Abstract
Transgenic exploitation of bacterial degradative genes in plants has been considered a favorable strategy for degrading organic pollutants in the environment. The aromatic ring characteristic of these pollutants is mainly responsible for their recalcitrance to degradation. In this study, a Plesiomonas-derived chlorocatechol 1,2-dioxygenase (TfdC) gene (tfdC), capable of cleaving the aromatic ring, was introduced into Arabidopsis (Arabidopsis thaliana). Morphology and growth of transgenic plants are indistinguishable from those of wild-type plants. In contrast, they show significantly enhanced tolerances to catechol. Transgenic plants also exhibit strikingly higher capabilities of removing catechol from their media and high efficiencies of converting catechol to cis,cis-muconic acid. As far-less-than-calculated amounts of cis,cis-muconic acid were accumulated within the transgenic plants, existence of endogenous TfdD- and TfdE-like activities was postulated and, subsequently, putative orthologs of bacterial tfdD and tfdE were detected in Arabidopsis. However, no TfdC activity and no putative orthologs of either tfdC or tfdF were identified. This work indicates that the TfdC activity, conferred by tfdC in transgenic Arabidopsis, is a key requirement for phytoremoval and degradation of catechol, and also suggests that microbial degradative genes may be transgenically exploited in plants for bioremediation of aromatic pollutants in the environment.
Decades of modern agricultural practices and industrializations have resulted in the extensive accumulation of organic pollutants in soil and water. Aromatic compounds, such as 2,4-dichlorophenoxyacetic acid, 3-chlorobenzoate, and chlorocatechol, are among the most hazardous due to their (or their derivatives') potential carcinogenesis and recalcitrance to degradation (Neilson et al., 1991; Capasso et al., 1995). On the other hand, the increasing accumulation of these compounds in the environment has prompted native bacteria (Pseudomonas, Alcaligenes, Ralstonia) to develop capabilities of using them as carbon and/or energy sources. Intensive studies have led to the identification and elucidation of a modified ortho-degradation pathway (Fig. 1; Perkins et al., 1990; Laemmli et al., 2000). Generally, the aromatic compounds are first converted to their respective chlorocatechols. These chlorocatechols are then sequentially converted to the 3-oxoadipate through the actions of four enzymes encoded by tfdC, tfdD, tfdE, and tfdF, respectively. The 3-oxoadipate is further funneled to the 3-oxoadipate pathway, leading to the production of succinate and acetyl-CoA, which can be incorporated into the fundamental metabolic and/or catabolic pathways (Perkins et al., 1990; Reineke, 1998). Cleavage of the aromatic ring in the ortho-position, between the hydroxyl groups (Harayama and Rekik, 1989), to form cis,cis-muconic acids is a rate-limiting step for the degradation of chlorocatechols and reduction of their toxic activity. Chlorocatechol 1,2-dioxygenase (TfdC; EC 1.13.11), encoded by the tfdC gene, catalyzes the reaction. It has been shown that the chlorocatechol oxidation reaction is the key step in the 3-chlorobenzoate degradation pathway of bacteria (Perez-Pantoja et al., 2003).
Figure 1.
Proposed degradation pathways of chlorinated aromatics. Enzymes catalyzing the reactions are as follows: TfdA, 2,4-dichlorophenoxyacetic acid α-ketoglutarate dioxygenase; TfdB, chlorophenol hydroxylase; TfdC, chlorocatechol 1,2-dioxygenase; TfdD, chloromuconate cycloisomerase; TfdE, dienelactone hydrolase; TfdF, maleylacetate reductase.
The tfdC was first cloned from plasmid pJP4 of JMP134, which can degrade and utilize chloroaromatics (Perkins et al., 1990). In our previous study, a similar tfdC gene was identified and cloned from Plesiomonas, which was isolated from the activated sludge collected from a wastewater-recycling factory and can use chlorobenzene as the sole source of carbon and energy. After being cloned into expression vector and expressed in Escherichia coli, the expected 33-kD protein was observed on the SDS-PAGE and a significant TfdC activity was detected from its cell extracts (Luo and Li, 1998; Ma et al., 2000).
Bioremediation is considered to be a cost-effective, environmentally friendly way for the cleanup of contaminated soils and surface water. However, persistence of the aromatic pollutants in the environment suggests the less effectiveness of natural-microorganism-promoted biodegradations in situ, probably because their efficiency is largely dependent on the suitability of environmental conditions in maintaining the growth of microorganisms. Phytoremediation has therefore received increasing attention in recent years. Plants are used instead for remedial purposes not only because they generally have extensive root systems, larger biomass, and consequently higher decontamination efficiency, but also because they are highly differentiated and protective organisms capable of tolerating adverse environmental factors effectively. It is also important that they can improve and/or provide conditions favorable for microbial degradation. Besides, as a renewable energy resource and a local climate regulator, plants have potential economic and environmental benefits (Kramer, 2005; Newman and Reynolds, 2005; Peuke and Rennenberg, 2005; Pilon-Smits, 2005).
Using genetic engineering to increase/create the remedial capacity of plants has been increasingly explored as a sustainable technology (Rugh et al., 1996; French et al., 1999; Hannink et al., 2001; Dhankher et al., 2002; Wang et al., 2004). A preliminary work exploring the potential of a chlorocatechol dioxygenase gene (cbnA) in phytoremediation was reported in rice (Oryza sativa), and its conversion of catechol to muconic acid was observed in transgenic calli and leaf tissues. However, the remediation potential was not demonstrated at the plant level (Shimizu et al., 2002).
To fully explore the potential of chlorocatechol dioxygenase gene in phytoremediation of soil contaminated with aromatic compounds, we modified the tfdC gene from Plesiomonas spp. and transferred it into Arabidopsis (Arabidopsis thaliana). Our results show that transgenic plants expressing the modified tfdC gene obtained a striking ability of tolerating, removing, and biodegrading catechol, implying that the tfdC gene has the potential of being explored for phytoremediation of sites contaminated with aromatic compounds.
RESULTS
Construction of Transgenic Arabidopsis
To efficiently express the tfdC gene from Plesiomonas in the plant, a plant consensus start sequence AAATGA was introduced using PCR method. The modified gene was subsequently inserted into the binary vector pPZPY122 (Hajdukiewicz et al., 1994) under the control of cauliflower mosaic virus 35S promoter, and introduced into Arabidopsis via Agrobacterium (LBA4404)-mediated transformation (Clough and Bent, 1998). Empty vector transgenic (VT) plants were also constructed to be used as a control. Twenty-seven tfdC transgenic (CT) plants (T1) were initially identified from 29 putative transgenic plants regenerated on half-strength Murashige and Skoog (MS) agar plates containing gentamicin by PCR. All the transgenic plants were phenotypically indistinguishable from wild-type plants when they were grown either on half-strength MS agar plates or in soil in the growth room, suggesting that the insertion of the tfdC gene in these plants caused no visible morphological effects.
Relative Toxicities of Catechol and cis,cis-Muconic Acid to Arabidopsis
Relative toxicities of catechol and cis,cis-muconic acid to the wild type as well as to transgenic Arabidopsis were determined on agar-solidified, half-strength MS plates. Seeds of the wild type were germinated on half-strength MS agar plates containing either 0.0, 0.1, 0.2, 0.3, 0.4, and 0.5 mm catechol, or 0.0, 0.1, 0.2, 0.3, 0.4, and 0.5 mm cis,cis-muconic acid. On plates containing even 0.1 mm catechol, some wild-type seeds were either not able to germinate or died shortly after germination, whereas on plates containing 0.5 mm cis,cis-muconic acid, they could still develop into seedlings (Fig. 2). This significantly differential toxic effect implies that the tolerance to catechol can be used as a valid indicator of the function of active tfdC gene in transgenic Arabidopsis. However, at the higher concentrations of cis,cis-muconic acid (above 0.4 mm), a visible toxicity to root development was also observed (Fig. 2, right). At 0.1 and 0.2 mm catechol, T2 transgenic lines, although segregating, generally showed enhanced tolerances compared to wild-type plants (data not shown). Four transgenic lines, CT1-2, CT1-5, CT1-16, and CT1-19, were chosen for further analyses.
Figure 2.
Relative toxicities of catechol and cis,cis-muconic acid to Arabidopsis. Morphologies of wild-type plants on half-strength MS agar plates containing 0.1 (left), 0.2 (middle), and 0.5 mm (right) catechol or cis,cis-muconic acid.
Stable Expressions of the tfdC Gene and TfdC Activities in Transgenic Plants
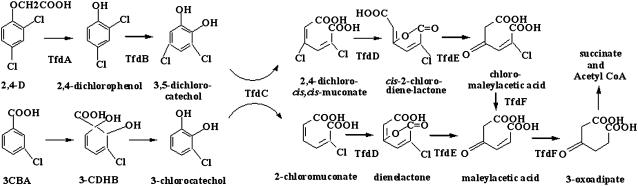
Expressions of the tfdC gene were determined in the T3 plants of homozygous transgenic lines using reverse transcription (RT)-PCR. An RT-PCR product of approximately 800 bp, corresponding to the tfdC coding sequence, was detected in all four CT lines analyzed, whereas no signals were detected in VT plants (Fig. 3A). With ACT11 as a reference, relatively higher expression levels were detected in CT1-5 and CT1-19 compared to those in CT1-2 and CT1-16.
Figure 3.
Expression levels of tfdC and activities of TfdC in transgenic Arabidopsis. A, RT-PCR-amplified tfdC fragments from the plants of VT and CT lines (CT1-2, CT1-5, CT1-16, CT1-19) with ACT11 as a reference. B, RT-PCR-amplified tfdC fragments from roots (R), rosette leaves (RL), cauline leaves (CL), and stems (S) of the transgenic line CT1-2. C, Activities of TfdC in the VT and CT plants. Catechol, Added substrate catechol in the reaction mixtures but no crude enzyme. The data represent mean ± sd (n = 3).
To determine the tissue-specific expression pattern, RT-PCR analysis was performed with total RNAs extracted from roots, rosette leaves, cauline leaves, and stems of CT1-2 plants. Transcripts of tfdC were detected in all the tissues examined, with the highest being observed in the stems and the lowest in the roots (Fig. 3B).
TfdC activities in the CT and VT lines were assayed with crude enzyme extracts from their seedlings. The TfdC activity was expressed as the amount of cis,cis-muconic acid produced per milligram of protein. All the four CT lines showed significantly higher TfdC activities during the period of incubation. In contrast, after incubating even for 30 min, no detectable amount of cis,cis-muconic acid was found in the treatment with the enzyme extract prepared from the VT plants (Fig. 3C), indicating that there is no significant intrinsic TfdC activity in Arabidopsis. Nevertheless, significantly differential TfdC activities were observed in different CT lines, which can be well correlated to the transcript levels of the tfdC in these lines.
Enhanced Tolerances of Homozygous CT Plants to Catechol on Plates
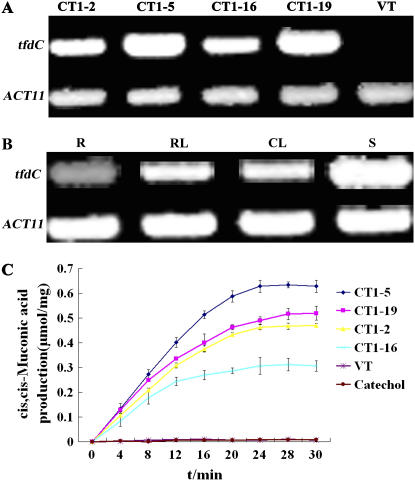
Forty seeds from each of VT and CT lines were sown on half-strength MS agar plates containing 0.0, 0.1, and 0.2 mm catechol, respectively, and grown vertically for 2 weeks. On the control medium without catechol, both CT and VT plants appeared equally healthy and produced similar biomasses (about 4 mg/plant). As the concentration of catechol in the medium increased, however, the CT plants generally exhibited significantly higher tolerances, showing greener and broader leaves as well as higher growth rates (Fig. 4A). On the other hand, growth of the VT plants was retarded even at 0.1 mm, with their average fresh weight being about 60% of those of the CT plants (Fig. 4B). At 0.2 mm, however, VT plants were severely inhibited, grew too small and abnormal so that the weights of individual plants could not be determined accurately. Not surprisingly, at higher concentrations of catechol, growth and development of the CT plants were also inhibited (Fig. 4A). These observations indicated that the inhibitions on the growth and development of VT as well as CT plants were solely due to the presence of catechol.
Figure 4.
Enhanced catechol tolerances of CT plants on plates. A, VT and CT T3 plants germinated and grown vertically for 2 weeks on half-strength MS agar plates containing 0.0, 0.1, or 0.2 mm catechol. B, Fresh weight of 2-week-old VT and CT plants grown on half-strength MS agar plates containing 0.0 and 0.1 mm catechol. C, Root length of 2-week-old VT and CT plants grown on half-strength MS agar plates containing 0.0, 0.1, and 0.2 mm catechol. The data represent mean ± sd (n = 10). Statistical analysis of differences in CT lines with respect to VT was performed using two-tailed t test. Significant difference is denoted with one asterisk (P < 0.05) or two (P < 0.01).
Efficient root and shoot formation is a crucial characteristic of transgenic plants designed for potential application in phytoremediation. Both VT and CT plants developed similar root systems with extensive secondary root branches and root hairs on the control medium without catechol, with the longest reaching about 4.5 cm. On the medium containing 0.1 mm catechol for 2 weeks, transgenic plants displayed minimal sign of phytotoxic effects, producing extensive root branches and showing a normal shoot development, especially CT1-16 and CT1-19, while VT seedlings exhibited a stunted development, producing few root hairs. At 0.2 mm catechol, although the shoot development of CT plants was affected, they could still develop gradually to later stages and produced secondary roots and visible root hairs. The average root length of CT plants was 0.6 cm. At the same concentrations, however, shoot development of the VT plants was halted at the two-leaf stage, and neither secondary roots nor visible root hairs were observed. The average root length of VT plants was about 40% of those of CT plants (Fig. 4, A and C). In addition, the CT plants examined also showed an enhanced tolerance to catechol in soil in the growth room (see Supplemental Fig. S1).
Higher Catechol Removal Efficiencies of CT Plants
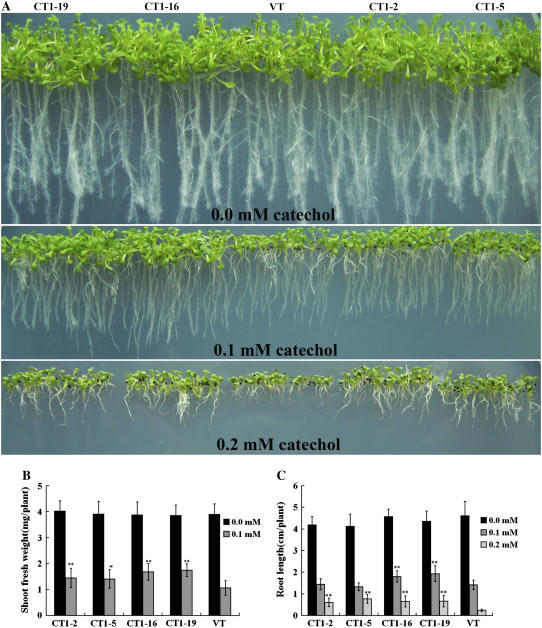
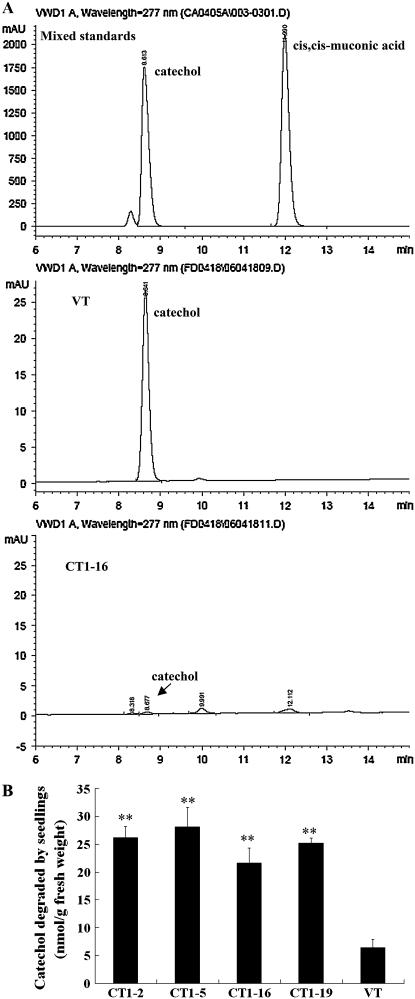
Fifty seeds from each of the VT and CT lines were germinated and grown in flasks containing half-strength MS liquid medium with 0.0, 0.1, and 0.2 mm catechol, respectively, on a shaker for 2 weeks. In the medium without catechol, both CT and VT plants grew healthy and produced similar fresh weights of about 2.8 g per flask. At 0.1 mm, however, the growth of VT plants was significantly inhibited, with the average fresh weight being about 0.4 g per flask, about 30% of those of CT1-16 and CT1-19 and about 50% of those of CT1-2 and CT1-5. At 0.2 mm, only some of the VT plantlets could survive the incubation, producing a final fresh weight of 0.055 g. In contrast, the seeds of the CT lines germinated well and the seedlings showed a relatively normal morphology and growth rate, producing an average final fresh weight of approximately 0.2 g (Fig. 5, A and B).
Figure 5.
Morphologies of VT and CT lines in catechol-containing liquid medium. A, Fifty seeds of VT and CT plants in flasks were allowed to grow for 2 weeks in half-strength MS liquid medium containing 0.0, 0.1, and 0.2 mm catechol. B, Fresh weights of seedlings in the individual flasks in the above treatment. The data represent mean ± sd (n = 3). Statistical analysis of differences in CT lines with respect to VT was performed using two-tailed t test. Significant difference is denoted with one asterisk (P < 0.05) or two (P < 0.01).
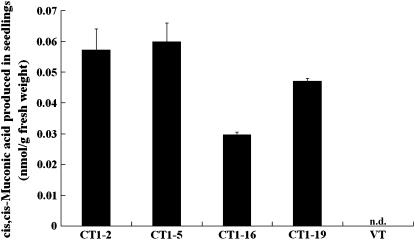
Concentrations of catechol and its catabolite, cis,cis-muconic acid, in the medium were then determined by HPLC. Catechol and cis,cis-muconic acid were identified by referring to their respective retention time (Fig. 6A, top). After 2-week incubation, a sharp contrast on catechol concentration was detected in the media of VT (Fig. 6A, middle) and CT (Fig. 6A, bottom) plants. According to the amount of catechol detected in the medium and that initially added, the degraded proportions were calculated. CT plants generally showed 3 to 5 times higher degradation rates to those of VT plants (Fig. 6B). No detectable amount of cis,cis-muconic acid was found in both media. A further HPLC analysis revealed that no detectable amounts of catechol were accumulated in the extracts of either CT or VT plants. However, cis,cis-muconic acid was found to accumulate significantly in the seedlings of CT plants (CT1-2, 0.057 ± 0.007 nmol g−1 fresh weight; CT1-5, 0.060 ± 0.006 nmol g−1 fresh weight; CT1-16, 0.030 ± 0.001 nmol g−1 fresh weight; CT1-19, 0.047 ± 0.001 nmol g−1 fresh weight; Fig. 7).
Figure 6.
HPLC analysis of removal of catechol in media. A, HPLC analyses of catechol and cis,cis-muconic acid in the media; after seedlings were grown in half-strength MS liquid media containing 0.2 mm catechol for 2 weeks, concentrations of catechol and cis,cis-muconic acid in VT (middle) and CT media (bottom) were determined by HPLC; mixed standards (top). B, Catechol degradation rates after 2-week incubation. Catechol amounts degraded were calculated by deducting the amount of the catechol left in medium from the amount of catechol added initially into medium, and degradation rates were expressed as nmol g−1 fresh weight. The data represent mean ± sd (n = 3). Statistical analysis of differences in CT lines with respect to VT was performed using two-tailed t test. Significant difference is denoted with two asterisks (P < 0.01).
Figure 7.
HPLC analysis of plant extracts. After seedlings were grown in half-strength MS liquid media containing 0.2 mm catechol for 2 weeks, extractions were performed. Concentrations of cis,cis-muconic acid were quantitatively determined by HPLC. The data represent mean ± sd (n = 3); n.d., not detected.
Far-Less-Than-Calculated Amounts of cis,cis-Muconic Acid Retained in the CT Plants
Approximately 5 nmol catechol was initially added into the medium of CT plants, but only trace levels were detected either in the medium or within the CT plants at the end of incubation. Theoretically, equivalent amounts of cis,cis-muconic acid should have been produced and retained in the CT plants (no cis,cis-muconic acid was detected in the media previously). Unexpectedly, only 0.01 nmol cis,cis-muconic acid (about 0.2% of the amount of catechol disappeared) was detected within the plants (Table I). To explain the discrepancy, we assumed the existence of endogenous TfdD-, TfdE-, and/or TfdF-like activities. A search of the Arabidopsis protein database against the amino acid sequences of bacterial TfdC, TfdD, TfdE, and TfdF, by WU-BLAST 2.0, revealed the existence of expected putative orthologs of tfdD and tfdE. An experiment was also carried out to measure the activities of TfdD and TfdE in the cell extracts of Arabidopsis using spectrophotometric methods. Our preliminary results indicated that the cell extracts did have the activities of both TfdD and TfdE. In the proceeding cause of enzymatic reaction, dienelactone, the catalyzed product of cis,cis-muconic acid by TfdD, was first accumulated and then declined, which was the characteristic reflection of the sequential functioning of TfdD and TfdE in the extracts (see Supplemental Fig. S2). However, according to the P-value criterion (P < 0.01), no putative orthologs of tfdC and tfdF have been identified in the database (Table II).
Table I.
Amounts of catechol degraded and cis,cis-muconic acid detected within VT and CT plants
Amounts of catechol degraded were calculated by deducting the amount of the catechol left in medium at the end of incubation from the amount of catechol added initially into medium, and the amount of cis,cis-muconic acid accumulated in plants was calculated according to HPLC data. The data represent mean ± sd (n = 3).
| Lines | Catechol Degraded (A) | cis,cis-Muconic Acid Detected in Plants (B) | A/B |
|---|---|---|---|
| nmol | nmol | % | |
| CT1-2 | 4.698 ± 0.011 | 0.0102 ± 0.0004 | 0.22 |
| CT1-5 | 4.891 ± 0.005 | 0.0104 ± 0.0002 | 0.21 |
| CT1-16 | 4.937 ± 0.001 | 0.0091 ± 0.0000 | 0.18 |
| CT1-19 | 4.909 ± 0.002 | 0.0092 ± 0.0002 | 0.19 |
| VT | 0.358 ± 0.130 | n.d.a |
Not detected.
Table II.
Identification of putative orthologs of bacterial tfdC, tfdD, tfdE, and tfdF in Arabidopsis by BLAST
Arabidopsis protein database was searched against the amino acid sequences of the bacterial TfdC, TfdD, TfdE, and TfdF, which are located sequentially on the ortho-pathway, by WU-BLAST 2.0. Candidates were picked up according to the P-value criterion (P < 0.01).
| Enzyme | AGI Code | Description | P Value |
|---|---|---|---|
| TfdC | NFa | ||
| TfdD | AT3G18270.1 | Mandelate racemase/muconate lactonizing enzyme family protein | 1.40E-11 |
| AT1G68900.1 | Mandelate racemase/muconate lactonizing enzyme | 0.0019 | |
| TfdE | AT2G32520.1 | Dienelactone hydrolase family protein | 1.00E-08 |
| AT3G23570.1 | Dienelactone hydrolase family protein | 0.0011 | |
| TfdF | NF |
Not found.
DISCUSSION
Catechol and its catabolites have cellular toxicities to bacteria and animals, as stated by a fact sheet of U.S. Environmental Protection Agency (http://www.epa.gov/ttn/atw/hlthef/pyrocate.html). However, to our best knowledge, there have been no data showing their toxicities to plants so far. In this study, a significant toxic effect of catechol to Arabidopsis was demonstrated. It not only severely inhibited the germination and growth of the wild-type Arabidopsis, but also significantly stunted the development of both the roots and shoots of the wild-type plants germinated otherwise in the absence of catechol. The toxicity of catechol is considered a consequence of its chemical properties and chemical reactions with biomolecules. Many reactions can occur between catechol and biomolecules (DNA and proteins) or membranes, causing them to break, to be inactivated, and/or to be destructed (Schweigert et al., 2001).
Fortunately, a significantly lower toxicity of cis,cis-muconic acid to Arabidopsis was identified and, consequently, a screening approach was used in analyzing transgenic plants in this study (Fig. 2). In the case of either an equal or even higher toxicity identified, an otherwise complicated, labor-intensive approach would have to be adopted to analyze the functional effect of the transgenic tfdC gene. This work implies that single microbial degradative genes can be transgenically exploited to analyze the phytodegradation of aromatic pollutants straightforward, provided that the immediate products are less toxic.
Expression of the tfdC gene was detected in all the tissues examined, including the root (Fig. 3B). However, higher expression levels were generally detected in tissues above the ground, with the highest found in the stem and the lowest in the root, which shares a general tendency with Shimizu's observation (Shimizu et al., 2002). Importantly, CT plants produced in this study can tolerate significantly higher concentrations of catechol than VT plants, producing higher biomass and more normal root in the presence of catechol (Figs. 4 and 5). Even more importantly, they can remove catechol from their surrounding media (Fig. 6) and degrade it much more efficiently than VT plants (Fig. 7). In addition, it was found that seedlings germinated first in the absence of catechol and then transferred to catechol-containing plates grew much better than seedlings germinated and grown on plates containing catechol all the time (Fig. 2; Supplemental Fig. S3). A similar phenomenon also was observed by Hannink (Hannink et al., 2001). According to Hannink et al.'s understanding, either the inhibitory effect of catechol to wild-type seedlings is more significant at the early stage of plant development, or increased biomass or possibly elevated enzyme activities in older seedlings allows more effective detoxification of catechol. However, we also believe that both of the factors may contribute to the differential tolerances observed.
Biodegradations of organic pollutants can occur either in vivo (Hannink et al., 2001) or in vitro (Wang et al., 2004). To microorganisms, the complete in vivo degradation of organic pollutants may mean assimilation of the carbon and capture of the energy they contain, as in the case of Plesiomonas, from which the tfdC gene was isolated. In this study, based on the observation that almost all the added catechol was removed but no cis,cis-muconic acid was detected in the media of CT seedlings (Fig. 6), we also consider that the ring cleavage, catalyzed by the exogenous TfdC, mainly took place in vivo. Furthermore, no catechol was found to accumulate in vivo, but significant amounts of cis,cis-muconic acid were detected in the CT seedlings (Fig. 7). These results together indicate that catechol was absorbed into the CT plant and degraded efficiently. A similar situation also was observed in the case of 2,4,6-trinitrotoluene phytodegradation (Hannink et al., 2001). No matter whether the immediate catabolites are toxic or not, it is a favorable characteristic to trap catabolites within the plants for phytoremedial purpose.
Stoichiometrically, a reduction in the nmol of catechol should equal an increase in the nmol of cis,cis-muconic acid in the liquid culture system. Far less amounts of cis,cis-muconic acid retained in the CT plants (Table I) prompt us to postulate that cis,cis-muconic acid was either gradually degraded further by endogenous TfdD-, TfdE-, and/or TfdF-like activities (Fig. 1), or sequestered within the plant in a form that may not be easily extractable. Considering the fact that cis,cis-muconic acid has been detected in the extracts of CT plants, we assume that the former was likely the case. The assumption is supported by the identification of putative orthologs of bacterial tfdD and tfdE in the Arabidopsis genome (Table II) and by the demonstration of the existence of TfdD- and TfdE-like activities in the cell extracts of Arabidopsis plants. In a broader sense, it is also plausible for the existence of these orthologs in Arabidopsis since plants have versatile detoxification systems to counter the phytotoxicity of the wide variety of natural and synthetic chemicals (Coleman et al., 1997). The finding that no obvious putative orthologs of tfdC and tfdF exist in the Arabidopsis genome supports our observation of no endogenous TfdC activity. These results together demonstrate that the TfdC activity conferred by tfdC in transgenic Arabidopsis is the key requirement for phytodegradation of catechol.
In conclusion, this study clearly shows that the bacterial tfdC gene can be manipulated to cleave the aromatic ring of a simple organic pollutant, catechol, in the model plant Arabidopsis, and that the catechol can be efficiently removed and degraded from the media of transgenic plants. An immediate future work would be to engineer its upstream genes, tfdA and tfdB, on the ortho-pathway (Fig. 1), as well as tfdC, ideally combined with pollutant (chlorobenzene, for example)-responsive cis-elements on the bacterial plasmid, into Arabidopsis to test the feasibility of phytodegrading more complex aromatic pollutants.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All the wild-type and transgenic Arabidopsis (Arabidopsis thaliana) lines used in this study are in the Columbia-0 background. When grown in plates, seeds were surface sterilized, placed in the dark at 4°C for 2 d, and then sown on plates containing half-strength MS salts with 2% (w/v) Suc. When grown in soil pots, seeds were sown in square pots (10-cm sides) with soil (peat soil:vermiculite:pearlite [v/v/v] = 3:9:0.5; purchased from Shanghai Institute of Landscape Science) presoaked with plant nutrient medium. All the plants were grown in a growth room at 22°C ± 2°C and with approximately 100 μmol s−1 m−2 light intensity under standard long-day conditions (16 h light/8 h dark).
Construction of Binary Vectors
The tfdC gene was isolated from the pL1 plasmid of Plesiomonas using PCR method with a forward primer (5′-ACGGAGGCAAAGTGAACAAAAGAG-3′) and a backward primer (5′-ACTGCTTCAATCGCGTCAATCTTC-3′), and modified using the forward 5′-ACGGAGGCAAAATGAACAAAAGAG-3′ (base changes underlined) and the same backward. The gene was modified to contain the plant consensus start sequence. PCR products were purified with glass beads and then ligated into pGEM-T Easy vector (Promega). The tfdC fragment was cleaved with BamHI and SacI, subcloned into BamHI-SacI sites of a plant transformation vector containing cauliflower mosaic virus 35S promoter pPZPY122, and designated pPZPY122/tfdC. Constructs were verified by sequencing. All DNA manipulations were performed according to standard protocols (Sambrook et al., 1989).
Transformation of Arabidopsis
Construct pPZPY122/tfdC was introduced into the Agrobacterium tumefaciens LBA4404 strain by electroporation. Arabidopsis (Columbia-0) was transformed using floral-dipping method. T1 seeds were screened for gentamicin resistance on half-strength MS agar plates containing 90 mg L−1 gentamicin. Genomic DNAs were isolated from transgenic and wild-type plants according to protocol (ftp://ftp.arabidopsis.org/home/tair/Protocols/compleat_guide). PCR was used to screen for putative T1 transgenic plants, with the conditions described previously. T2 homozygous transgenic lines were selected based on the segregation analysis of gentamicin resistances of their self-fertilized T3 seedlings. The homozygosis of T2 transgenic lines was checked again by growing T3 seedlings on media containing 0.1 mm catechol. T3 homozygous seeds were used for all subsequent analyses.
Total RNA Extraction and RT-PCR
Total RNAs were extracted from both transgenic and wild-type seedlings using TRIzol reagent (Invitrogen). First-strand cDNA synthesis was performed with 2 μg of total RNA using a first-strand cDNA synthesis kit (Shenergy Biocolor), and the products were quantitatively equalized for RT-PCR using ACT11 cDNA as a reference. The RT-PCR reactions were performed with the following primers: tfdC-1 (5′-ACGGAGGCAAAGTGAACAAAAGAG-3′), tfdC-2 (5′-ACTGCTTCAATCGCGTCAATCTTC-3′), ACT11-1 (5′-GATTTGGCATCACACTTTCTACAATG-3′), and ACT11-2 (5′-GTTCCACCACTGAGCACAATG-3′). Amplified fragments were separated on a 1.2% agarose gel, cloned in pGEM-T (Promega), and then verified by sequencing.
Extraction of TfdC from Seedlings
Fresh leaves were homogenized in a Waring blender with precooled 0.05 m phosphate buffer, pH 7.0, and the homogenate was filtered through six layers of gauze. The filtrate was then centrifuged at 13,000g for 30 min at 4°C. Crude enzyme was quantified by Coomassie Brilliant Blue G250 (Spector, 1978) and used for activity assay.
Measurement of cis,cis-Muconic Acid with Spectrophotometer
Reaction mixtures contained 3 μmol EDTA, 0.3 μmol catechol, 26.1 μmol sodium phosphate, pH 7.0, and cell exacts (0.3 mg of total protein) in a final volume of 3 mL. Reactions proceeded at 25°C for 30 min. The increase in A260 was used as a measure of the accumulation of cis,cis-muconic acid. When 1 μmol catechol changes to cis,cis-muconic acid, A260 increases 5.6. TfdC activity was expressed as amount of cis,cis-muconic acid produced per milligram of protein (Spain and Nishino, 1987).
Tests of Plant Tolerances
For test on plates, VT and CT plant seeds were sown on half-strength MS agar plates containing either catechol or cis,cis-muconic acid, which were dissolved in water and filtrated (0.22-μm filter; Millipore) before added to plates. About 40 seeds of each line were placed on plates under aseptic conditions. After grown vertically for 2 weeks, the longest root length and fresh weight were measured for each treatment of approximately 10 seedlings.
For test in soil, VT and CT plant seeds were germinated in soil soaked with plant nutrient medium containing 0, 1, 2, 3, 4, and 5 mm catechol, respectively. After 2 weeks, the plantlets were sprayed with 0, 1, 2, 3, 4, and 5 mm catechol solutions every 3 d for 3 weeks before photos were taken.
Measurements of Catechol and cis,cis-Muconic Acid by HPLC
VT and CT plant seeds (50 per flask) were surface sterilized, germinated in 25 mL of half-strength MS liquid medium containing catechol, and grown for 3 weeks with rotary shaking at 120 rpm, 23°C. Samples were taken from the media and seedlings. The medium samples were directly analyzed with Agilent 1100 HPLC using an Agilent Eclipse XDS-C18 column. Methanol-water containing 1% (v/v) acetic acid was used as the eluant at a flow rate of 0.8 mL/min. The column effluent was monitored simultaneously at 277 nm by a diode array detector. A 10-μL aliquot of each sample was injected onto the column. The retention times of known standards were used to identify catechol and cis,cis-muconic acid. Catechol was purchased from Sinopharm Chemical Reagent. cis,cis-Muconic acid was purchased from Sigma-Aldrich (Fluka).
Extraction of Catechol and Its Metabolites from Plants
The seedling samples were homogenized in a Waring blender with 200 μL of precooled methanol and then sonicated for 10 min. After centrifugation at 13,000g for 30 min at 4°C, the supernatant was filtered (0.45-μm filter; Millipore). Ten milliliters of each sample was then analyzed by HPLC using the conditions described above to determine the concentration of catechol and cis,cis-muconic acid in the seedlings. The peak areas were converted to molar amounts according to the standard curves of the standards and normalized to the wet weight of samples.
Measurement of TfdD- and TfdE-Like Enzymatic Activities
Preparation of the cell extracts of Arabidopsis plants and assay of TfdD- and TfdE-like enzymatic activities were carried out as reported (Schmidt and Knackmuss, 1980; Kuhm et al., 1990; Schlomann et al., 1990).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number NC_005912 for tfdC.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Enhanced tolerances of CT plants in soil.
Supplemental Figure S2. Assay of TfdD- and TfdE-like activities.
Supplemental Figure S3. Enhanced tolerances of CT plants in plates.
Supplementary Material
Acknowledgments
We thank Mr. Wenli Hu, Lingjian Wang, and Dr. Xiaoya Chen of the Institute of Plant Physiology and Ecology for their help in HPLC analysis.
This work was supported by the National Natural Science Foundation of China (project no. 39770478).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Benke Kuai (bkkuai@fudan.edu.cn).
The online version of this article contains Web-only data.
References
- Capasso R, Evidente A, Schivo L, Orru G, Marcialis MA, Cristinzio G (1995) Antibacterial polyphenols from olive oil mill waste waters. J Appl Bacteriol 79: 393–398 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coleman JOD, Blake-Kalff MMA, Davies TGE (1997) Detoxification of xenobiotics by plants chemical modification and vacuolar compartmentation. Trends Plant Sci 2: 144–151 [Google Scholar]
- Dhankher OP, Li YJ, Rosen BP, Shi J, Salt D, Senecoff JF, Sashti NA, Meagher RB (2002) Engineering tolerance and hyperaccumulation of arsenic in plants by combining arsenate reductase and gamma-glutamylcysteine synthetase expression. Nat Biotechnol 20: 1140–1145 [DOI] [PubMed] [Google Scholar]
- French CE, Rosser SJ, Davies GJ, Nicklin S, Bruce NC (1999) Biodegradation of explosives by transgenic plants expressing pentaerythritol tetranitrate reductase. Nat Biotechnol 17: 491–494 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Hannink N, Rosser SJ, French CE, Basran A, Murray JAH, Nicklin S, Bruce NC (2001) Phytodetoxification of TNT by transgenic plants expressing a bacterial nitroreductase. Nat Biotechnol 19: 1168–1172 [DOI] [PubMed] [Google Scholar]
- Harayama S, Rekik M (1989) Bacterial aromatic ring-cleavage enzymes are classified into two different gene families. J Biol Chem 264: 15328–15333 [PubMed] [Google Scholar]
- Kramer U (2005) Phytoremediation: novel approaches to cleaning up polluted soils. Curr Opin Biotechnol 16: 133–141 [DOI] [PubMed] [Google Scholar]
- Kuhm AE, Schlomann M, Knackmuss H, Pieper DH (1990) Purification and characterization of dichloromuconate cycloisomerase from Alcaligenes eutrophus JMP 134. Biochem J 266: 877–883 [PMC free article] [PubMed] [Google Scholar]
- Laemmli CM, Leveau JHJ, Zehnder AJB, van der Meer JR (2000) Characterization of a second tfd gene cluster for chlorophenol and chlorocatechol metabolism on plasmid pJP4 in Ralstonia eutropha JMP134(pJP4). J Bacteriol 182: 4165–4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo RX, Li SP (1998) Degradation characteristics of chlorobenzene by Plesiomonas sp. L1. China Environ Sci 18: 272–275 [Google Scholar]
- Ma ZH, Luo RX, Xia YF, Wang WD, Chen WJ, Kuai BK (2000) Cloning of a new catechol 1,2-dioxygenase gene (tfdC) from Plesiomonas and its expression in the E. coli. Acta Microbiol Sin 40: 580–585 [PubMed] [Google Scholar]
- Neilson AH, Allard AS, Hynning PA, Remberger M (1991) Distribution, fate and persistence of organochlorine compounds formed during production of bleached pulp. Toxicol Environ Chem 30: 3–41 [Google Scholar]
- Newman LA, Reynolds CM (2005) Bacteria and phytoremediation: new uses for endophytic bacteria in plants. Trends Biotechnol 23: 6–8 [DOI] [PubMed] [Google Scholar]
- Perez-Pantoja D, Ledger T, Pieper DH, Gonzalez B (2003) Efficient turnover of chlorocatechols is essential for growth of Ralstonia eutropha JMP134(pJP4) in 3-chlorobenzoic acid. J Bacteriol 185: 1534–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins EJ, Gordon MP, Caceres O, Lurquin PF (1990) Organization and sequence-analysis of the 2,4-dichlorophenol hydroxylase and dichlorocatechol oxidative operons of plasmid Pjp4. J Bacteriol 172: 2351–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peuke AD, Rennenberg H (2005) Phytoremediation. EMBO Rep 6: 497–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon-Smits E (2005) Phytoremediation. Annu Rev Plant Biol 56: 15–39 [DOI] [PubMed] [Google Scholar]
- Reineke W (1998) Development of hybrid strains for the mineralization of chloroaromatics by patchwork assembly. Annu Rev Microbiol 52: 287–331 [DOI] [PubMed] [Google Scholar]
- Rugh CL, Wilde HD, Stack NM, Thompson DM, Summers AO, Meagher RB (1996) Mercuric ion reduction and resistance in transgenic Arabidopsis thaliana plants expressing a modified bacterial merA gene. Proc Natl Acad Sci USA 93: 3182–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schlomann M, Schmidt E, Knackmuss H (1990) Different types of dienelactone hydrolase in 4-fluorobenzoate-utilizing bacteria. J Bacteriol 172: 5112–5118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E, Knackmuss H (1980) Chemical structure and biodegradability of halogenated aromatic compounds. Conversion of chlorinated muconic acids into maleoylacetic acid. Biochem J 192: 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweigert N, Zehnder AJB, Eggen RIL (2001) Chemical properties of catechols and their molecular modes of toxic action in cells, from microorganisms to mammals. Environ Microbiol 3: 81–91 [DOI] [PubMed] [Google Scholar]
- Shimizu M, Kimura T, Koyama T, Suzuki K, Ogawa N, Miyashita K, Sakka K, Ohmiya K (2002) Molecular breeding of transgenic rice plants expressing a bacterial chlorocatechol dioxygenase gene. Appl Environ Microbiol 68: 4061–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain JC, Nishino SF (1987) Degradation of 1,4-dichlorobenzene by a Pseudomonas sp. Appl Environ Microbiol 53: 1010–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector T (1978) Refinement of the Coomassie blue method of protein quantitation. Anal Biochem 86: 142–146 [DOI] [PubMed] [Google Scholar]
- Wang GD, Li QJ, Luo B, Chen XY (2004) Ex planta phytoremediation of trichlorophenol and phenolic allelochemicals via an engineered secretory laccase. Nat Biotechnol 22: 893–897 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.