to their individual sites and, from dominance and recessive behavior, deduce that certain CesA alleles exclude others from accessing each site. Constructs that swapped or deleted N-terminal domains were stably transformed into the wild type and into the temperature-sensitive mutants rsw1 (Ala-549Val in CesA1) and rsw5 (Pro-1056Ser in CesA3). Dominant-positive behavior was assayed as root elongation at the restrictive temperature and dominant-negative effects were observed at the permissive temperature. A protein with the catalytic and C-terminal domains of CesA1 and the N-terminal domain of CesA3 promoted growth only in rsw1 consistent with it accessing the CesA1 site even though it contained the CesA3 N-terminal domain. A protein having the CesA3 catalytic and C-terminal domains linked to the CesA1 N-terminal domain dramatically affected growth, but only in the CesA3 mutant. This is consistent with the operation of the same access rule taking this chimeric protein to the CesA3 site. In this case, however, the transgene behaved as a genotype-specific dominant negative, causing a 60% death rate in rsw5, but giving no visible phenotype in wild type or rsw1. We therefore hypothesize that possession of CesA3WT protects Columbia and rsw1 from the lethal effects of this chimeric protein, whereas the mutant protein (CesA3rsw5) does not.
Cellulose, a crystalline β-1,4-glucan, forms the microfibrils prominent in most plant cell walls. Its deposition in primary walls during cell expansion is critical for determining cell and organ shape (Arioli et al., 1998), whereas cellulose deposited in secondary walls after expansion ceases dominates the final mechanical properties of plant organs (Turner and Somerville, 1997). Cellulose synthesis in higher plants has been linked to rosette terminal complexes (RTCs; Brown et al., 1996), structures seen in the plasma membrane by freeze etch electron microscopy. If each RTC synthesizes one microfibril, it must simultaneously extend over 30 glucan chains in that microfibril (Newman et al., 1996). Electron microscopy resolves only six subunits, suggesting each contains multiple glycosyltransferases.
CesA proteins were identified as the likely glycosyltransferases for cellulose synthesis by their presence in cellulose-rich cotton fiber (Pear et al., 1996), their association with cellulose-deficient mutants (Arioli et al., 1998; Taylor et al., 1999, 2000, 2003; Fagard et al., 2000; Burn et al., 2002a; Ellis et al., 2002; Caño-Delgado et al., 2003), and their location in RTCs (Kimura et al., 1999). They are encoded by multigene families in all plants studied to date with 10 members in Arabidopsis (Arabidopsis thaliana). Their protein structure is well conserved, with clusters of two and six predicted transmembrane domains (TMDs) dividing the protein into three major cytoplasmic domains. The N-terminal domain comprises some 180 to 280 amino acid residues and includes a highly variable region (HVR1) and a Cys-rich region suggested to form two zinc-binding RING-finger domains (Kurek et al., 2002). It self-associates in yeast two-hybrid assays, and disulfide bonds between some of the Cys residues dimerize CesA proteins expressed in yeast (Saccharomyces cerevisiae). TMDs 1 and 2 separate the N-terminal domain from a central domain of about 600 residues with TMDs 3 to 8 in turn separating the central domain from a C-terminal sequence of about 20 residues. The central domain contains the D,D,D,QxxRW signature characteristic of many glycosyltransferases (Coutinho et al., 2003) and is accepted to be catalytically active (Vergara and Carpita, 2001). Its structure is highly conserved between CesA paralogs, with the exception of about 70 residues lying between the second and third Asp residues of the D,D,D,QxxRW signature. Here, the sequence differs between paralogs but is well conserved between orthologs, leading Vergara and Carpita (2001) to name it the class-specific region (CSR). Mutations inhibiting cellulose synthesis commonly occur in the central catalytic domain, but one (Chen et al., 2005) occurs among TMDs 3 to 8 where mutations conferring resistance to the herbicide isoxaben also occur (Scheible et al., 2001; Desprez et al., 2002).
The phenotypes of Arabidopsis mutants suggest that CesAs 1, 3, and 6 make primary wall cellulose (Arioli et al., 1998; Fagard et al., 2000; Burn et al., 2002a; Ellis et al., 2002; Caño-Delgado et al., 2003), whereas CesAs 4, 7, and 8 make secondary wall cellulose (Turner and Somerville, 1997; Taylor et al., 1999, 2000, 2003). The strong phenotypes resulting from mutating just one CesA gene are readily explained if three CesAs form a multienzyme complex in which each CesA has a unique function so that a mutation in one CesA inhibits the whole complex. The important studies of Turner and colleagues extended such genetic arguments by showing that CesAs 4, 7, and 8 physically interact (Taylor et al., 2003) and that lack of one CesA stopped the other two CesAs from reaching the plasma membrane in vivo (Gardiner et al., 2003). This is strong evidence that CesAs 4, 7, and 8 must form a multienzyme complex to make secondary wall cellulose and that, if any other CesAs are expressed in the same cells, they cannot replace any of those CesAs in the complex.
The mutants linking three CesAs to primary wall cellulose synthesis suggest that a similar three-CesA complex could exist, but physical evidence for interaction of CesAs is missing. A multienzyme complex making primary wall cellulose can explain important features of isoxaben resistance and sensitivity if a few isoxaben-sensitive (wild-type) subunits of CesAs 3 and 6 allow isoxaben to inhibit the entire complex (Scheible et al., 2001; Desprez et al., 2002; Robert et al., 2004). This has led to several detailed models for the complex, which all envisage CesAs 1 and 3 occupying separate, noninterchangeable sites. To begin exploring what equips CesAs 1 and 3 to occupy those separate sites and so makes them functionally noninterchangeable, we swapped and deleted their N-terminal domains to look for parts of the proteins that were essential and parts that could be changed. To test the activity of these chimeric proteins, we stably transformed them into rsw1 (mutated in CesA1; Arioli et al., 1998) and rsw5 (mutated in CesA3). The temperature-sensitive phenotypes allow us to see dominant-positive effects at the mutants' restrictive temperature and dominant-negative effects at the permissive temperature. rsw5 is of interest in its own right because it carries a mutation in the C-terminal domain of CesA3, which demonstrates that short domain is important for cellulose production.
RESULTS
rsw5, a Temperature-Sensitive Allele of CesA3 Mutated in the C-Terminal Domain
rsw5 was identified as a radial-swelling mutant using the screen of Baskin et al. (1992). It is strongly temperature sensitive but, as discussed below, has a more significant phenotype at the permissive temperature (18°C–20°C) than rsw1. rsw5 was shown (Peng, 1999) to make less cellulose than the wild type at the restrictive temperature (30°C), but did not accumulate the readily extracted glucan found in the seedling shoots of rsw1 (Arioli et al., 1998), rsw2/acw1 (Lane et al., 2001; Sato et al., 2001), and rsw3 (Burn et al., 2002b). It was mapped by standard methods to a 200-kb interval on chromosome 5 and the gene encoding AtCesA3 (At5g05170) was sequenced as a candidate gene. A single-nucleotide substitution (C > T) at position 4658 of the genomic coding sequence changed Pro-1056Ser in the conceptual translation. As described below, At5g05170 cDNA and genomic DNA complement the mutant phenotype, confirming the link between the phenotype and the mutated gene.
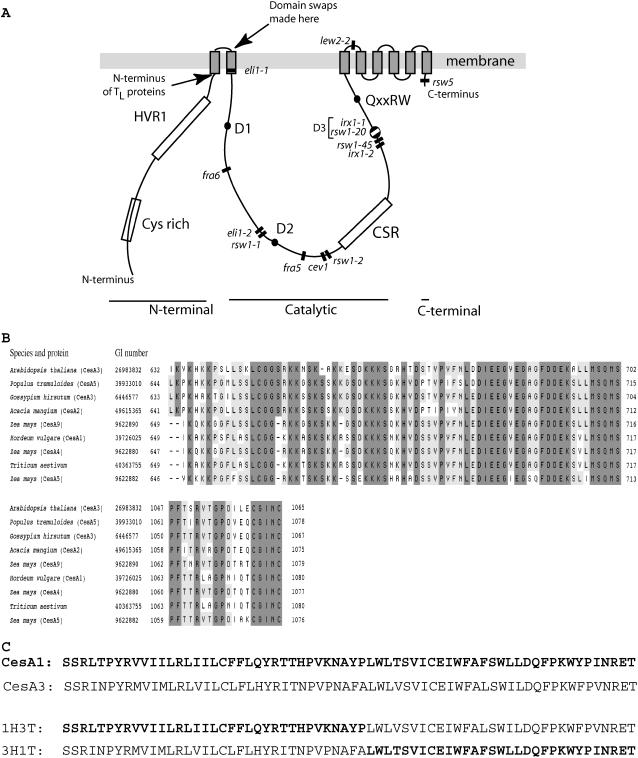
The Pro-1056Ser change in CesA3rsw5 lies in the predicted C-terminal domain (residues 1044–1065) following TMD8. All previous missense mutations that inhibit cellulose synthesis occur in the central catalytic domain or in the region containing TMDs 3 to 8 (Fig. 1A). Other mutations in or between these TMDs confer isoxaben resistance on CesAs 3 and 6, but do not inhibit cellulose synthesis (Heim et al., 1991; Scheible et al., 2001; Desprez et al., 2002). The C-terminal domain is well conserved in CesAs 2, 5, 6, and 9 of Arabidopsis (Desprez et al., 2002), but the other AtCesAs show only limited sequence similarity. The whole C-terminal domain is, however, well conserved in putative orthologs of AtCesA3 (Fig. 1B) defined, following Vergara and Carpita (2001), by having a highly similar CSR lying between the second and third Asp residues in the D,D,D,QxxRW signature (Fig. 1A).
Figure 1.
A, Diagram of the CesA domain structure showing the sites of domain swaps to make 3H1T and 1H3T, the site of the N terminus of TL deletion proteins, and the sites of previously reported amino acid substitutions inhibiting cellulose synthesis. Mutations inhibiting cellulose synthesis (Arioli et al., 1998; Taylor et al., 2000, 2003; Beeckman et al., 2002; Ellis et al., 2002; Gillmor et al., 2002; Caño-Delgado et al., 2003; Zhong et al., 2003; Chen et al., 2005) are shown by bars and cluster in or adjacent to the central catalytic domain. The exception is the rsw5 allele of CesA3 located in the short C-terminal sequence following TMD8. Mutations are drawn from all CesAs and so are placed relative to major protein features rather than by precise residue numbers. B, Sequence alignments showing that the C-terminal domain is conserved in putative CesA3 orthologs that share similar CSRs (Vergara and Carpita, 2001). A BLAST search with the CSR of AtCesA3 was used to identify putative orthologs in diverse species (top block). C-terminal sequences of the same proteins were then aligned by a BLAST search (bottom block) using the 18 C-terminal amino acids of AtCesA3. (Note that the AtCesA3 CSR given in figure 2 of Vergara and Carpita [2001] is incorrect.) C, Sequences of AtCesA1 and AtCesA3 around the site of the head-tail junction in chimeric proteins showing high level of conservation. The sequence of the chimeric proteins is shown below with the contributions from CesA1 identified in bold letters.
The CesA1 Catalytic and C-Terminal Domains Partially Complement rsw1 Even When Linked to the CesA3 N-Terminal Domain
To explore what makes CesAs 1 and 3 noninterchangeable, we constructed cDNAs encoding chimeric proteins in which the N-terminal sequences extending to just beyond TMD1 (Fig. 1A) were swapped between CesAs 1 and 3 to give proteins referred to as 3H1T and 1H3T. In this shorthand, H denotes head (the N-terminal cytoplasmic domain and TMD1), T denotes tail (the remainder of the protein onward from the start of TMD2), and the number denotes the CesA from which it is derived. 3H1T has residues 1 to 286 of CesA3 followed by residues 303 onward of CesA1; 1H3T has residues 1 to 302 of CesA1 followed by residues 287 onward of CesA3 (Fig. 1C). As expected, eight TMDs were predicted for both chimeric proteins. We also made constructs in which the N-terminal domain up to the start of TMD1 was deleted (Fig. 1A) from CesA1 and CesA3, leaving a long form of the tail (1TL and 3TL). 1TL had residues 277 onward from CesA1; 3TL had residues 261 onward from CesA3. Plants of rsw1, mutated Ala-549Val in CesA1, were then transformed with a genomic construct covering the CesA1 coding sequence and promoter region, with the CesA1 and CesA3 cDNAs, with the two chimeric cDNAs (3H1T and 1H3T), and with the deletion constructs (1TL and 3TL). All transgenes, except for the genomic constructs, were expressed by the cauliflower mosaic virus 35S promoter. To estimate transgene function in vivo, we measured root elongation over 2 d after T1 seedlings were transferred to the mutant's restrictive temperature, conditions where elongation was strongly inhibited in the mutant. Inhibited root elongation is a well-documented accompaniment to radial swelling (Baskin et al., 1992; Sugimoto et al., 2001) and is technically much simpler to measure than swelling. Table I and Figure 2 summarize the results.
Table I.
Summary of the effects of transgenes on wild-type, rsw1, and rsw5 plants
–, No effect; DP, dominant positive giving partial complementation at the restrictive temperature; DN, dominant negative detected at the permissive temperature.
| Protein Encoded by Transgene | Genotype Transformed
|
||
|---|---|---|---|
| Wild Type | rsw1 | rsw5 | |
| CesA1 (1H1T) | – | DP | – |
| CesA3 (3H3T) | – | – | DP |
| 3H1T | – | DP | – |
| 1H3T | – | – | DN |
| 1TL | – | – | – |
| 3TL | – | – | – |
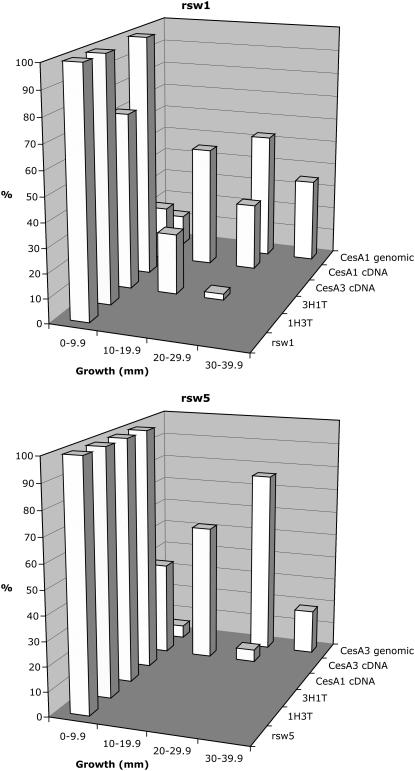
Figure 2.
Histograms showing root elongation increments in T1 seedlings of rsw1 and rsw5 that had been transformed with genomic and cDNA constructs encoding wild-type and chimeric proteins. The position of the tip of the longest root on each seedling was marked immediately before transfer from permissive to restrictive temperature and elongation measured 2 d later. The graphs plot the percentage of the total population (n > 30) that showed various ranges of elongation. Genomic constructs support greater elongation than cDNA constructs and, of the combinations where the transgene is not an allele of the mutated gene, only 3H1T in rsw1 is active.
The CesA1 genomic construct restored elongation of rsw1 to levels close to those seen in the wild type, whereas the CesA1 cDNA supported lower, but still substantial, elongation (Fig. 2A). CesA3 cDNA and the 1H3T cDNA were completely ineffective, but the 3H1T construct promoted root elongation in some rsw1 seedlings, indicating its capacity to partially replace CesA1 in vivo. 1TL did not promote root elongation (data not shown), confirming that the N-terminal region from CesA3 was needed to make 3H1T functional in vivo. rsw1 is very similar to the wild type when grown at its permissive temperature (Baskin et al., 1992; Williamson et al., 2001) and all transformants visually resembled nontransformed plants after growth to maturity at the permissive temperature, indicating that none of the constructs acted as a dominant negative in rsw1 at its permissive temperature. Table I summarizes the results.
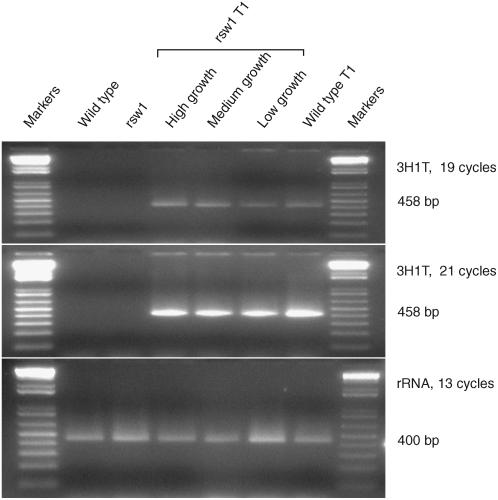
We confirmed expression of chimeric cDNAs by reverse transcription (RT)-PCR using one primer drawn from each CesA encoding part of the chimeric protein. The primer pair amplified a product spanning the transition site in the chimeric mRNA, but would not amplify a product from any endogenous gene. The 3H1T primers, for example, amplified no product from the wild type or rsw1, but amplified the predicted 458-bp fragment from T1 seedlings containing the 3H1T cDNA (Fig. 3). To investigate why transformants carrying the 3H1T construct showed some variation in growth response, we conducted semiquantitative RT-PCR on pooled plants assessed as showing small, medium, or large growth increments. Expression of 3H1T was highest in plants showing the largest growth response and lowest in those showing the smallest (Fig. 3, rsw1 T1 lanes).
Figure 3.
Demonstration that 3H1T chimeric cDNA is expressed only in T1 seedlings carrying the transgene and that the level of expression broadly correlates with the observed root elongation. Multiple RT-PCR incubations were set up using primers amplifying a 458-bp fragment from the chimeric 3H1T mRNA and tubes withdrawn after varying numbers of cycles to ensure amplification was not saturating. The top and middle panels show there is no 3H1T expression in wild-type and rsw1 seedlings unless they have been transformed with the chimeric cDNA (rsw1 T1 and wild-type T1 lanes). rsw1 transformants were pooled according to whether they showed high, medium, or low growth in the root elongation assay and used to show that, after 19 cycles, the level of 3H1T expression broadly correlates with their observed growth increment. Differences are lost by saturation after 21 cycles. A 400-bp fragment of 18S rRNA (third image) provides a loading control and, given that loading is highest in the low-growth rsw1 transformants, emphasizes the low expression of 3H1T in those seedlings.
The CesA3 Catalytic and C-Terminal Domains Linked to the CesA1 N-Terminal Domain Show a Dominant-Negative Effect But Only in rsw5
The preceding results show that the central catalytic and/or C-terminal domains rather than the N-terminal domains of CesA1 are essential to provide a protein able to partially replace CesA1rsw1 (the mutated CesA1 in rsw1) at its restrictive temperature. To explore whether the same rule applied to making proteins able to replace CesA3rsw5, we transformed rsw5 (Pro-1056Ser in CesA3) with the same cDNAs (original and chimeric), with the two deletion constructs 1TL and 3TL, and with a CesA3 genomic construct. The results (Fig. 2B; Table I) show that only CesA3 (cDNA and genomic) supported elongation and, in particular, that the CesA3 catalytic and C-terminal domains did not promote elongation when linked to the CesA1 N-terminal domain in 1H3T. There were again no effects of 1TL and 3TL (data not shown). Observing the subsequent growth of transformants showed that 1H3T in fact had a strong, dominant-negative effect on plants returned to the permissive temperature and grown to maturity. This led to high mortality rates with deaths beginning shortly after seedling exposure to the restrictive temperature for the root elongation assay and continuing to occur even in nearly mature plants. As a result, only about 37% of T1 seedlings set seed, whereas about 90% was typical in all other T1 populations. Mature T1 plants of rsw5 transformed with 1H3T showed wide variations in bolt height and other morphological features (Fig. 4A). In contrast, a 3H1T transgene caused no changes to the morphology or viability of rsw5 transformants (Fig. 4B). rsw5 without a transgene was noticeably shorter than the wild type (Fig. 4C), reflecting the persistence of a significant phenotype at the permissive temperature. Complementation with CesA3 cDNA produced a clear increase in height (Fig. 4D), although still not equaling that of the wild type (Fig. 4C, two left-hand pots).
Figure 4.
Dominant-negative effects of 1H3T in rsw5 plants. T1 seedlings were returned to the permissive temperature after measuring seedling root elongation at the restrictive temperature, planted four per pot, and photographed at maturity. A, rsw5 transformed with 1H3T showing that many deaths had reduced the number of survivors and that their sizes were highly variable. B, rsw5 transformed with 3H1T appearing indistinguishable from rsw5 plants without the transgene shown on the right in C. C, Wild type (left) and rsw5 (right) without transgenes showing the significant phenotype of rsw5 at the permissive temperature. D, rsw5 transformed with CesA3 cDNA has a height that is intermediate between rsw5 without the transgene and the wild type seen in C.
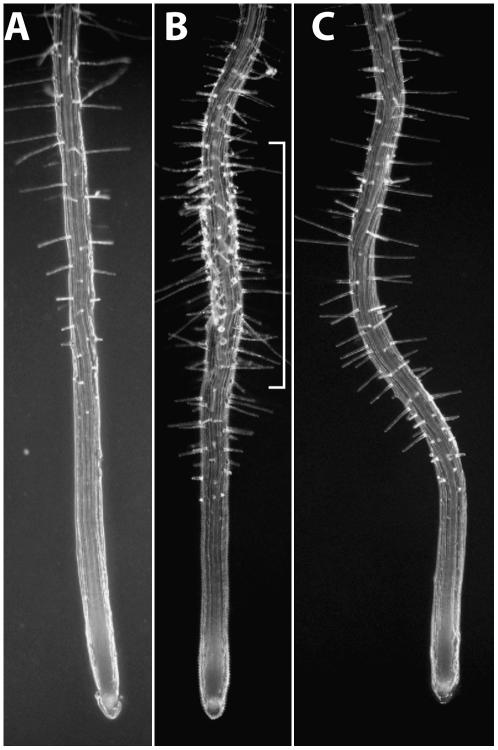
Suspecting that death and stunting seen at the permissive temperature of rsw5 containing 1H3T probably reflected impaired cellulose synthesis, we looked for dominant-negative effects of 1H3T on seedling root growth at the permissive temperature and, in particular, whether this would cause root radial swelling, a readily visible consequence of impaired cellulose synthesis and normally only seen at the restrictive temperature. We plated approximately 100 seeds collected from each of eight different T1 plants and grew them without kanamycin selection on agar plates at the permissive temperature. Abnormalities were mild, but some seedlings showed localized root swelling with signs of swollen root hair bases (Fig. 5), traits associated with reduced cellulose synthesis. A random selection of the T2 seedlings grown to maturity at the permissive temperature again showed a high mortality rate (data not shown) even though, by necessity, they were the progeny of the minority of T1 plants that survived to maturity.
Figure 5.
Dominant-negative effects of 1H3T on rsw5 seedling roots at the permissive temperature and seen in the T2 generation. A to C, Seedling roots of wild type (A), rsw5 transformed with 1H3T (B), and rsw5 with no transgene (C). All were grown without kanamycin selection on agar plates at the permissive temperature. Note the swollen region of the root (bracket) of rsw5 transformed with 1H3T, indicative of a dominant-negative effect. In B and C, the more closely spaced root hairs and the shorter elongation zone before the root hairs begin to emerge and reflect the reduced growth rate in rsw5 that is seen even at the permissive temperature.
Effects of Transgenes on Wild-Type Plants
Wild-type Columbia plants were also transformed with all constructs. By visual inspection, T1 plants showed no growth or morphological abnormalities indicative of dominant-negative or any other effects.
DISCUSSION
We describe a temperature-sensitive allele (rsw5) of CesA3 that shows the importance of the short C-terminal domain for cellulose production by CesA proteins. We used this CesA3 mutant and our CesA1 mutant (rsw1) to investigate the requirements for CesA function. Swapping or deleting the N-terminal domains of CesA1 and CesA3 produced proteins that, by visual phenotype, were functional, nonfunctional, or showed dominant-negative properties in rsw1 and rsw5. In contrast, all transgene proteins appeared nonfunctional in the wild type. In analyzing those transgene experiments, we will argue that the mutant proteins CesA1rsw1 and CesA3rsw5, but not wild-type proteins, open the CesA1 and CesA3 sites, respectively, in the cellulose synthase complex to transgene proteins. Only certain transgene proteins can occupy each of those sites, and we interpret our data to indicate that it is the source of the catalytic and/or C-terminal domains rather than the source of the N-terminal domain that allows a chimeric CesA to occupy either the CesA1 or the CesA3 site.
The C-Terminal Domain of CesA3 Is Important for Cellulose Production
The short C-terminal domain lying beyond TMD8 has not figured prominently in previous discussions of CesA structure and function. The properties of the rsw5 mutant show that a Pro-1056Ser change in the C-terminal domain of CesA3 reduces cellulose production. The C-terminal domain is therefore functionally important in cellulose production and the N-terminal domain (comprising more than 20% of CesA amino acid sequences) is conspicuous for currently lacking any mutations that inhibit cellulose synthesis. Sequence analysis of the C-terminal domain shows substantial differences between some CesA paralogs in Arabidopsis, although CesAs 2, 5, 6, and 9 show considerable similarity (see the alignment in Desprez et al., 2002). These C-terminal similarities of CesAs 2, 5, 6, and 9 are in keeping with the position of these CesAs in dendrograms showing relatedness for the entire sequence (Robert et al., 2004). We show that CesA3 orthologs, defined by possessing similar CSRs in the central catalytic domain (Vergara and Carpita, 2001), also possess similar C-terminal sequences. Conservation of the C-terminal sequence in CesA3 orthologs in both monocot and dicot plants suggests that the properties of this region have an ancient origin and is consistent with a conserved function in cellulose synthesis.
Differences between cDNA and Genomic Constructs in Promoting Root Elongation
We used a root growth assay to assess in vivo CesA function, an approach directly comparable to Fagard et al.'s use of a hypocotyl elongation assay to compare the strength of CesA6 alleles (Fagard et al., 2000). The first result of applying the growth assay to assess transgene function was an appreciation that cDNAs expressed behind the 35S promoter were less effective than genomic constructs in promoting root elongation. The variability in growth of individual T1 seedlings carrying a cDNA correlates in broad terms with differences in transgene mRNA levels. Variation in transgene expression between different transgenic lines has been noted previously (Swinburne et al., 1992) and could result from the transgene insertion occurring at a different site in the genome for each T1 seedling (Bechtold et al., 2003). The 35S promoter expresses indicator genes, such as green fluorescent protein, in root cells of the elongation and meristematic zones (Ridge et al., 1999), where mutations in CesAs 1 and 3 initiate radial swelling. Our results suggest that expression levels from cDNA constructs are lower than from genomic constructs in those cells that are critical for the growth phenotype.
Interpreting the Effects of Chimeric and Deletion Constructs in Terms of a Multisubunit Cellulose Synthase
We seek a framework to interpret our results that covers both dominant-positive and dominant-negative effects, accounts for the specificity regarding genotype seen with the dominant-negative effect, and is consistent with the interpretation of other relevant data, such as the number of glucan chains in microfibrils and the genetics of isoxaben resistance. Consider first the common basis for dominant-negative effects and how they relate to some likely properties of cellulose synthase.
Dominant-negative proteins are impaired function variants that also inhibit active proteins when the two coexist in the same cell. They commonly do this by entering and inactivating a multisubunit complex that may also contain the active (usually, but not invariably, the wild-type) protein (Gilbert, 2000; Alberts et al., 2001). A multisubunit cellulose synthase with specific sites for CesA1 and CesA3 is seemingly required to account for the noninterchangeability of CesA1 and CesA3, to provide enough glycosyltransferases to elongate microfibrils with >30 glucan chains, and to account for isoxaben resistance (see introduction). Such a complex therefore offers a plausible site of action for a dominant-negative CesA. Finer details of the cellulose synthase complex envisaged in several models (Scheible et al., 2001; Desprez et al., 2002; Robert et al., 2004) are not relevant to these experiments.
We hypothesize only two further properties for the cellulose synthase complex to interpret our results: (1) particular domains within the tails of CesAs 1 and 3 are required to access the CesA1 and CesA3 site; and (2) entry of a particular protein to each site may be restricted if other proteins preferentially occupy it. The latter would be seen as an aspect of the dominant/recessive effects familiar in genetics. Consider the two postulates in turn.
Particular Domains within the Tails of CesAs 1 and 3 Are Required to Access Each CesA Site
The chimeric protein 3H1T can partially replace CesA1 in the root growth assay using rsw1 at its restrictive temperature. This provides strong evidence that, like the more effective CesA1 itself, 3H1T must occupy the CesA1 site in cellulose synthase to partially complement the rsw1 root phenotype. Therefore, the domains fitting 3H1T to enter the CesA1 site will lie in those parts of the chimeric protein derived from CesA1 (i.e. the tail, comprising TMD2 onward). At first sight, the dominant-negative effect of 1H3T is less easily interpreted. However, we believe it can be interpreted within exactly the same framework if we recall that 1H3T only shows its massive dominant-negative effect (60% mortality) with rsw5 (the CesA3 mutant) and shows no visual phenotype with rsw1 or the wild type. In exactly the way that 3H1T depended on a CesA1 mutation to show dominant-positive activity (and so allowed us to infer that it enters the CesA1 site), we argue that 1H3T's dependence on a CesA3 mutation to show its dominant-negative activity supports the view that 1H3T is occupying the CesA3 site when it produces its dominant-negative effects. This is, of course, wholly consistent with the conclusion drawn from 3H1T that the residues determining the site occupied are in the tail that, in the case of 1H3T, comes from CesA3.
A requirement in both arguments (and one to which we return below) is that wild-type CesA1 and CesA3 proteins are dominant over mutant or chimeric CesAs. In the case of 3H1T, dominance means CesA1WT occupies the CesA1 site and effectively excludes 3H1T. (If it did not, we might expect to see a weak dominant-negative effect in the wild type because 3H1T only partially complements rsw1 at the restrictive temperature.) In the case of 1H3T, wild-type CesA3 occupies the CesA3 site and excludes 1H3T so that we see no dominant-negative effects in the wild type or rsw1 but massive effects in rsw5, where CesA3rsw5 does not exclude 1H3T from the complex.
An alternative to this view of the divergent effects of 1H3T in rsw5, rsw1, and the wild type is that 1H3T magnifies the permissive temperature phenotype of rsw5 by a double-mutant effect, but does not act similarly on rsw1 because it lacks a permissive temperature phenotype (or, at most, shows a very weak one under certain illumination conditions). We reject this hypothesis because there is no intrinsic obstacle to rsw1 showing a permissive temperature phenotype in a double-mutant situation. Moreover, it can do so even when the second mutant lacks its own permissive temperature phenotype. This can be illustrated with rsw3, a cellulose-deficient mutant defective in glucosidase II, an enzyme of the endoplasmic reticulum quality control pathway (Burn et al., 2002b). The rsw1rsw3 double mutant shows a permissive temperature phenotype for stem growth rate and cell length, a phenotype that neither rsw1 nor rsw3 shows as single mutant (Burn et al., 2002b; Table I). The likely basis for this phenotype in the double mutant is the subthreshold effect present in each single mutant combining to generate an above-threshold effect in the double mutant. We suggest, therefore, that there is nothing in the properties of permissive temperature phenotypes of rsw1 and rsw5 that provide an a priori reason why 1H3T should produce such different effects in them and so return to our hypothesis to explain the differences.
That hypothesis is, of course, a double-mutant hypothesis, but one that provides a very specific mechanism to explain how mutant CesA proteins (missense or chimeric) interact to produce either a massive phenotype (60% lethal) or one so mild as to be visually undetectable. To reiterate, we hypothesize that the effect seen with 1H3T reflects the chimeric protein potentially occupying the CesA3 site and CesA3WT protecting that site in a way that CesA3rsw5 cannot. As a result, the double-mutant 1H3T in rsw5 shows a frequently lethal phenotype, whereas both the other double mutant (3H1T in rsw1) and the single mutant (1H3T in the wild type) enjoy the protective effect of CesA3WT at the CesA3 site and so show no visible phenotype. We do not believe it is important that we may have missed a mild 1H3T phenotype in the wild type or rsw1. The phenotype of rsw1rsw3 shows that single-mutant phenotypes are not essential to see double-mutant phenotypes and any single-mutant phenotypes discovered would be readily accommodated within our existing model by hypothesizing that the protection afforded by CesA3WT was incomplete (but still much greater than that afforded by CesA3rsw5). This would allow some 1H3T to enter the CesA3 site and produce a mild phenotype.
We found no evidence from positive or negative changes to growth to suggest that either 1TL or 3TL enters the cellulose synthase complex. This suggests that entry requires a head domain, although the activity of chimeric proteins suggests that the head's source is not decisive for entry to the complex. It is striking that head interchanges can occur even though the head domains of CesAs 1 and 3 show only moderate sequence similarity. CesA1 has a 21-residue extension at the N terminus that CesA3 lacks. From there onward, 56% of residues are conserved, but levels of conservation are particularly low in the HVR1 region (Fig. 1A), which is flanked by regions showing higher conservation. Consider now the second postulate in our hypothesis.
Entry of a Particular Protein to Each Site May Be Restricted If Other Proteins Preferentially Occupy It
The proposed order for the CesA1 site is:
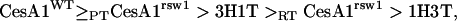 |
where > denotes “is dominant to” and subscripts PT and RT refer to the mutant's permissive and restrictive temperatures, respectively. It is based on the following observations: (1) Wild-type CesA1 is always dominant; (2) rsw1 at its permissive temperature is little different from the wild type (CesA1WT ≥ PTCesA1rsw1) and is unaffected by 3H1T (PTCesA1rsw1 > 3H1T); (3) 3H1T partially complements rsw1 grown at its restrictive temperature (3H1T > RTCesA1rsw1); and (4) 1H3T does not complement rsw1 grown at its restrictive temperature (RTCesA1rsw1 > 1H3T).
The proposed order for the CesA3 site:
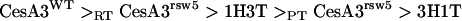 |
transposes the positions of the two chimeric genes and places 1H3T above PTCesA3rsw5. This order is consistent with observations that (1) the wild type is always dominant; (2) 1H3T does not complement rsw5 at its restrictive temperature (RTCesA3rsw5 > 1H3T) but is a dominant negative with rsw5 at its permissive temperature (1H3T > PTCesA3rsw5); and (3) 3H1T has no effect in the presence of any of these proteins.
Why Is 3H1T a Dominant Positive When 1H3T Is a Dominant Negative?
We have taken dominant effects, whether positive or negative, as evidence that the transgene-encoded protein must have entered and affected the cellulose synthase complex, but the issue arises as to why one chimeric protein (3H1T) is a dominant positive and the other (1H3T) is a dominant negative. As noted earlier, transgene effects depend on a particular combination of CesA proteins encoded by the endogenous genes and by the transgenes, either or both of which could contribute to the differences we see between 1H3T and 3H1T. First, differences in the capacities of the head regions could cause differences in the properties of the two chimeric genes. Specifically, 3H could meet the requirements for 3H1T to replace CesA1rsw1 at the restrictive temperature (dominant positive), whereas 1H may be unable to meet the functional requirements to allow 1H3T to replace CesA3rsw5 at the restrictive temperature. Its impaired functionality then shows up as a dominant negative at the permissive temperature. However, we cannot discount a second possibility, that properties specific to CesA1rsw1 and CesA3rsw5 influence the way the chimeric proteins perform, because we argue that the chimeric proteins can only enter the synthase sites when the mutant proteins allow them to. For example, disassembly of RTCs in rsw1 at its restrictive temperature (Arioli et al., 1998) might dissociate CesA3WT from CesA1rsw1, thus maximizing the chances of 3H1T interacting with CesA3WT and showing its partial functionality. The assembly state of the RTCs in rsw5 is unknown and if, for example, it is not as severe as in rsw1, 1H3T may not be offered such a good opportunity to interact with CesA1WT if CesA1WT and CesA3rsw5 do not fully dissociate at the restrictive temperature. Further understanding will require either additional temperature-conditional rsw alleles to see if the effects of the chimeric proteins depend on the particular rsw allele used or physicochemical characterization of the CesA protein complexes themselves.
In conclusion, the site of the amino acid substitution in rsw5 provides evidence that the small C-terminal domain of AtCesA3 performs an essential function in cellulose synthesis, a conclusion reinforced by the conservation of amino acid sequence we detected in putative AtCesA3 orthologs. Studies of chimeric CesA proteins point to the identity of the catalytic and/or C-terminal domains being most important for determining which of the noninterchangeable CesA1 and CesA3 sites a protein can access. Our interpretation of these results rests on specific hypotheses about the existence and properties of CesA complexes making primary wall cellulose, and our current work is directed to testing those predictions by isolating CesA complexes from wild-type, mutant, and transformed plants.
MATERIALS AND METHODS
Plant Material
rsw1 and rsw5 were isolated during a screen for temperature-sensitive radial-swelling mutants (Baskin et al., 1992). rsw1 is mutated in At4g32410 so that there is an Ala-549Val substitution in CesA1 (Arioli et al., 1998). In all experiments using mutants, the permissive temperature was 18°C to 20°C and the restrictive temperature was 30°C.
General Molecular Methods
Basic molecular biology procedures were performed essentially as described (Sambrook and Russell, 2001). TMDs were the consensus values given at Aramemnon, the plant membrane protein database (http://aramemnon.botanik.uni-koeln.de/index.ep). Consensus values are given after the protein is analyzed by 16 different TMD prediction programs. Sequence analysis of CSR and C-terminal domains was carried out using the BLAST facility at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/BLAST).
Molecular Analysis of rsw5
The fast DNA kit and protocol (Q-BIOgene) were used to isolate DNA from pooled plants in a F2-mapping population from crossing rsw5 (in the Arabidopsis [Arabidopsis thaliana] Columbia ecotype) with Landsberg erecta. This was probed with a series of cleaved-amplified polymorphic sequence markers (Konieczny and Ausubel, 1993; Lukowitz et al., 2000) to place the mutant gene on chromosome 5. Fine mapping with cleaved-amplified polymorphic sequence markers accessed from The Arabidopsis Information Resource (TAIR) database (http://www.arabidopsis.org) restricted the gene to about 200 kb between SNP13521 and SNP13537. At5g05170 encoding CesA3 was the obvious candidate gene. It was amplified by PCR from the mutant line and several products sequenced to verify the mutation.
Genomic and cDNA Constructs of CesA1 and CesA3
A genomic CesA3 clone containing the coding sequence and 1,050 bp of upstream sequence was constructed by PCR using the primers 5′-GTCGGCTAGCGAAGAGAAAGTG-3′ and 5′-AAGAGCTAGCGAGGATGATTGAAGATG-3′ to amplify from Columbia DNA. This was digested with NheI and ligated into the XbaI site of the binary vector pOCA28, a spectomycin-resistant derivative of pOCA18 (Olszewski et al., 1988). The cauliflower mosaic virus 35S-driven CesA3 cDNA clone was constructed by cleaving a pUNI 51 clone of the cDNA (GenBank BT002335; stock C104938 at the Arabidopsis Biological Resource Center [ABRC]) with SfiI, blunting the fragment prior to ligation into the SmaI site of pART7, and then ligating the NotI fragment into pART27 (Gleave, 1992). The CesA1 genomic clone has been described (Arioli et al., 1998). The CesA1 cDNA (Burn et al., 2002a) was cloned into the XhoI site of pART7 and thence into the NotI site of pART27. All steps involving PCR were performed with either high-fidelity Taq DNA polymerase (Invitrogen) or Vent Taq DNA polymerase (New England BioLabs).
Chimeric and Truncated Genes
Chimeric genes were made from CesA1 and CesA3 cDNAs by overlapping extension PCR (Horton et al., 1989). Briefly, fragments from the genes that are to be recombined are generated separately by PCR using primers designed so that the ends of the products contain complementary sequences where they are to be joined. When these PCR products are mixed, denatured, and reannealed, the strands having the matching sequences at their 3′ ends overlap and act as primers for each other. Extension of this overlap by DNA polymerase produces a full-length molecule in which the original sequences originating from the different cDNAs are spliced together. Finally, this novel fusion molecule is amplified by conventional primers matching the sequences at its ends. Primer pairs used to amplify the two products to form 3H1T were 5′-ATGGAATCCGAAGGAGAAACC-3′ and 5′-CAACCACAAAGCAAAGGCATTTGGCACTGG-3′ to amplify 3H and 5′-AAATGCCTTTGCTTTGTGGTTGACCTCGGTTATC-3′ and 5′-CCGGTTCACTGGGGTTTGATG-3′ to amplify 1T. Primer pairs used to amplify the two products to form 1H3T were 5′-TCTCTGTGTGTCGGTGGCTGCGAT-3′ and 5′-CAGCCATAGAGGATATGCATTTTTCACAGG-3′ to amplify 1H and 5′-TGCATATCCTCTATGGCTGGTCTCTGTGA-3′ and 5′-TAGTCGACGGCCCATGAGG-3′ to amplify 3T. For each chimeric construct, the first of the primers used to amplify the head and the second of the primers used to amplify the tail were used in the final PCR step. The molecules were cloned into pART7 and then into pART27.
Constructs encoding the truncated CesA proteins 1TL and 3TL that lacked residues from the N terminus to the start of TMD1 (Fig. 1A) were made by PCR amplification from the appropriate cDNAs with one primer designed to incorporate the normal C-terminal stop codon and the second to create a novel start codon just before the bases encoding the amino acids forming TMD1. Primers used with CesA1 to make 1TL were 5′-ATGGTGATTATTCTCCGGCTTATC-3′and 5′-CCGGTTCACTGGGGTTTGATG-3′, whereas the primers used with CesA3 to make 3TL were 5′-ATGGTTATTATGCTGCGGCTTGTTATC-3′ and 5′- TAGTCGACGGCCCATGAGG-3′.
Semiquantitative RT-PCR Detection of Chimeric Gene Expression
To assay expression of chimeric mRNA, we selected primer pairs that would amplify across the point where the sequence changed from one CesA to the other. 3H1T message was detected with forward 5′-GTTATTATGCTGCGGCTTGTTATC-3′ and reverse 5′-ATTCTTTGCAAACTCTGCGG-3′; 1H3T message was detected with forward 5′-GTGATTATTCTCCGGCTTATC-3′ and reverse 5′-TGGTGCACGAGGCTCTATGC-3′. A product from 18S rRNA provided a loading control after amplification with forward 5′-TTGTGTTGGCTTCGGGATCGGAGTAAT-3′ and reverse 5′-TGCACCACCACCCATAGAATCAAGAA-3′. Total RNAs were extracted with total RNA isolation reagent (Advanced Biotechnology) and RT-PCRs were carried out by a SuperScript one-step RT-PCR system with platinum Taq DNA polymerase (Invitrogen). Conditions for RT were 20 min at 48°C and 2 min at 94°C. Conditions for PCR were 15 s at 94°C; 30 s at 55°C; 30 s at 68°C; run PCR for required number of cycles plus an extra 7 min at 72°C.
Transformation, Selection, and Root Elongation Assay
All constructs were fully sequenced before plants were transformed by floral dipping (Clough and Bent, 1998). Kanamycin selection was conducted on horizontal agar plates grown at the permissive temperature with 16-h days. Twelve-day-old seedlings were transferred to vertical, nonselective agar plates for 2 d at the same temperature, after which the position of the tip on the longest root on each seedling was marked on the outside of the dish. After 2 d at the restrictive temperature, the agar plates were scanned and growth over the 2 d measured on the scanned image. Sample sizes were a minimum of 30 plants. Plates were returned to permissive temperature for 2 d before seedlings were transferred to soil and grown to seed set at the same temperature. Maturing plants were observed for developmental abnormalities and death rates and were photographed at maturity.
Acknowledgments
We thank the Arabidopsis Biological Resource Center for supplying CesA3 cDNA.
This work was supported by the Australian Research Council through the Discovery Program (DP0208889) and by Bayer Cropscience and the Australian Research Council through the Linkage Program (LP0211640).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Richard E. Williamson (richard.williamson@anu.edu.au).
References
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2001) Molecular Biology of the Cell, Ed 4. Garland, New York
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R, et al (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279: 717–720 [DOI] [PubMed] [Google Scholar]
- Baskin TI, Betzner AS, Hoggart R, Cork A, Williamson RE (1992) Root morphology mutants in Arabidopsis thaliana. Aust J Plant Physiol 19: 427–437 [Google Scholar]
- Bechtold N, Jolivet S, Voisin R, Pelletier G (2003) The endosperm and the embryo of Arabidopsis thaliana are independently transformed through infiltration by Agrobacterium tumefaciens. Transgenic Res 12: 509–517 [DOI] [PubMed] [Google Scholar]
- Beeckman T, Przemeck GK, Stamatiou G, Lau R, Terryn N, De Rycke R, Inzé D, Berleth T (2002) Genetic complexity of cellulose synthase A gene function in Arabidopsis embryogenesis. Plant Physiol 130: 1883–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RM Jr, Saxena IM, Kudlicka K (1996) Cellulose biosynthesis in higher plants. Trends Plant Sci 1: 149–156 [Google Scholar]
- Burn JE, Hocart CH, Birch RJ, Cork AC, Williamson RE (2002. a) Functional analysis of the cellulose synthase genes CesA1, CesA2 and CesA3 in Arabidopsis thaliana. Plant Physiol 129: 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burn JE, Hurley UA, Birch RJ, Arioli T, Cork A, Williamson RE (2002. b) The cellulose-deficient mutant rsw3 is defective in a gene encoding a putative glucosidase II, an enzyme processing N-glycans during ER quality control. Plant J 32: 949–960 [DOI] [PubMed] [Google Scholar]
- Caño-Delgado A, Penfield S, Smith C, Catley M, Bevan M (2003) Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J 34: 351–362 [DOI] [PubMed] [Google Scholar]
- Chen Z, Hong X, Zhang H, Wang Y, Li X, Zhu JK, Gong Z (2005) Disruption of the cellulose synthase gene, AtCesA8/IRX1, enhances drought and osmotic stress tolerance in Arabidopsis. Plant J 43: 273–283 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coutinho PM, Deleury E, Davies GJ, Henrissat B (2003) An evolving hierarchical family classification for glycosyltransferases. J Mol Biol 328: 307–317 [DOI] [PubMed] [Google Scholar]
- Desprez T, Vernhettes S, Fagard M, Refregier G, Desnos T, Aletti E, Py N, Pelletier S, Höfte H (2002) Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol 128: 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Karafyllidis I, Wasternack C, Turner JG (2002) The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Höfte H (2000) PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12: 2409–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner JC, Taylor NG, Turner SR (2003) Control of cellulose synthase complex localization in developing xylem. Plant Cell 15: 1740–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SF (2000) Developmental Biology, Ed 6. Sinauer Associates, Sunderland, MA
- Gillmor CS, Poindexter P, Lorieau J, Palcic MM, Somerville C (2002) α-Glucosidase I is required for cellulose biosynthesis and morphogenesis in Arabidopsis. J Cell Biol 156: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP (1992) A versatile binary vector system with a T-DNA organizational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Heim DR, Skomp JR, Waldron C, Larrinua IM (1991) Differential response to isoxaben of cellulose biosynthesis by wild-type and resistant strains of Arabidopsis thaliana. Pestic Biochem Physiol 39: 93–99 [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77: 61–68 [DOI] [PubMed] [Google Scholar]
- Kimura S, Laosinchai W, Itoh T, Cui XJ, Linder CR, Brown RM Jr (1999) Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. Plant Cell 11: 2075–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 4: 403–410 [DOI] [PubMed] [Google Scholar]
- Kurek I, Kawagoe Y, Jacob-Wilk D, Doblin M, Delmer D (2002) Dimerization of cotton fiber cellulose synthase catalytic subunits occurs via oxidation of the zinc-binding domains. Proc Natl Acad Sci USA 99: 11109–11114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DR, Wiedemeier A, Peng L, Höfte H, Vernhettes S, Desprez T, Hocart CH, Birch RJ, Baskin TI, Burn JE, et al (2001) Temperature sensitive alleles of RSW2 link the KORRIGAN endo-1,4-β-glucanase to cellulose synthesis and cytokinesis in Arabidopsis thaliana. Plant Physiol 126: 278–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible W-R (2000) Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol 123: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman RH, Davies LM, Harris PJ (1996) Solid-state C-13 nuclear magnetic resonance characterization of cellulose in the cell walls of Arabidopsis thaliana leaves. Plant Physiol 111: 475–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski NE, Martin FB, Ausubel FM (1988) Specialized binary vector for plant transformation: expression of the Arabidopsis thaliana AHAS gene in Nicotiana tabacum. Nucleic Acids Res 16: 10765–10782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM (1996) Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA 93: 12637–12642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L (1999) Characterisation of cellulose synthesis in Arabidopsis thaliana. PhD thesis. Australian National University, Canberra, Australia
- Ridge RW, Uozumi Y, Plazinski J, Hurley UA, Williamson RE (1999) Developmental transitions and dynamics of the cortical ER of Arabidopsis cells seen with green fluorescent protein. Plant Cell Physiol 40: 1253–1261 [DOI] [PubMed] [Google Scholar]
- Robert S, Mouille G, Höfte H (2004) The mechanism and regulation of cellulose synthesis in primary walls: lessons from cellulose-deficient Arabidopsis mutants. Cellulose 11: 351–364 [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sato S, Kato T, Kakegawa K, Ishii T, Liu YG, Awano T, Takabe K, Nishiyama Y, Kuga S, Nakamura Y, et al (2001) Role of the putative membrane-bound endo-1,4-β-glucanase KORRIGAN in cell elongation and cellulose synthesis in Arabidopsis thaliana. Plant Cell Physiol 42: 251–263 [DOI] [PubMed] [Google Scholar]
- Scheible W-R, Eshed R, Richmond T, Delmer D, Somerville C (2001) Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proc Natl Acad Sci USA 98: 10079–10084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Williamson RE, Wasteneys GO (2001) Wall architecture in the cellulose deficient rsw1 mutant of Arabidopsis thaliana: microfibrils but not microtubules lose their transverse alignment before microfibrils become unrecognizable in the mitotic and elongation zones of roots. Protoplasma 215: 172–183 [DOI] [PubMed] [Google Scholar]
- Swinburne J, Balcells L, Scofield SR, Jones JD, Coupland G (1992) Elevated levels of Activator transposase mRNA are associated with high frequencies of Dissociation excision in Arabidopsis. Plant Cell 4: 583–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100: 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Laurie S, Turner SR (2000) Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12: 2529–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR (1999) The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11: 769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SR, Somerville CR (1997) Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9: 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara CE, Carpita NC (2001) β-d-glycan synthases and the CesA gene family: lessons to be learned from the mixed-linkage (1→3),(1→4)β-d-glucan synthase. Plant Mol Biol 47: 145–160 [PubMed] [Google Scholar]
- Williamson RE, Burn JE, Birch R, Baskin TI, Arioli T, Betzner AS, Cork A (2001) Morphology of rsw1, a cellulose-deficient mutant of Arabidopsis thaliana. Protoplasma 215: 116–127 [DOI] [PubMed] [Google Scholar]
- Zhong R, Morrison WH III, Freshour GD, Hahn MG, Ye ZH (2003) Expression of a mutant form of cellulose synthase AtCesA7 causes dominant negative effect on cellulose biosynthesis. Plant Physiol 132: 786–795 [DOI] [PMC free article] [PubMed] [Google Scholar]