Abstract
etr1-1 is a dominant ethylene receptor gene in Arabidopsis (Arabidopsis thaliana) and confers ethylene insensitivity. The truncated etr1-1(1-349) protein is capable of repressing ethylene responses, whereas etr1(1-349) is not, lending support to a hypothesis that the dominant etr1-1(1-349) could convert wild-type receptors to an ethylene-insensitive state. Assuming that etr1-1(1-349) and etr1(1-349) would share the same signaling mechanism, we hypothesize that the etr1(1-349) protein is capable of repressing ethylene responses when not bound with ethylene. In this study, we show that both etr1(1-349) and etr1-1(1-349) are capable of receptor signal output, which is primarily dependent on subfamily I receptors. The etr1(1-349) and etr1-1(1-349) clones were individually transformed to mutants and the resulting phenotypes were scored. Each of those transgenes restored the rosette growth and flower fertility of etr1-7 ers1-2 to a similar extent. In contrast, neither etr1(1-349) nor etr1-1(1-349) was capable of signal output in etr1-7 ers1-3. The ERS1 transcript was detectable in ers1-2 but not in ers1-3, implying that ETR1 N-terminal signaling is subfamily I dependent. Loss of the subfamily II receptor genes did not perturb etr1-1(1-349)-mediated ethylene insensitivity. Possible roles of subfamily I receptors and disulfide linkages in ETR1 receptor signal output mediated through the N terminus are discussed.
Ethylene is a simple gaseous hormone important to the regulation of plant growth and development, including seed germination, responses to pathogen and stress, fruit ripening, senescence, and abscission. Genetic studies on mutants exhibiting altered responses to ethylene in Arabidopsis (Arabidopsis thaliana) have presented a linear signal transduction pathway involving genes encoding five ethylene receptors (Chang et al., 1993; Hua et al., 1995, 1998; Sakai et al., 1998), a mitogen-activated protein kinase kinase kinase homolog CTR1(Kieber et al., 1993; Gao et al., 2003; Huang et al., 2003), a putative metal ion transporter EIN2 (Alonso et al., 1999), a subset of transcription factors including EIN3 and EILs (Chao et al., 1997; Solano et al., 1998), and the immediate targets of EIN3, ERF1 and EDFs (Alonso et al., 2003).
Other studies identify components regulating this linear signaling pathway, including two F-box proteins, a copper transporter protein, and a previously unidentified membrane protein RTE1 (Resnick et al., 2006). Accumulation of the EIN3 protein is regulated by the F-box proteins EBF1 and EBF2 via ubiquitin-mediated protein degradation (Guo and Ecker, 2003; Potuschak et al., 2003; Gagne et al., 2004). The copper transporter protein RAN1 is important to copper binding in Arabidopsis ethylene receptors (Hirayama et al., 1999), and the strong ran1-3 allele confers constitutive ethylene response and lethality (Woeste and Kieber, 2000). RTE1 may play a role in Arabidopsis ETR1 signaling. Loss-of-function rte1 mutants suppress the dominant etr1-2 mutation and phenotypically mimic the loss-of-function etr1-7 mutant. Overexpression of RTE1 leads to ethylene insensitivity, which is substantially weakened by the etr1-7 mutation (Resnick et al., 2006).
Arabidopsis ethylene receptor proteins are structurally similar to prokaryotic and yeast (Saccharomyces cerevisiae) two-component modules, which have signal input and output domains and exhibit His-kinase activity. Among the five Arabidopsis ethylene receptors, ETR1 and ERS1 have the conserved amino acid residues and signature motifs required for His-kinase activity and both belong to subfamily I receptors (Chang et al., 1993; Hua et al., 1995; Gamble et al., 1998). Subfamily II receptors, including ETR2, EIN4, and ERS2, do not carry most of those conserved residues and those signature motifs are largely missing (Hua et al., 1998; Sakai et al., 1998). His-kinase activity has been demonstrated for ETR1 and ERS1 (Gamble et al., 1998; Moussatche and Klee, 2004), whereas Ser-Thr kinase activity has been shown for ERS1, ETR2, EIN4, and ERS2 (Moussatche and Klee, 2004), regardless of their identities in receptor classification. Another distinct feature between subfamily I and II receptors is the lack of a putative signal peptide on the N terminus of subfamily I receptor proteins. Functional significance about receptor classification is unclear.
ETR1, ETR2, and EIN4 receptors are hybrid receptors on which a receiver domain follows the kinase domain, whereas the receiver domain is lacking in ERS1 and ERS2. Subfamily I receptors, ETR1 and ERS1, play a unique role in receptor signal output, and the loss-of-function mutations of both subfamily I genes result in severe constitutive ethylene response (Hall and Bleecker, 2003; Wang et al., 2003). Ectopic expression of the wild-type subfamily II receptor genes, ETR2, EIN4, and ERS2, fails to rescue the etr1-7 ers1-2 mutant phenotype, indicating that roles of subfamily I receptors cannot be replaced by subfamily II (Wang et al., 2003).
The ETR1 receptor is the most characterized ethylene receptor protein and exhibits characteristics found in the prokaryotic His-kinase, including His-kinase activity and structural similarity. Most prokaryotic His-kinases form a noncovalent homodimer through the dimerization domain in the His-kinase core upon autophosphorylation (Stock et al., 2000). The ETR1 protein may exist as a covalently linked homodimer through the disulfide linkages on Cys-4 and Cys-6, and mutations on Cys-4 and Cys-6 disrupt ETR1 dimerization (Schaller et al., 1995). Oligomerization of the prokaryotic His-kinases via noncovalent interactions has been demonstrated, and it is hypothesized that Arabidopsis ethylene receptors may also oligomerize and function as a cluster (O'Malley et al., 2005).
Roles of ETR1 His-kinase and receiver domains in ETR receptor signaling have been dissected in several studies. Although His-kinase activity has been demonstrated for the ETR1 receptor protein (Gamble et al., 1998), the mutant etr1 protein lacking His-kinase activity is able to rescue the etr1-7 ers1-2 mutant phenotype, indicating that ETR1 receptor activity can be independent of canonical His-kinase activity (Wang et al., 2003). Moreover, the ETR1 His-kinase domain appears to be dispensable because the kinase domain-lacking etr1-1(1-349) variant can cause ethylene insensitivity (Gamble et al., 2002).
Mechanisms by which the dominant etr1-1(1-349) may mediate receptor signal output have been proposed. The etr1-1(1-349) portion itself could be capable of repressing ethylene responses (Gamble et al., 2002), or etr1-1(1-349) could convert a full-length receptor to a signaling state (Gamble et al., 2002). However, evidence for heterodimerization between ETR1 and receptors of other identities is lacking. Because the ETR1 receiver forms a noncovalent dimer, noncovalent association between receptors may be important to its signaling (Muller-Dieckmann et al., 1999; Gamble et al., 2002). On the other hand, the truncated etr1(1-349) isoform is not sufficient to rescue the etr1-6 etr2-3 ein4-4 loss-of-function mutant phenotype and it is interpreted that etr1(1-349) fails to repress ethylene responses. It is thus hypothesized that the truncated etr1-1(1-349) protein alone might not be able to repress ethylene responses, but is capable of converting other wild-type receptors to a signaling state (Qu and Schaller, 2004).
Although etr1(1-349) does not rescue the etr1-6 etr2-3 ein4-4 mutant phenotype, it does not exclude that etr1(1-349) would be capable of receptor signal output. In other words, in the air, etr1(1-349) might still be capable of receptor signal output, but it might not be sufficient to compensate for the triple mutations, assuming etr1(1-349) and etr1-1(1-349) adopt the same signaling mechanism. Alternatively, the dominant etr1-1(1-349) might acquire a novel signaling mechanism that is not adopted by etr1(1-349). To dissect ethylene receptor signaling, examining etr1(1-349)-mediated signaling and its dependence on wild-type receptors will be essential.
In this study, we examined the effects of loss of wild-type receptors on ETR1 N-terminal signaling to elucidate possible mechanisms by which the ETR1 N terminus may mediate receptor signal. Because only subfamily I receptor genes can rescue the subfamily I null mutant phenotype (Wang et al., 2003), identity of the subfamily I receptor signal would be different from that of subfamily II. Thus, examining the identity of ETR1 N-terminal signaling would help the study of how its signal is mediated to repression of ethylene responses. Our data show that etr1(1-349) is capable of repressing ethylene responses and that the etr1(1-349)- and etr1-1(1-349)-mediated signaling is primarily subfamily I receptor dependent. Besides, loss of disulfide linkages does not abolish ETR1 receptor signal output. Possible roles of wild-type subfamily I receptors and covalent interaction in ETR1 N-terminal-mediated signaling are discussed.
RESULTS
etr1(1-349) Has Minor Effects on the Growth of etr1-7 etr2-3 ein4-4
It has been interpreted that etr1(1-349) is not sufficient to repress ethylene responses because it fails to rescue the etr1-6 etr2-3 ein4-4 mutant phenotype (Qu and Schaller, 2004). Because the growth of etr1-6 etr2-3 ein4-4 can be restored by ETR1 to wild type-like, it is likely that ETR1 also compensates for ETR2 and EIN4 signaling in the triple mutant. Thus, it does not rule out that etr1(1-349) could be able to repress ethylene responses but unable to compensate for the etr2-3 and ein4-4 mutations.
In an effort to examine whether etr1(1-349) can signal, the etr1(1-349) clone was transformed to etr1-7 etr2-3 ein4-4. Among 28 individual transformation lines, nine were randomly picked and characterized. Phenotypes of etr1-7 etr2-3, etr1-7 ein4-4, etr2-3 ein4-4, and etr1-7 etr2-3 ein4-4 were compared with the resulting transformants. etr1-7 etr2-3 ein4-4 had the shortest primary root and hypocotyl among those mutants. Hypocotyl lengths of etr1-7 etr2-3 and etr2-3 ein4-4 were not statistically different (P > 0.05) and slightly longer than that of etr1-7 ein4-4 by 1.0 ± 0.44 mm (for a 95% confidence interval). In this experiment, hypocotyl lengths of the etr2-3 ein4-4 and etr1-7 etr2-3 ein4-4 seedlings were compared with that of T:etr1(1-349) etr1-7 etr2-3 ein4-4. If etr1(1-349) could complement the etr1-7 mutation, the resulting transformed etr1-7 etr2-3 ein4-4 would be phenotypically similar to etr2-3 ein4-4.
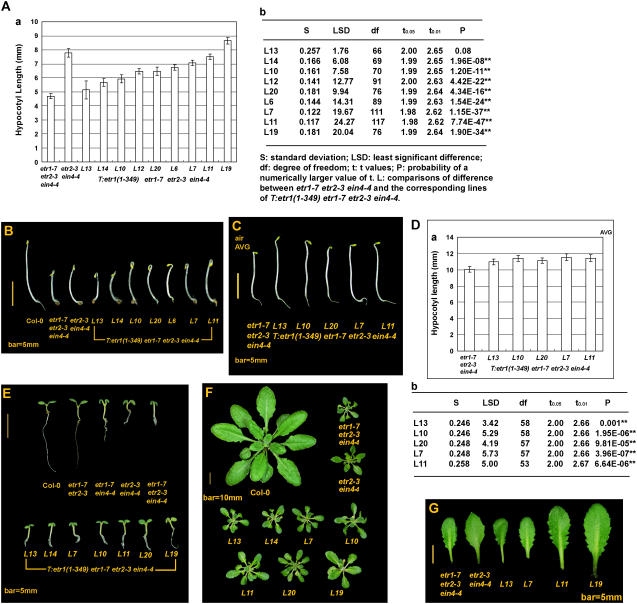
etr1-7 etr2-3 ein4-4 exhibited a constitutive seedling triple-response phenotype when germinated in air (Hua and Meyerowitz, 1998). Eight of nine independent transformation lines were statistically longer than etr1-7 etr2-3 ein4-4 (P < 0.05) and seven were shorter than etr2-3 ein4-4 (P < 0.05; Fig. 1, A, a, and B). One line (line 11) had a hypocotyl length not different from that of etr2-3 ein4-4 (P > 0.05); another line (line 19) was longer (P < 0.01). None of those transformants exhibited ethylene insensitivity (data not shown). The hypocotyl length measurements were next subjected to AVOVA followed by lsd for pairwise comparisons. The F value was 102.54 > F(0.05) = 1.91. Consistent with the t test, those transformants were longer than etr1-7 etr2-3 ein4-4 as analyzed by lsd, except for line 13 (Fig. 1A, b).
Figure 1.
Phenotype and hypocotyl measurement of the etr1(1-349)-transformed etr1-7 etr2-3 ein4-4. A, Hypocotyl measurements of nine individual lines (a). Analyzed by lsd, eight individual lines are longer than etr1-7 etr2-3 ein4-4 (b). B, Phenotype of dark-grown seedlings of etr2-3 ein4-4, etr1-7 etr2-3 ein4-4, wild-type, and transformation lines. C, Phenotype of dark-grown seedlings germinated in the presence of AVG. D, Measurements (a) and comparisons (b) of hypocotyl lengths of transformation lines treated with AVG. E, Phenotype of light-grown transformation lines in comparison to mutants defective in multiple receptor genes. F, Adult phenotypes of etr1-7 etr2-3 ein4-4 and transformation lines in comparison to that of the wild type and etr2-3 ein4-4. G, Leaf morphology of the etr1(1-349)-transformed etr1-7 etr2-3 ein4-4 is identical to that of the etr1-7 etr2-3 ein4-4 mutant, but larger. Error bar indicates the 95% interval of a mean. Sizes of the scale bars are as indicated.
Root length of five etr1(1-349)-transformed etr1-7 etr2-3 ein4-4 lines was examined and compared by ANOVA with untransformed lines. The F value was 1.05 < F(0.05) = 3.11 (P > 0.05). This result indicates that there is no difference in root growth between transformed and untransformed etr1-7 etr2-3 ein4-4.
Being statistically different, we next calculated the extent of hypocotyl growth restored by the etr1(1-349) transgene. For the 95% confidence interval, those eight independent transformation lines were 0.98 ± 0.86 to 3.97 ± 0.84 mm longer than etr1-7 etr2-3 ein4-4 (see Supplemental Table S1a). As a comparison, etr2-3 ein4-4 was 3.09 ± 1.30 mm longer than etr1-7 etr2-3 ein4-4 (½α = 0.025).
Because the endogenous ethylene production of etr1-7 etr2-3 ein4-4 and those transformation lines could affect seedling phenotypes, the ethylene biosynthesis inhibitor l-α-(2-aminoethoxyvinyl)Gly (AVG) was next included to block endogenous ethylene production. As a control, the ethylene-overproducing mutant eto1-1 was treated with AVG and the seedling was long (data not shown), indicating that endogenous ethylene biosynthesis is blocked by the AVG used in this experiment.
The AVG-treated etr1-7 etr2-3 ein4-4 and transformation lines exhibited a longer seedling hypocotyl and primary root (Fig. 1C). Five individual transformation lines scored (Fig. 1, C and D) were all longer than the AVG-treated etr1-7 etr2-3 ein4-4 in a range of 0.91 ± 0.49 to 1.48 ± 0.52 mm (½α = 0.025; see Supplemental Table S1b). Analyzed by ANOVA and lsd, those five transformation lines were all longer than the untransformed mutant after AVG treatment (Fig. 1D, b; P < 0.01), consistent with t test. These results indicate that hypocotyl elongation of etr1(1-349)-transformed etr1-7 etr2-3 ein4-4 was caused by the transgene, but not affected by endogenous ethylene production.
Although our data suggest that etr1(1-349) has minor effects on hypocotyl elongation in etr1-7 etr2-3 ein4-4, it did not imply which mutations could be complemented. We next compared phenotypes of light-grown seedlings of etr1-7 ein4-4, etr2-3 ein4-4, and etr1-7 etr2-3 (Fig. 1E). The light-germinated etr1-7 etr2-3 ein4-4 seedling carried small and epinastic cotyledons and had the shortest primary root. The etr1-7 ein4-4 seedling was phenotypically similar to etr1-7 etr2-3 ein4-4, but its primary root was longer. In comparison to etr1-7 etr2-3 ein4-4, the etr2-3 ein4-4 seedling carried a longer primary root and the cotyledons were larger and less epinastic. In contrast to those double mutants, etr1-7 etr2-3 was phenotypically similar to the wild-type seedling with well-expanded and developed cotyledons and an elongated primary root.
The etr1(1-349) transgene partially restored the seedling growth of etr1-7 etr2-3 ein4-4 to various extents in those light-grown seedlings. Those transformation lines carried expanded and less epinastic cotyledons and had a longer primary root than etr1-7 etr2-3 ein4-4. Some individual lines were not visibly distinguishable from the etr2-3 ein4-4 seedlings (Fig. 1E).
When grown in soil, etr1-7 etr2-3 ein4-4, etr2-3 ein4-4, and those transformation lines were initially small and indistinguishable. Over time, etr2-3 ein4-4 and those transformation lines became larger than etr1-7 etr2-3 ein4-4. However, rosette sizes of those transformants were still much smaller than the wild type (Fig. 1F). The rosette leaf of etr2-3 ein4-4 was sharp at the leaf tip, whereas etr1-7 etr2-3 ein4-4 had an oval leaf tip. The leaf shape of those transformation lines was phenotypically similar to that of etr1-7 etr2-3 ein4-4, but larger (Fig. 1G). The floral phenotypes of etr1-7 etr2-3 ein4-4, characteristic of a protruding pistil, were identical to those transformation lines (data not shown).
These results indicate that the etr1(1-349) transgene does not rescue the etr1-7 mutation in the triple-mutant background nor compensate for other mutations, but has minor effects on the growth recovery of etr1-7 etr2-3 ein4-4.
etr1-7 ers1-2 Growth and Fertility Are Restored by etr1(1-349) and etr1-1(1-349)
Our data suggest that etr1(1-349) partially restored the growth of etr1-7 etr2-3 ein4-4, but did not complement any mutations nor cause morphological changes. To further verify whether etr1(1-349) represses ethylene responses in the air, we next examined etr1(1-349) signaling in etr1-7 ers1-2, a mutant severely defective in growth and fertility (Hall and Bleecker, 2003; Wang et al., 2003).
etr1(1-349) and etr1-1(1-349) clones were individually transformed to etr1-7/etr1-7 ers1-2/+ and etr1-7 ers1-2 homozygous transformants were obtained in the primary (T1) and the following (T2 and higher) generations. Stable transformants were obtained and repeatedly analyzed in the T4 and T5 generations. The etr1-7 transformant was also obtained due to segregation of the ers1-2 allele.
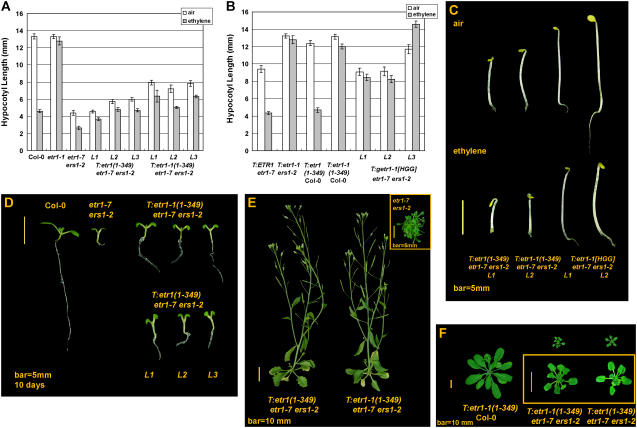
Figure 2, A and C, shows the measurement and phenotype of the dark-grown etr1-7 ers1-2 seedling carrying etr1(1-349) or etr1-1(1-349). The etr1(1-349)-transformed seedling was longer than etr1-7 ers1-2 in a range of 1.32 ± 0.42 to 1.55 ± 0.37 mm (½α = 0.025) in the air. The etr1-1(1-349)-transformed etr1-7 ers1-2 seedling was 2.91 ± 0.45 to 3.52 ± 0.42 mm longer than etr1-7 ers1-2 in air (½α = 0.025). When germinated in ethylene, the etr1-1(1-349)-transformed etr1-7 ers1-2 was longer than etr1-7 ers1-2 in a range of 2.85 ± 0.22 to 4.25 ± 0.35 mm (½α = 0.025). Ethylene treatment caused a shortening of the etr1-1(1-349)-transformed etr1-7 ers1-2 seedling in a range of 1.15 ± 0.25 to 2.19 ± 0.48 mm (½α = 0.025; see Supplemental Table S2a). For three independent lines scored, the ethylene-grown T:etr1-1(1-349) etr1-7 seedlings were 9.2 ± 0.16, 7.16 ± 0.18, and 7.50 ± 0.26 mm in hypocotyl length (α = 0.05), shorter than the air-grown seedlings by 2.39 ± 0.46 to 4.73 ± 0.28 mm (½α = 0.025; >65 degrees of freedom [df]), suggesting ethylene responses in the absence of ETR1.
Figure 2.
Phenotypes and hypocotyl measurements of etr1(1-349)- and etr1-1(1-349)-transformed etr1-7 ers1-2. A, Hypocotyl lengths of the etr1(1-349)- and etr1-1(1-349)-transformed etr1-7 ers1-2 seedlings are longer than the untransformed seedlings. B, etr1-1(1-349) confers strong ethylene insensitivity in Col-0. getr1-1[HGG] rescues the etr1-7 ers1-2 mutant phenotype. ETR1 and etr1(1-349) do not cause ethylene insensitivity. C, Seedling phenotypes of dark-grown transformants. D, Phenotypes of light-grown seedlings. etr1(1-349)- and etr1-1(1-349)-transformed etr1-7 ers1-2 seedlings are larger than etr1-7 ers1-2. E, Rosette phenotypes at the 7-week stage; the transformants are substantially larger than the untransformed mutant. F, Rosette phenotypes at the 4-week stage. Error bar indicates the 95% confidence of a mean.
As comparisons, the full-length etr1-1, ETR1, and getr1-1[HGG] clones, of which getr1-1[HGG] encodes a kinase-dead etr1-1 isoform, were individually transformed to etr1-7 ers1-2. The seedling hypocotyl measurements and phenotype are shown (Fig. 2, B and C). The etr1-1-transformed etr1-7 ers1-2 seedling was ethylene insensitive and carried a long hypocotyl. The ETR1 transgene rescued the etr1-7 ers1-2 mutant phenotype and the seedling was long in the air but short in ethylene. Measured from three independent lines, getr1-1[HGG] rescued the etr1-7 ers1-2 mutant phenotype and conferred ethylene insensitivity. Ectopic expression of the ETR1 and etr1(1-349) genes did not lead to ethylene insensitivity.
When germinated under light, the etr1-7 ers1-2 mutant carried small and compact cotyledons and its hypocotyl and primary root were short. With the etr1-1(1-349) or etr1(1-349) transgene, the etr1-7 ers1-2 seedling phenotype was partially rescued and the cotyledons became larger and expanded. Its hypocotyl and primary root became longer. There was little visible difference between the etr1(1-349)- and etr1-1(1-349)-transformed etr1-7 ers1-2 seedlings, except that the latter was longer in the primary root (Fig. 2D).
Adult phenotypes of those transformants were also examined (Fig. 2, E and F). The rosette of the etr1-7 ers1-2 mutant is small and compact and the flower is sterile (Hall and Bleecker, 2003; Wang et al., 2003). The etr1-7 ers1-2 mutant was initially indistinguishable from the etr1(1-349)- or etr1-1(1-349)-transformed etr1-7 ers1-2 (data not shown). Over time, transformation lines gradually became larger than etr1-7 ers1-2 in rosette size and the flower was fertile. Figure 2E shows that there was no visible difference between the etr1(1-349)- and etr1-1(1-349)-transformed etr1-7 ers1-2 7 weeks after germination. At this developmental stage, those transformants were even larger than ctr1-1 (data not shown). In contrast, the 7-week-old etr1-7 ers1-2 was still small and compact in rosette leaves (Fig. 2E).
These data indicate both etr1(1-349) and etr1-1(1-349) are capable of repressing ethylene responses and partially restoring the etr1-7 ers1-2 growth. For N-terminal signaling, His-kinase activity can be dispensable, but the kinase domain is important.
Analyses of the ers1-2 and ers1-3 Alleles
Both etr1(1-349) and etr1-1(1-349) were capable of repressing ethylene responses in etr1-7 ers1-2. The N terminus could itself repress ethylene responses or be dependent on subfamily II receptors. To test these hypotheses, it is important to examine whether etr1-7 ers1-2 is a null mutant and whether the subfamily II triple mutations would mask ETR1 N-terminal signaling.
The ers1-2 mutation is once demonstrated to give rise to a mosaic transcript consisting of the ERS1 and T-DNA sequences (Wang et al., 2003). Northern-blot analysis detects an extremely weak hybridization signal in ers1-2 (Zhao et al., 2002), but it is not known whether it is correctly spliced and polyadenylated. We hypothesized that the ers1-2 allele is not a null and that etr1(1-349)/etr1-1(1-349) could signal in the presence of the remaining ERS1 protein. In this study, we analyzed ers1-2 and another T-DNA insertional mutant, ers1-3 (G.E. Schaller, unpublished data).
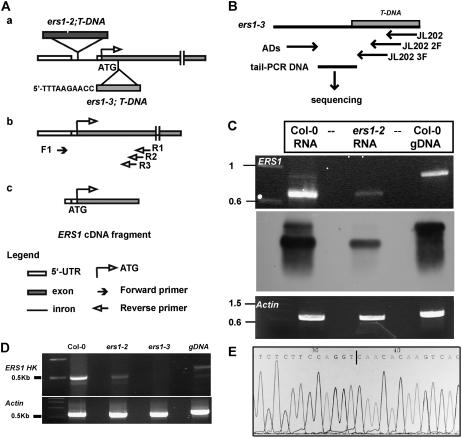
Figure 3A depicts reverse transcription (RT)-PCR analysis of the ERS1 transcript across the T-DNA insertional site in ers1-2. The wild-type ERS1 transcript was detectable by RT-PCR in the wild type and ers1-2. The RT-PCR product was then subjected to Southern hybridization and sequencing. Southern hybridization detected the ERS1 fragment (Fig. 3C) and sequence analysis (Fig. 3E) showed that the 5′-ERS1 transcript of ers1-2 was identical to that of wild-type ERS1, indicating that the intron and T-DNA sequences are correctly spliced. These results suggest that ers1-2 may have the wild-type ERS1 transcript.
Figure 3.
Analyses of the ers1-2 and ers1-3 mutations. A, Schematic illustration of the ers1-2 and ers1-3 mutations and RT-PCR detection for the ERS1 transcript. ers1-2 mutation and flanking sequence of the T-DNA insertion site in ers1-3 are as indicated (a). Correctly spliced ERS1 transcript and primers used for RT-PCR are shown (b). Expected RT-PCR fragment generated from the ERS1 mRNA is shown (c). B, TAIL-PCR analysis of the T-DNA flanking sequence in ers1-3. C, RT-PCR and Southern hybridization analyses detect the wild-type ERS1 transcript across the T-DNA insertion site in ers1-2. D, RT-PCR analysis does not detect the polyadenylated ERS1 transcript in ers1-3. E, Sequence of the ERS1 transcript across the T-DNA insertion site in ers1-2. Vertical line indicates the T-DNA insertion site. gDNA, Genomic DNA used as a template for PCR. The actin transcript was an internal control for RT-PCR.
The flanking sequence of the T-DNA insertion site in ers1-3 was verified by thermal asymmetric interlaced (TAIL)-PCR (Fig. 3B), which showed that T-DNA interrupts the second exon of ERS1 (Fig. 3A). This result was further confirmed by direct sequencing of the PCR product generated from sequence-specific primers on T-DNA and ERS1 (data not shown).
We next examined the existence of the polyadenylated ERS1 transcript in ers1-2 and ers1-3. When oligo(dT)20 was primed for RT, RT-PCR amplified the ERS1 transcript from the RNA isolated from ers1-2, but not from ers1-3 (Fig. 3D). RNA isolated from the wild type gave a stronger RT-PCR amplification than from ers1-2. Using wild-type genomic DNA as a control, the PCR product was larger due to the intron sequence. This result indicates that the RT-PCR fragment was amplified from the ERS1 transcript, but not from genomic DNA. An internal control for RT-PCR analysis was included and amplified the actin transcript both in ers1-2 and ers1-3 (Fig. 3, C and D). These results suggest that ers1-3 does not have a polyadenylated ERS1 transcript and is a strong allele, whereas ers1-2 is leaky.
Signaling of etr1(1-349)/etr1-1(1-349) Is Masked in etr1-7 ers1-3
Our results showed that the ers1-2 mutation is leaky, and signaling of etr1(1-349) and etr1-1(1-349) in etr1-7 ers1-2 could be dependent on the remaining ERS1. Effects of the loss-of-function mutations of both subfamily I genes on ETR1 N-terminal signaling were next examined in etr1-7 ers1-3.
T:etr1(1-349) etr1-7, a sibling of the etr1(1-349)-rescued etr1-7 ers1-2 transformant derived from the same T:etr1(1-349) etr1-7/etr1-7 ers1-2/+ parent, was genetically crossed with ers1-3. T:etr1(1-349) etr1-7 ers1-3 individuals were identified among F2 and F3 progeny. T:etr1-1(1-349) etr1-7 ers1-3 plants were obtained by a genetic cross of T:etr1-1(1-349) etr1-7 and ers1-3 in which T:etr1-1(1-349) etr1-7 exhibited ethylene insensitivity.
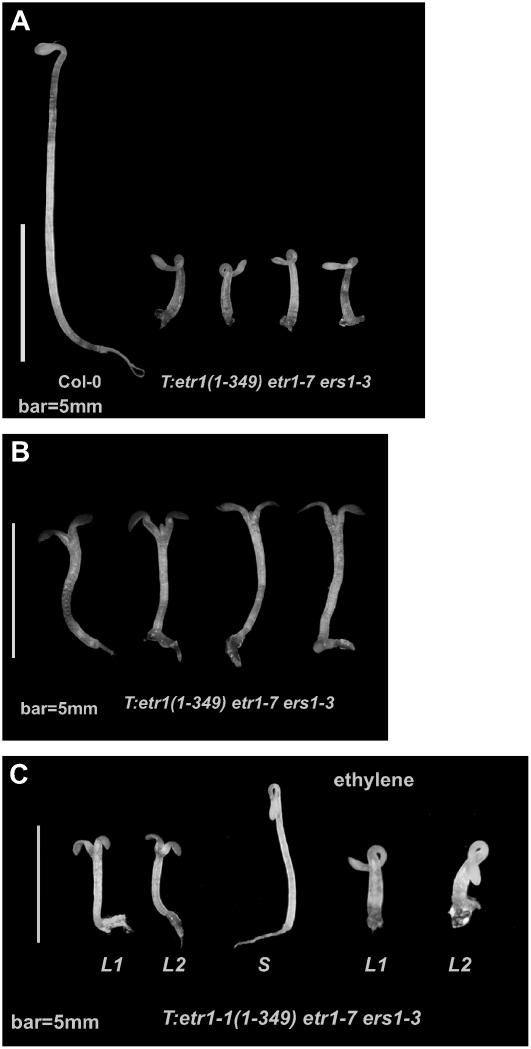
When germinated in the dark, T:etr1(1-349) etr1-7 ers1-3 was short (2.42 ± 0.41 mm; α = 0.05) and exhibited little primary root growth (Fig. 4A). This was similar to the etr1-7 ers1-3 mutant (2.60 ± 0.32 mm; α = 0.05) and there was no difference in their seedling hypocotyl lengths (P > 0.05; 44 df). Two T:etr1-1(1-349) etr1-7 ers1-3 lines were examined (Fig. 4C) and the hypocotyl lengths of the dark-grown seedlings (2.75 ± 0.20 and 2.84 ± 0.16 mm; α = 0.05) were statistically the same as that of etr1-7 ers1-3 (P > 0.01; 49 and 51 df for these two lines). Analyzed by ANOVA, there was no statistical difference in the hypocotyl length of etr1-7 ers1-3, T:etr1-1(1-349) etr1-7 ers1-3, and T:etr1(1-349) etr1-7 ers1-3; the F value was 2.33 < F(0.05) = 3.12 (P > 0.05). These data suggest that neither the etr1-1(1-349) nor the etr1(1-349) transgene was able to alter etr1-7 ers1-3 hypocotyl growth.
Figure 4.
Phenotype of the etr1(1-349)- and etr1-1(1-349)-transformed etr1-7 ers1-3 mutant. Seedling phenotypes of light-grown (A) and dark-grown (B) T:etr1(1-349) etr1-7 ers1-3. C, Phenotypes of light-grown (left) and dark-grown (right) T:etr1-1(1-349) etr1-7 ers1-3 seedlings. S, Sibling of T:etr1-1(1-349) etr1-7 ers1-3 segregated from the same T:etr1-1(1-349) etr1-7 ers1-3/+ parent.
Light-grown T:etr1(1-349) etr1-7 ers1-3 and T:etr1-1(1-349) etr1-7 ers1-3 seedlings were phenotypically identical to etr1-7 ers1-3; they carried a short hypocotyl and the cotyledons were small and epinastic (Fig. 4, B and C). The etr1-7 ers1-3 rosette was much smaller and shorter than etr1-7 ers1-2; it exhibited early senescence and only carried a few small and underdeveloped leaves (data not shown). The adult transformants carrying etr1(1-349) or etr1-1(1-349) exhibited the etr1-7 ers1-3 adult phenotype (data not shown).
The T:etr1-1(1-349) etr1-7 ers1-3/+ sibling was partially insensitive to ethylene (Fig. 4C) and etr1(1-349) and etr1-1(1-349) rescued the etr1-7 ers1-2 siblings, suggesting that the transgenes were functional in the etr1-7 ers1-2 isogenic background but not in etr1-7 ers1-3.
Ethylene Insensitivity Conferred by etr1-1(1-349) Can Be Subfamily II Independent
Essential roles of subfamily I receptors in ETR1 N-terminal signaling were shown in our study. We next examined the roles of subfamily II receptors in ETR1 N-terminal signaling.
The etr1-1(1-349) clone was transformed to etr2-3 ein4-4 ers2-3, which lacks wild-type subfamily II receptors. Dominance and ethylene insensitivity caused by etr1-1(1-349) was scored based on the seedling triple-response assay and adult phenotype. etr1(1-349) signaling was not studied because it is not dominant and may not compensate for any mutation.
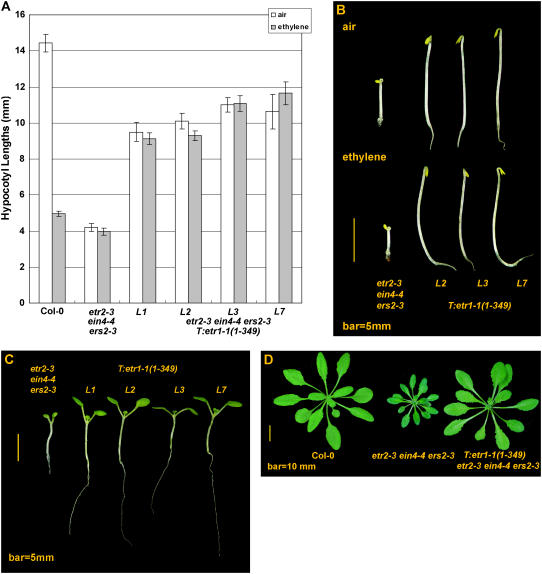
etr2-3 ein4-4 ers2-3 seedlings exhibited constitutive ethylene response in the air (Hua and Meyerowitz, 1998). Scored from four independent transformation lines, the etr1-1(1-349) transgene conferred ethylene insensitivity and restored hypocotyl length of the ethylene-grown etr2-3 ein4-4 ers2-3 seedling (Fig. 5, A and B). Those transformants were longer than etr2-3 ein4-4 ers2-3 in a range of 5.16 ± 0.40 to 7.11 ± 0.5 mm (½α = 0.025; see Supplemental Table S3) in ethylene. Grown in air, hypocotyl lengths were longer than the untransformed lines in a range of 5.52 ± 0.58 mm to 7.05 ± 0.45 mm (½α = 0.025; Fig. 5A; see Supplemental Table S3). Seedlings of a same line showed no statistical difference in hypocotyl length when germinated in air and ethylene (P > 0.05), except for one line that was merely 0.81 ± 0.52 mm shorter in ethylene (½α = 0.025; 58 df), suggesting that the mutation background has little effect on etr1-1(1-349)-mediated ethylene insensitivity. Although the constitutive ethylene response phenotype of etr2-3 ein4-4 ers2-3 was rescued by the dominant etr1-1(1-349), those transformation lines were shorter than the wild type when grown in the air (P < 0.05; see Supplemental Table S3).
Figure 5.
Subfamily II triple mutations have little effect on etr1-1(1-349)-mediated ethylene insensitivity. A, Hypocotyl measurements of etr1-1(1-349)-transformed etr2-3 ein4-4 ers2-3 lines. B, Phenotypes of dark-grown seedlings. C, Light-grown transformants resemble wild type. D, etr1-1(1-349) rescues the etr2-3 ein4-4 ers2-3 mutant phenotype but does not restore growth comparable to that of the wild type. Error bar indicates the 95% confidence interval of a mean.
The light-grown etr2-3 ein4-4 ers2-3 seedling had small cotyledons and was short in hypocotyl length and primary root. In four independent transformation lines examined, etr1-1(1-349) rescued the seedling phenotype and both the hypocotyl and primary root were long (Fig. 5C). The adult phenotype of etr2-3 ein4-4 ers2-3 was rescued by etr1-1(1-349) and resembled the wild type, but smaller in rosette size (Fig. 5D).
These results suggest that ethylene insensitivity conferred by etr1-1(1-349) is not altered in the absence of subfamily II receptors.
Loss of Subfamily I Receptors Masks the etr1(1-349)/etr1-1(1-349) Signaling Elevated by Ag(I)
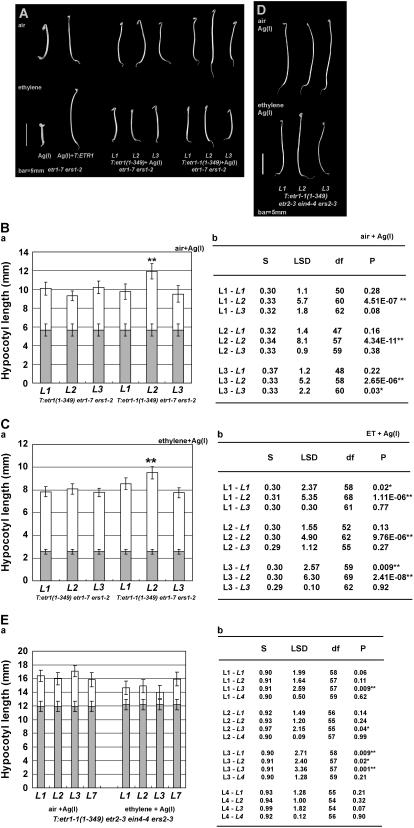
Being capable of repressing ethylene responses, the ETR1 receptor signal could be initiated and mediated through the N terminus. The silver ion Ag(I) has been demonstrated to bind ETR1 and cause ethylene insensitivity (Rodriguez et al., 1999). We next examined whether Ag(I) would elevate receptor signal output through the ETR1 N terminus and possible roles of wild-type receptors in ETR1 N-terminal signaling induced by Ag(I).
Silver nitrate caused different degrees of repression of ethylene responses in mutants lacking subfamily I and subfamily II receptors (see Supplemental Fig. S2). Loss of subfamily II had little effect on Ag(I)-induced ethylene insensitivity (Cancel and Larsen, 2002), whereas Ag(I) only restored the hypocotyl growth of etr1-7 ers1-2 by 1.24 ± 0.63 mm.
Germinated in air with silver nitrate, for three individual transformation lines scored, T:etr1(1-349) etr1-7 ers1-2 was 3.65 ± 0.54 to 4.55 ± 0.66 mm longer than etr1-7 ers1-2, and T:etr1-1(1-349) etr1-7 ers1-2 was 3.84 ± 0.88 to 6.27 ± 0.84 mm longer (½α = 0.025; >41 df; Fig. 6, A and B). ANOVA indicated that etr1(1-349)- and etr1-1(1-349)-transformed lines were different in hypocotyl lengths [F = 16.57 and F(0.05) = 2.27]. Analyzed by lsd, except for line 2 of T:etr1-1(1-349) etr1-7 ers1-2, which was itself longer than others, there was no difference between etr1(1-349)- and etr1-1(1-349)-transformed etr1-7 ers1-2 lines (α = 0.01; Fig. 6B, b). When subfamily II receptors are lacking, for four individual lines scored, T:etr1-1(1-349) etr2-3 ein4-4 ers2-3 was longer than etr2-3 ein4-4 ers2-3 by 3.91 ± 1.02 to 5.16 ± 0.84 mm in the presence of Ag(I) (Fig. 6, D and E; ½α = 0.025; >54 df). These results indicate that, in air, Ag(I) treatment induces hypocotyl elongation in those mutants and transformation lines and that both etr1(1-349) and etr1-1(1-349) are Ag(I) responsive.
Figure 6.
Receptor signal output mediated by the ETR1 N terminus is elevated by Ag(I). A, Seedling phenotype of the etr1(1-349)- and etr1-1(1-349)-transformed etr1-7 ers1-2 lines germinated in the presence of silver nitrate. B and C, Measurement and comparison of the seedling hypocotyl length of transformation lines. Hypocotyl length of seedling germinated in air with Ag(I) treatment (B, a) and in ethylene with Ag(I) treatment (C, a) is shown. lsd analyses for seedlings germinated in air with Ag(I) (B, b) and in ethylene with silver nitrate (C, b) are shown. D, Seedling phenotype of etr1-1(1-349)-transformed et2-3 ein4-4 ers2-3 lines germinated in the presence of silver nitrate. E, Hypocotyl measurement of etr1-1(1-349)-transformed etr2-3 ein4-4 ers2-3 lines (a), and comparison of seedling hypocotyl length of transformation lines germinated in air and ethylene with Ag(I) treatment (b). For lsd analyses in B and C, transformation lines carrying etr1(1-349) are represented as L and those carrying etr1-1(1-349) are represented as L. For lsd in E, air-grown lines are represented as L and ethylene-grown lines are represented as L. Ln-Ln indicates a paired comparison. When a paired comparison is statistically highly significant (P ≤ 0.01), it is marked with double asterisks. Gray bars, Hypocotyl lengths of the untransformed and Ag(I)-treated lines; white bars, amount of hypocotyl elongation caused by a transgene and Ag(I).
Germinated in ethylene, the T:etr1(1-349) etr1-7 ers1-2 seedling was longer than etr1-7 ers1-2 by 5.22 ± 0.38 to 5.53 ± 0.46 mm (½α = 0.025; >55 df), and the T:etr1-1(1-349) etr1-7 ers1-2 seedling was 5.19 ± 0.44 to 6.95 ± 0.54 mm longer than etr1-7 ers1-2 (Fig. 6, A and C). Analyzed by lsd, etr1(1-349)- and etr1-1(1-349)-transformed etr1-7 ers1-2 lines were similar in hypocotyl length, except for line 2 of etr1-1(1-349) etr1-7 ers1-2 (α = 0.01; Fig. 6C, b). The T:etr1-1(1-349) etr2-3 ein4-4 ers2-3 lines were 1.8 ± 0.91 to 3.71 ± 1.02 mm longer than etr2-3 ein4-4 ers2-3 in the presence of Ag(I) and ethylene (Fig. 6, D and E; ½α = 0.025; >54 df), and they were not shorter than the Ag(I)-treated wild type (data not shown). Hypocotyl lengths of the majority of Ag(I)-treated T:etr1-1(1-349) etr2-3 ein4-4 ers2-3 lines were not altered by ethylene treatment, as analyzed by lsd (Fig. 6E; α = 0.01).
These results indicate that the etr1-1/etr1 N terminus is Ag(I) responsive and that Ag(I) has a stronger effect on hypocotyl elongation than the etr1-1 mutation. Although Ag(I) treatment elevates ETR1 N-terminal signaling, transformation lines are ethylene responsive in the absence of subfamily I receptors (see Supplemental Table S2b). In contrast, loss of subfamily II receptors has little effect on Ag(I)-induced ethylene insensitivity.
Effects of subfamily I receptors on ETR1 N-terminal signaling induced by Ag(I) were further examined in etr1-7 ers1-3. For those lines obtained from the genetic cross, T:etr1(1-349) etr1-7 ers-3 and T:etr1-1(1-349) etr1-7 ers1-3 were not Ag(I) responsive (data not shown). Analyzed by one-way ANOVA, seedling hypocotyl lengths of the Ag(I)-treated and nontreated etr1-7 ers-3 and T:etr1-1(1-349) etr1-7 es1-3 were all statistically identical [F = 2.28 < F(0.05) = 2.46; P > 0.05]. The measurement was next analyzed by two-way ANOVA. Neither Ag(I) nor etr1-1(1-349) alone was able to alter the seedling hypocotyl length of etr1-7 ers1-3 (P > 0.05; detailed data not shown), and combination of Ag(I) treatment and etr1-1(1-349) had no effect on hypocotyl growth [F = 0.60 < F(0.05) = 4.15; P > 0.05]. These data indicate that Ag(I)-induced repression of ethylene responses is subfamily I dependent.
Receptor Signaling Mediated by etr1-1(1-349) Can Be Noncovalent
It is hypothesized that etr1-1(1-349) signal output could be mediated by itself through covalent dimerization with ETR1 or through noncovalent interactions with other receptors (Gamble et al., 2002). To further study how the ETR1 N terminus would repress ethylene responses, we next explored these possibilities by preventing the disulfide bonds and examining the signaling of various etr1 variants.
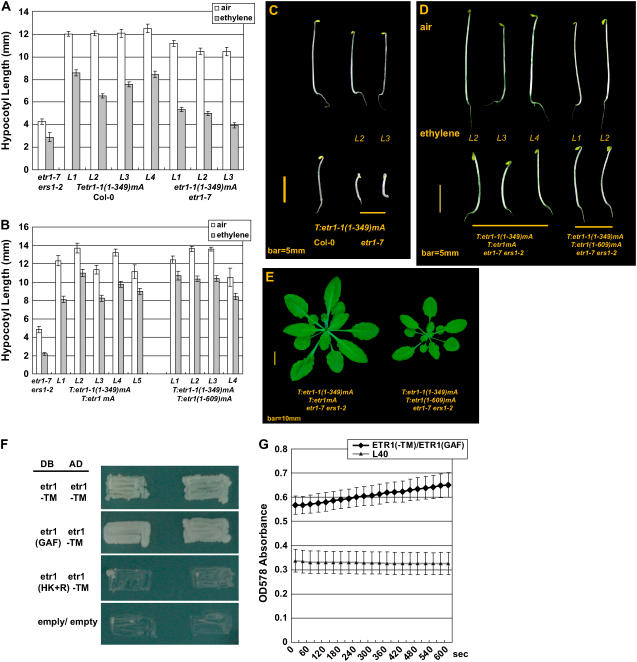
ETR1 and etr1-1 clones were mutated to etr1mA, etr1(1-609)mA, and etr1-1(1-349)mA, of which each encodes an etr1 variant whose disulfide-forming residues, Cys-4 and Cys-6, were replaced with Ala. Each of those clones was transformed to the wild type and etr1-7 ers1-2 and receptor signaling was scored based on the seedling and adult phenotypes. Besides, etr1-1(1-349)mA and etr1mA were coexpressed in etr1-7 ers1-2 for analysis of possible noncovalent receptor signal output.
Figure 7, A and C, shows the phenotype and hypocotyl measurement of dark-grown seedlings of the etr1-1(1-349)mA-transformed wild type and etr1-7. Ethylene treatment resulted in shortening of the seedling hypocotyl in those transformants (P < 0.05; see Supplemental Table S4a). For four individual lines scored, the etr1-1(1-349)mA-transformed wild type was longer than the untransformed seedling (4.96 ± 0.16 mm; α = 0.05) by 1.59 ± 0.30, 2.60 ± 0.35, 3.49 ± 0.33, and 3.64 ± 0.33 mm (½α = 0.025) when germinated in ethylene. In ethylene, the etr1-1(1-349)mA-transformed etr1-7 was longer than etr1-7 (3.48 ± 0.15 mm; α = 0.05) by 1.85 ± 0.24, 1.53 ± 0.20, and 0.47 ± 0.26 mm for three independent lines examined (½α = 0.025), and one line was not statistically different from etr1-7 (P > 0.05). T:etr1-1(1-349)mA etr1-7 lines were not statistically longer than the wild type when treated with ethylene (data not shown). etr1-1(1-349)mA did not rescue the seedling and rosette phenotypes of the etr1-7 ers1-2 mutant and the flower was sterile in six independent transformation lines examined (data not shown). The etr1mA and etr1(1-609)mA transgenes rescued the etr1-7 ers1-2 adult and seedling phenotypes, and did not confer ethylene insensitivity (see Supplemental Fig. S1A). Phenotypes of those adult transformants are shown (see Supplemental Fig. S1B). These data indicate that, in the presence of subfamily I receptors, these disulfide-free etr1 variants are capable of repressing ethylene responses without covalent dimerization.
Figure 7.
etr1mA and etr1(1-609)mA elevate the signaling mediated by etr1-1(1-349)mA in etr1-7 ers1-2. A and C, Hypocotyl lengths (A) and seedling phenotypes (C) of seedlings expressing the disulfide-deficient etr1-1(1-349)mA variant. B and D, Hypocotyl lengths (B) and seedling phenotypes (D) of seedlings coexpressing etr1-1(1-349)mA and etr1mA or etr1(1-609)mA in etr1-7 ers1-2. E, Adult phenotypes of the coexpression lines. F and G, Yeast cell growth on selection medium (F) and enzyme kinetics of the reporter protein β-galactosidase (G) in a yeast two-hybrid assay. DB, DNA-binding domain fusions (lexA); AD, activation domain fusions (GAL4); etr1(HK+R), etr1(293-729) fragment. Error bar, 95% confidence interval of a mean.
Possible noncovalent interactions between ETR1 and etr1-1(1-349) were next examined by coexpressing etr1-1(1-349)mA and etr1mA. Scored from five independent etr1-7 ers1-2 lines coexpressing etr1-1(1-349)mA and etr1mA, both air- and ethylene-germinated seedlings had a longer hypocotyl than etr1-7 ers1-2 (Fig. 7, B and D). The air-grown seedling was longer than the ethylene-treated seedling in a range of 2.15 ± 0.85 to 4.21 ± 0.64 mm (½α = 0.025; see Supplemental Table S4b), suggesting weak ethylene responses. The severe etr1-7 ers1-2 adult phenotype was rescued and flower fertility was restored (Fig. 7E). We next examined whether the ETR1 C terminus could elevate the etr1-1(1-349)mA signaling without the receiver domain, and etr1(1-609)mA was coexpressed with etr1-1(1-349)mA in etr1-7 ers1-2. Scored from four independent lines, growth of the dark-germinated etr1-7 ers1-2 seedlings expressing both transgenes was restored in air and ethylene (Fig. 7, B and D). The seedling hypocotyl in ethylene was shorter than in the air in a range of 1.74 ± 0.64 to 3.27 ± 0.42 mm (½α = 0.025; see Supplemental Table S4b), suggesting weakened signaling in response to ethylene. Rosette growth and flower fertility (data not shown) of etr1-7 ers1-2 were also restored; however, the rosette was smaller than the etr1-7 ers1-2 mutant, which coexpressed the etr1-1(1-349)mA and etr1mA transgenes (Fig. 7E). As comparisons, ETR1 and etr1-1 were individually transformed to etr1-7 ers1-2 and the mutant phenotype was rescued (see Supplemental Fig. S1B). This result indicates that the roles of the ETR1 receiver domain in ETR1 N-terminal signaling could be minor.
Relative hypocotyl lengths of coexpression lines in response to ethylene and different concentrations of 1-aminocyclopropane-1-carboxylic acid (ACC) were next scored for degrees of ethylene insensitivity (see Supplemental Fig. S3, A and B). T:etr1-1(1-349)mA etr1-7 ers1-2 lines were very short and there was little room for hypocotyl shortening under ethylene treatment; relative lengths of those lines would be less meaningful and thus not scored. Lines coexpressing etr1-1(1-349)mA with etr1mA or etr1(1-609)mA exhibited larger relative hypocotyl lengths than those expressing etr1-1(1-349)mA in the wild type and etr1-7. Relative hypocotyl lengths were smallest when etr1-1(1-349)mA was expressed in etr1-7. These data indicate that loss of ETR1 would weaken the degree of ethylene insensitivity conferred by etr1-1(1-349)mA. Both etr1mA and etr1(1-609)mA are able to elevate the signaling of etr1-1(1-349)mA in etr1-7 ers1-2. Moreover, the etr1-1(1-349)mA variant can signal noncovalently.
It is hypothesized that etr1-1(1-349) would convert wild-type receptors to signaling status (Qu and Schaller, 2004). Our results suggest that ETR1 is important to noncovalent etr1-1(1-349)mA signaling. We next explored the possibility of noncovalent interaction between the ETR1 N terminus and ETR1 by yeast two-hybrid analysis. It has been shown that the GAF domain is capable of dimerization, which is important to protein function or enzyme activity (Ho et al., 2000; Martinez et al., 2005). If ETR1 signaling can be covalent-free, the GAF domain could be a candidate involved in noncovalent signaling of etr1(1-349)/etr1-1(1-349). The transmembrane (TM) domain-lacking proteins etr1(129-729) and etr1(129-349), designated as etr1(−TM) and etr1(GAF), respectively, were subjected to yeast two-hybrid assay. etr1(−TM) was able to interact with etr1(−TM) and etr1(GAF). When the GAF domain of the etr1(−TM) protein was removed, the resulting etr1(293-729) protein failed to interact with etr1(−TM) and yeast failed to grow on the selection medium (Fig. 7F). The yeast two-hybrid reporter, β-galactosidase, had an enzyme activity of 5.24 ± 2.49 (μmol of chlorophenolred-β-d-galactopyranoside [CPRG] hydrolyzed to chloramphenicol red and d-Gal per minute by crude protein extract from 1 OD600 of yeast) based on its kinetics (Fig. 7G). As a comparison, the ERS1(261-613)-CRT1(53-568) interaction was weaker (data not shown). Plasmids carrying no fusion protein gave a background β-galactosidase activity of −1.24 ± 0.32. Weak β-galactosidase activity may indicate a transient protein association.
DISCUSSION
etr1(1-349) Protein Is Capable of Receptor Signaling
The truncated ETR1 N terminus has been interpreted to be incapable of repressing ethylene responses because the mutant phenotype of etr1-6 etr2-3 ein4-4 was rescued by ETR1 and etr1(1-603), but not by etr1(1-349) (Qu and Schaller, 2004). However, the restored growth of etr1-6 etr2-3 ein4-4 could be a result of gain of function conferred by the ETR1 and etr1(1-603) transgenes. In other words, if the ETR1/etr1(1-603) transgenes specifically complement the etr1-6 mutation, the resulting transformant would resemble etr2-3 ein4-4 instead of the wild type. Our data show that etr2-3 ein4-4 was much shorter and smaller than the wild type in the seedling and rosette stages. It is likely that the ectopic expression of ETR1/etr1(1-603) may mask the etr2-3 and ein4-4 mutations while complementing etr1-6.
We first showed that the etr1(1-349) transgene partially restored the growth of etr1-7 etr2-3 ein4-4. The dark-grown seedling was not phenotypically distinguishable from etr2-3 ein4-4 and etr1-7 ein4-4. Germinated under the light, some independent transformation lines resembled etr2-3 ein4-4. However, the adult transformants resembled etr1-7 etr2-3 ein4-4 but were larger, in agreement with a previous study (Qu and Schaller, 2004). Based on these data, the etr1(1-349) transgene more likely does not compensate any of those mutations nor cause morphological changes, but simply elevates receptor signal strength.
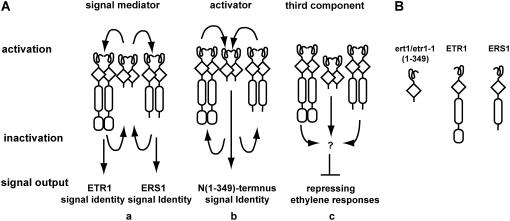
etr1(1-349)-mediated receptor signal output was further examined in the severe etr1-7 ers1-2 mutant, which can only be rescued by subfamily I receptor genes (Wang et al., 2003). etr1(1-349) substantially restored the rosette growth and flower fertility of etr1-7 ers1-2. Both the light-grown seedling and rosette phenotypes of the etr1(1-349)-transformed etr1-7 ers1-2 were not distinguishable from that of the etr1-1(1-349)-transformed mutant. This result further supports our interpretation that the truncated etr1(1-349) protein is capable of signaling. We hypothesize that the etr1(1-349)-mediated receptor signal exhibits subfamily I receptor identity because the etr1-7 ers1-2 mutant phenotype cannot be rescued by subfamily II (Fig. 8A).
Figure 8.
Possible roles of wild-type subfamily I receptors in ETR1 N-terminal-mediated receptor signal output. A, Wild-type ETR1 and ERS1 could function as a signal mediator (a) or an activator (b) of etr1(1-349)/etr1-1(1-349). Components regulated by subfamily I could mediate ETR1 N-terminal signaling (c). Signal output of the ETR1 N terminus could be mediated through an activation-inactivation cycle; conversion to signaling status could be mediated through direct or indirect interactions between receptors. B, Schematic structures of the components.
Subfamily I and Subfamily II Receptors Have Different Roles in the Signaling of etr1(1-349)/etr1-1(1-349)
The dominant etr1-1(1-349) signaling is weakened in etr1-7, implying that ETR1 has a role in N-terminal-mediated signaling. The requirement of ERS1 for ETR1 N-terminal signaling is supported by the results that N-terminal signaling was completely blocked in etr1-7 ers1-3 but not in etr1-7 esr1-2, in which no polyadenylated ERS1 transcript was detectable in ers1-3. These data suggest that subfamily I receptors are required for ETR1 N-terminal-mediated signal output and that subfamily II receptors do not substitute the roles of subfamily I in the etr1(1-349)/etr1-1(1-349) signaling.
Although subfamily II receptors appear less essential to etr1-1(1-349) signaling, the etr2-3 ein4-4 ers2-3 transformants were shorter and smaller than wild type in the seedling and adult stages. These results may imply that the dominant etr1-1(1-349) signaling could be partially masked by the subfamily II triple mutations or that the etr1-1(1-349) signal does not compensate for the subfamily II signal due to distinct signal identities. Alternatively, the etr1-1(1-349) expression level could be low and not sufficient to rescue the mutant phenotype. It remains an open question as to why etr1-1(1-349) does not fully restore the growth of subfamily II null. Our results show that the degree of etr1-1(1-349)-mediated ethylene insensitivity is not perturbed by the loss of subfamily II receptor genes, implying that subfamily II receptors have little effect on etr1-1(1-349)-mediated ethylene insensitivity.
Effects of loss of wild-type receptors and Ag(I) on ETR1 N-terminal signaling were also examined in our studies. Our results show that etr1-1(1-349)-mediated signaling is elevated by Ag(I), suggesting that the etr1-1 mutation may not interfere with Ag(I) sensing in the presence of subfamily I receptors. Because silver nitrate treatment causes hypocotyl elongation in etr1(1-349)- and etr1-1(1-349)-transformed etr1-7 ers1-2 and etr2-3 ein4-4 ers2-3 lines, a fraction of the Ag(I)-induced receptor signaling through the ETR1 N terminus could be independent of wild-type receptors. However, etr1(1-349) and etr1-1(1-349) failed to repress ethylene responses in etr1-7 ers1-3 in the presence of Ag(I), indicating that Ag(I)-induced ethylene insensitivity through the ETR1 N terminus is subfamily I dependent. In contrast, loss of subfamily II receptors has little effect on Ag(I)-induced ethylene insensitivity. Because etr1-7 ers1-3 is not responsive to silver nitrate, Ag(I)-induced ethylene insensitivity would be subfamily I dependent. These results indicate that subfamily I and subfamily II may have different roles in ETR1 N-terminal-mediated and Ag(I)-induced receptor signaling.
Possible Roles of Subfamily I Receptors and Disulfide Linkages in ETR1 N-Terminal-Mediated Signaling
The property of the ethylene receptor signal is a mystery and unlikely to be quantitatively studied by biochemical approaches. Being essential to ETR1 N-terminal signaling, subfamily I receptors could either function as an activator or a signal mediator (Fig. 8A, a and b). Although etr1-1(1-349) signaling is dominant, it does not exclude the possibility that the dominant receptor activity is dependent on an activator. Moreover, the possibility that the ETR1 N terminus could mediate signal through components regulated by subfamily I receptors cannot be excluded (Fig. 8A, c).
Evidence supporting wild-type ETR1 being a signal mediator is that etr1-1(1-349) covalently dimerizes with ETR1 (Gamble et al., 2002) and that ETR1 and CTR1 physically interact (Clark et al., 1998) and colocalize (Gao et al., 2003). It is also hypothesized that etr1-1(1-349) would convert other wild-type receptors to signaling status (Gamble et al., 2002; Qu and Schaller, 2004). It is shown that only subfamily I receptors can rescue the etr1-7 ers1-2 double mutant (Wang et al., 2003), implying that the ETR1 N-terminal-mediated signal would exhibit the subfamily I receptor identity because it rescues the subfamily I mutant phenotype. Thus, ERS1 is very likely involved in mediating ETR1 N-terminal signaling. In this scenario, etr1(1-349)/etr1-1(1-349) would convert ETR1 and ERS1 to a signaling state and the resulting signal is individually sent by ETR1 and ERS1, giving rise to the ETR1 and ERS1 signal identities (Fig. 8A, a). Partial growth restored by etr1(1-349) in etr1-7 etr2-3 ein4-4 shown in a previous study (Qu and Schaller, 2004) and Figure 1 suggest a result of elevated ERS1 signaling, assuming etr1(1-349) enhances the ERS1 signaling state. Whether etr1(1-349) or etr1-1(1-349) would convert subfamily II receptors to signaling status remains unknown, although neither is able to repress ethylene responses in the absence of subfamily I receptors.
We found no evidence to rule out the possibility that the ETR1 N terminus could directly repress ethylene responses. Because ETR1 N-terminal signaling is subfamily I dependent, wild-type subfamily I receptors could act as an activator of the ETR1 N terminus (Fig. 8A, b).
Our results suggest the importance of subfamily I in ETR1 N-terminal signaling, implying a possibility of direct signaling between receptors. However, the property of the receptor signal is unknown and it would be challenging to demonstrate signaling between receptors. It is hypothesized that the covalent linkages through Cys-4 and Cys-6 on ETR1 are involved in ETR1 and ETR1 N-terminal signaling (Schaller et al., 1995; Gamble et al., 2002; Qu and Schaller, 2004). Possible roles of covalent dimerization in ETR1 N-terminal signaling were studied by preventing disulfide bond formation. Our results show that loss of the disulfide bonds does not significantly alter the signaling of etr1mA and etr1-1mA and that the truncated etr1-1(1-349)mA protein caused partial ethylene insensitivity. Thus, disulfide linkages appear to be dispensable to ETR1 N-terminal signaling. However, dominant signaling was weak in etr1-7 and not detectable in etr1-7 ers1-2. Disappearance of etr1-1(1-349)mA signaling in etr1-7 ers1-2 could be caused by a low protein level, altered receptor activity perturbed by the mutations, or subfamily I-dependent signaling.
Because etr1-1(1-349) signaling is weakened by the etr1-7 mutation and blocked by the etr1-7 ers1-3 mutations, it would be very likely that etr1-1(1-349)mA signaling is subfamily I dependent. This hypothesis is in agreement with the result that ectopic expression of etr1mA or etr1(1-609)mA elevated the dominant etr1-1(1-349)mA signaling in etr1-7 ers1-2, suggesting important roles of ETR1 in etr1-1(1-349)mA signaling. Although etr1-1(1-349)mA expression level and receptor activity in etr1-7 ers1-2 were not examined, our results show functional significance of etr1mA and etr1(1-609)mA to etr1-1(1-349)mA signaling.
In etr1-1(1-349)-transformed etr1-7 and etr1-7 ers1-2 lines, ethylene insensitivity conferred by etr1-1(1-349) was weakened by ethylene. Repression of ethylene responses was also reduced by ethylene in those coexpression lines. We hypothesize that an activation-inactivation cycle could be involved in signaling between etr1-1(1-349)/etr1(1-349) and wild-type subfamily I receptors (Fig. 8A). If subfamily I receptors would act as a signal mediator, etr1(1-349)/etr1-1(1-349) would convert ETR1 or ERS1 to a signaling state (Fig. 8A, a). Once the activated ETR1/ERS1 mediates the N-terminal signal, it returns to an inactive state and becomes activated again upon perceiving the etr1(1-349)/etr1-1(1-349) signal. If subfamily I receptors would act as an activator, ETR1 and ERS1 would activate the ETR1 N terminus to a signaling state (Fig. 8A, b). For either possibility, ethylene would inactivate wild-type subfamily I receptors during the signal relay, resulting in weakened receptor signal output, and signaling between the truncated and wild-type receptors could be direct or indirect. The GAF domain is capable of dimerization and activation of enzymatic activity in several organisms (Aravind and Ponting, 1997; Ho et al., 2000; Martinez et al., 2002). Yeast two-hybrid assay of etr1(GAF) and etr1(−TM) would suggest transient interaction through the ETR1 GAF domain. Further study will be required to investigate the possible roles of the GAF domain in inter-receptor signaling.
Based on our results, we hypothesize that both wild-type and dominant receptors might adopt a generalized signaling mechanism by which the receptor signal initiated in the N terminus is mediated to downstream components. A dominant receptor would act together with wild-type receptors, through the N terminus, and cause ethylene insensitivity. Wild-type receptors would integrate the receptor signal through the N terminus and repress ethylene responses. This hypothesis, however, does not exclude the possibility that each receptor would be capable of repressing ethylene responses directly.
MATERIALS AND METHODS
Plant Material and Growth
The etr1-7 ers1-2/+ mutant was from Bleecker (Wang et al., 2003). The etr2-3 ein4-4, etr1-7 ein4-4, etr1-7 etr2-3, etr1-7 etr2-3 ein4-4, and etr2-3 ein4-4 ers2-3 mutants were as described (Hua and Meyerowitz, 1998). ers1-3 was from the Arabidopsis Biological Resource Center (ABRC; stock no. CS 6373). Genotyping of the receptor genes was followed as described (Hua and Meyerowitz, 1998; Hall and Bleecker, 2003) or by sequencing. The ers1-3 mutation was genotyped according to G.E. Schaller (unpublished data). Arabidopsis (Arabidopsis thaliana) was grown at 22°C under 18-h-light and 6-h-dark cycles. For seed germination, Arabidopsis seeds were stratified for 3 d (72 h) at 4°C in the dark on 0.8% agar supplemented with one-half-strength Murashige and Skoog basal medium (Sigma) and then germinated at 22°C. For the seedling triple-response assay, the stratified seeds were germinated in the dark at 22°C for 80 h and the seedling hypocotyl length was measured. For the ethylene treatment, 20 μL L−1 of ethylene gas was included. For each measurement, at least 20 individual seedlings were scored, and the measurement was represented as the 95% confidence interval of a mean. The ethylene-treated etr1-7 ers1-2 seedling was short and not phenotypically distinguishable from the segregating siblings (etr1-7 and etr1-7 ers1-2/+). The etr1-7 ers1-2 mutant is sterile and kept as etr1-7 ers1-2/+ and it is unlikely to distinguish etr1-7 ers1-2 from its siblings under ethylene treatment. To measure the hypocotyl length of the ethylene-treated etr1-7 ers1-2, the seedlings (including etr1-7, etr1-7 ers1-2/+, and etr1-7 ers1-2) were individually measured and scored for the rosette phenotype and then confirmed by genotyping. The corresponding measurement of each etr1-7 ers1-2 individual was then scored. The same procedure was followed for the measurement and genotyping of etr1-7 ers1-3. For ACC treatment, seeds were placed on Murashige and Skoog- and ACC-containing agar, stratified for 3 d, and then germinated in the dark for 80 h at 22°C. For AVG treatment, seeds were germinated as described, except that the Murashige and Skoog-containing agar was included with 0.01 mm AVG.
Transgenes and Identification of Transformants
The etr1-1 and ETR1 cDNA clones were from Chang (1993). The ETR1 promoter and getr1-1[HGG] clones were from Bleecker (Wang et al., 2003). The mutant etr1mA, etr1(1-609)mA, and etr1-1(1-349)mA clones were created by mutagenesis as described below. All transgenes used in this study were driven by the native ETR1 promoter from Bleecker (Wang et al., 2003). All transgenes used in this work were derived from the etr1-1 and ETR1 cDNA clones, except for getr1-1[HGG], which was a genomic clone.
Replacement of ETR1 Cys-4 and Cys-6 with Ala-4 and Ala-6 was made by PCR, by which the primer set ETR1CA-F-NcoI (5′-CGCCATGGAAGTCGCCAATGCTATT-3′) and ETR1-R-BstB1 (5′-CACATGCCTTCCGGTTTCTT-3′) generated the (C4A;C6A) mutations. The resulting DNA fragment was subsequently used to swap with the wild-type ETR1 fragment to generate etr1mA. The etr1-1mA clone was created in the same way, except that the etr1-1 cDNA template was used for PCR.
The primer set ETR1-F-BstXI (5′-TAACCAAGTGTTTGGTACTAG-3′) and ETR1-R-BamHI-350 (5′-CAGGATCCTAAACCGCTAGGAAATC-3′) generated a BstXI/BamHI fragment. The truncated etr1(1-349) and etr1-1(1-349) clones were made by swapping the BstXI/BamHI fragment with the PCR-generated BstXI/BamHI fragment. etr1-1(1-349)mA was created the same way by which the PCR-generated fragment replaced the BstXI/BamHI fragment of the etr1-1mA clone.
DNA clones generated from site-directed mutagenesis and PCR were confirmed by sequencing. Transformation was followed as described (Clough and Bent, 1998). Plants transformed with the pCGN1547 vector were selected by kanamycin. Basta (glufosinate ammonium) was used to select for the pMLBart-transformed plants. For coexpression lines, the etr1-1(1-349)mA transgene was subcloned to the binary vector pCAMBIA1301 and transformed to the homozygous T:etr1mA etr1-7 ers1-2 and T:etr1(1-609)mA etr1-7 ers1-2 plants. The resulting T1 seedlings were selected by hygromycin (50 mg/L) on Murashige and Skoog-containing agar medium.
TAIL-PCR and Flanking Sequence Analysis of the T-DNA Insertion Site in ers1-3
The flanking sequence of the T-DNA insertion site in ers1-3 was determined by TAIL-PCR using combinations of T-DNA-specific primers and eight random primers (activation domains [ADs]) as described (Rohmer et al., 2003). The T-DNA-specific primers were designed according to the T-DNA sequence on ers1-2 (C.-K. Wen, unpublished data), except that JL202 has been published (Hall and Bleecker, 2003). The other two T-DNA-specific primers were JL202-2F (5′-ATAACGCTGCGGACATCTACATTT-3′) and JL202-3F (5′-ATGTAGATTTCCCGGACATGAAGCC-3′). The ers1-3 genomic DNA was individually amplified by JL202 and each of the eight ADs. The resulting DNA was reamplified using JL202-2F and each of the eight ADs. In the final PCR reaction, JL202-3F and each of the eight ADs reamplified the DNA from the second-round PCR amplification. The resulting DNA amplified by JL202-3F and AD6 was subjected to sequencing and the flanking sequences were determined. The flanking sequence was further confirmed by sequencing a PCR fragment generated by the primer set JL202 and ers1-3R (5′-TCGAGCATGTACTGCCATCTCAGCCTCTT-3′).
RT-PCR Analyses of ers1-2 and ers1-3
Arabidopsis total RNA was isolated as described (Wen and Chang, 2002). To generate the ERS1 cDNA fragment across the T-DNA insertion site by RT-PCR, DNase-treated total RNA (1 μg) was primed with R1 (5′-GACTCAAAGTATGAGAAAGC-3′) for first-strand cDNA synthesis, and the RNA template was removed by RNase H. The resulting cDNA was amplified by the primer set F1 (5′-GCTCCGCCGTCATGAATCC-3′) and R2 (5′-GAAGGCATCCACAACGCAC-3′). DNA generated from RT-PCR was subjected to Southern hybridization or a second round of PCR amplification. cDNA generated from the second-round amplification, using the primer set F1 and R3 (5′-TCTAATTCCATGAGTAAGCATCCTAACAT-3′), was purified from gel fractionation and subjected to sequencing. The primer set used for generating the actin cDNA fragment was actin-F1 [5′-TGGCATCA(T/C)ACTTTCTACAA-3′] and actin-R1 [5′-CCACCACT(G/A/T)AGCACAATGTT-3′]. The RT-PCR procedure was the same as described above.
To detect the polyadenylated transcript, first-strand cDNA was reverse transcribed with SuperScript II using oligo(dT)20 and treated with RNase H. The resulting cDNA was subjected to RT-PCR. The primer set ERS1-F(PstI) (5′-CTGATTCTGTCTGCAGA-3′) and ERS1-R(BamHI) (5′-GCGGATCCTCACCAGTTCCACGGTCT-3′) amplified the His-kinase-encoding region.
Southern Hybridization
DNA was fractionated on a 1.5% agarose gel in Tris-acetate EDTA buffer. Each DNA sample was spaced by an empty lane to avoid cross-contamination from the neighboring lanes. The gel was washed in 3 m NaCl containing 0.4 n NaOH for 1 h, and then washed again in 3 m NaCl containing 8 mm NaOH for 15 min. The washed gel was placed onto a nylon membrane (Hybond ECL; Amersham) and DNA was blotted for 3 h. Probe labeling and hybridization were followed according to the manufacturer's instructions (AlkPhos Direct; Amersham). The fluorescence hybridization signal was detected by Hyperfilm ECL (Amersham) for 8 min.
Statistics
Seedling hypocotyl length was represented as the 95% confidence interval of a mean according to the t0.05 value, df, and sd. df is determined as n − 1, in which n is the sample size (n ≥ 20 in this study). When a comparison involves two means, df is (n1 − 1) + (n2 − 1). The sign α indicates significance level or error rate, and α = 0.05 was used throughout this study to estimate the 95% confidence interval of a mean. The difference between two means was estimated as described (Steel and Torrie, 1981) and was represented as the 95% confidence interval. When ½α = 0.025 was specified, it indicates a two-tailed t test. The estimated population mean is μ. P value indicates the probability of a numerically larger value of t. When P > 0.05, it suggests that the difference between two means is not statistically significant (the null hypothesis; H0:μ1 = μ2). When P < 0.05, it suggests statistical difference (the alternative hypothesis; H1:μ1 ≠ μ2) and an asterisk is marked. When P < 0.01, it suggests that the difference is highly statistically significant and double asterisks are marked. Difference between two means is not determined (ND) when P > 0.05. When a difference between two means is represented as the 95% confidence interval, it implies P < 0.05 and is not specified. Multiple comparisons were made by ANOVA, followed by lsd. When lsd is greater than a t value of an α level (which was 0.05 or 0.01 in this study), significance is declared (Steel and Torrie, 1981).
Plasmids and Yeast Two-Hybrid Assay
The etr1(129-729) and etr1(129-343) clones are from C. Chang (unpublished data). The etr1(293-729) clone is as described (Clark et al., 1998).
Yeast (Saccharomyces cerevisiae) strain L40 (Clark et al., 1998) was transformed with the bait (the pLexA fusion in pBTM116) and prey (the GAL4 fusion in pACT II). The resulting transformed yeast colonies were patched on His-lacking medium for selection of the HIS3 reporter gene (Clark et al., 1998). For yeast growth on selection medium, agarose, instead of agar, was used because we noticed that agar gave background growth. The β-galactosidase activity of the yeast two-hybrid assay was measured as the following. Briefly, CPRG was hydrolyzed by the reporter protein β-galactosidase to chloramphenicol red and d-Gal. Hydrolyzed CPRG was measured by reading absorbance at OD578 every second for 600 s and at least 10 independent yeast clones were measured for each assay. The absorbance at OD578 was converted to the amount of CPRG hydrolyzed by β-galactosidase of yeast cells equivalent to 1 OD600. Enzyme activity was calculated based on the slope between 210 and 450 s after the reaction and represented as a 95% confidence interval of a mean.
Nomenclature
The mutant proteins, including the artificially mutagenized variants derived from the wild-type proteins, are in lower case. Genes that are artificially mutagenized from the wild-type clones are italicized in lower case. Wild-type proteins are capitalized and genes are italicized and capitalized.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The etr1mA and etr1(1-609)mA transgenes restore etr1-7 ers1-2 growth.
Supplemental Figure S2. Hypocotyl measurement of wild type and mutants in response to silver nitrate treatment.
Supplemental Figure S3. etr1-1(1-349)mA signaling can be elevated by etr1mA and etr1(1-609)mA.
Supplemental Table S1. Extent of seedling hypocotyl growth of etr1-7 etr2-3 ein4-4 restored by etr1(1-349).
Supplemental Table S2. etr1-1(1-349)-transformed etr1-7 ers1-2 lines are partially ethylene insensitive and responsive to silver nitrate.
Supplemental Table S3. Effects of subfamily II triple mutations on etr1-1(1-349) signaling.
Supplemental Table S4. Differences of seedling hypocotyl lengths between transformants germinated in air and ethylene.
Supplementary Material
Acknowledgments
We thank C. Chang for the etr1-1, ETR1, etr1(129-729), etr1(129-343), and etr1(293-729) clones and the receptor gene mutants; A. Bleecker for providing etr1-7 ers1-2/+, the ETR1 promoter, and the getr1-1[HGG] clone; G. Eric Schaller for sharing unpublished information for the genotyping of ers1-3; and our colleague H.X. Lin for advice on statistics.
This work was supported by the Chinese Academy of Sciences and the National Natural Sciences Foundation of China (grant no. 90408008 to C.-K.W. and grant no. 30421001).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Chi-Kuang Wen (cwen@sippe.ac.cn).
The online version of this article contains Web-only data.
References
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Aravind L, Ponting CP (1997) The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem Sci 22: 458–459 [DOI] [PubMed] [Google Scholar]
- Cancel JD, Larsen PB (2002) Loss-of-function mutations in the ethylene receptor ETR1 cause enhanced sensitivity and exaggerated response to ethylene in Arabidopsis. Plant Physiol 129: 1557–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM (1993) Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262: 539–544 [DOI] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144 [DOI] [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C (1998) Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci USA 95: 5401–5406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Gagne JM, Smalle J, Gingerich DJ, Walker JM, Yoo S-D, Yanagisawa S, Vierstra RD (2004) Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc Natl Acad Sci USA 101: 6803–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble RL, Coonfield ML, Schaller GE (1998) Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc Natl Acad Sci USA 95: 7825–7829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble RL, Qu X, Schaller GE (2002) Mutational analysis of the ethylene receptor ETR1. Role of the histidine kinase domain in dominant ethylene insensitivity. Plant Physiol 128: 1428–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Chen YF, Randlett MD, Zhao XC, Findell JL, Kieber JJ, Schaller GE (2003) Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complexes. J Biol Chem 278: 34725–34732 [DOI] [PubMed] [Google Scholar]
- Guo H, Ecker JR (2003) Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- Hall AE, Bleecker AB (2003) Analysis of combinatorial loss-of-function mutants in the Arabidopsis ethylene receptors reveals that the ers1 etr1 double mutant has severe developmental defects that are EIN2 dependent. Plant Cell 15: 2032–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Kieber JJ, Hirayama N, Kogan M, Guzman P, Nourizadeh S, Alonso JM, Dailey WP, Dancis A, Ecker JR (1999) RESPONSIVE-TO-ANTAGONIST1, a Menkes/Wilson disease-related copper transporter, is required for ethylene signaling in Arabidopsis. Cell 97: 383–393 [DOI] [PubMed] [Google Scholar]
- Ho YS, Burden LM, Hurley JH (2000) Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J 19: 5288–5299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Chang C, Sun Q, Meyerowitz EM (1995) Ethylene insensitivity conferred by Arabidopsis ERS gene. Science 269: 1712–1714 [DOI] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM (1998) Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94: 261–271 [DOI] [PubMed] [Google Scholar]
- Hua J, Sakai H, Nourizadeh S, Chen QG, Bleecker AB, Ecker JR, Meyerowitz EM (1998) EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell 10: 1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Li H, Hutchison CE, Laskey J, Kieber JJ (2003) Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J 33: 221–233 [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Martinez SE, Bruder S, Schultz A, Zheng N, Schultz JE, Beavo JA, Linder JU (2005) Crystal structure of the tandem GAF domains from a cyanobacterial adenylyl cyclase: modes of ligand binding and dimerization. Proc Natl Acad Sci USA 102: 3082–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez SE, Wu AY, Glavas NA, Tang XB, Turley S, Hol WG, Beavo JA (2002) The two GAF domains in phosphodiesterase 2A have distinct roles in dimerization and in cGMP binding. Proc Natl Acad Sci USA 99: 13260–13265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussatche P, Klee HJ (2004) Autophosphorylation activity of the Arabidopsis ethylene receptor multigene family. J Biol Chem 279: 48734–48741 [DOI] [PubMed] [Google Scholar]
- Muller-Dieckmann HJ, Grantz AA, Kim SH (1999) The structure of the signal receiver domain of the Arabidopsis thaliana ethylene receptor ETR1. Struct Fold Des 7: 1547–1556 [DOI] [PubMed] [Google Scholar]
- O'Malley RC, Rodriguez FI, Esch JJ, Binder BM, O'Donnell P, Klee HJ, Bleecker AB (2005) Ethylene-binding activity, gene expression levels, and receptor system output for ethylene receptor family members from Arabidopsis and tomato. Plant J 41: 651–659 [DOI] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P (2003) EIN3-dependent regulation of plant ethylene hormone signaling by two arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689 [DOI] [PubMed] [Google Scholar]
- Qu X, Schaller GE (2004) Requirement of the histidine kinase domain for signal transduction by the ethylene receptor ETR1. Plant Physiol 136: 2961–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick JS, Wen C-K, Shockey JA, Chang C (2006) REVERSION-TO-ETHYLENE SENSITIVITY1, a conserved gene that regulates ethylene receptor function in Arabidopsis. Proc Natl Acad Sci USA 103: 7917–7922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez FI, Esch JJ, Hall AE, Binder BM, Schaller GE, Bleecker AB (1999) A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science 283: 996–998 [DOI] [PubMed] [Google Scholar]
- Rohmer L, Kjemtrup S, Marchesini P, Dangl JL (2003) Nucleotide sequence, functional characterization and evolution of pFKN, a virulence plasmid in Pseudomonas syringae pathovar maculicola. Mol Microbiol 47: 1545–1562 [DOI] [PubMed] [Google Scholar]
- Sakai H, Hua J, Chen QG, Chang C, Medrano LJ, Bleecker AB, Meyerowitz EM (1998) ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA 95: 5812–5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Ladd AN, Lanahan MB, Spanbauer JM, Bleecker AB (1995) The ethylene response mediator ETR1 from Arabidopsis forms a disulfide-linked dimer. J Biol Chem 270: 12526–12530 [DOI] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel RGD, Torrie JH (1981) Principles and Procedures of Statistics. McGraw-Hill, New York
- Stock AM, Robinson VL, Goudreau PN (2000) Two-component signal transduction. Annu Rev Biochem 69: 183–215 [DOI] [PubMed] [Google Scholar]
- Wang W, Hall AE, O'Malley R, Bleecker AB (2003) Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc Natl Acad Sci USA 100: 352–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen CK, Chang C (2002) Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14: 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woeste KE, Kieber JJ (2000) A strong loss-of-function mutation in RAN1 results in constitutive activation of the ethylene response pathway as well as a rosette-lethal phenotype. Plant Cell 12: 443–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XC, Qu X, Mathews DE, Schaller GE (2002) Effect of ethylene pathway mutations upon expression of the ethylene receptor ETR1 from Arabidopsis. Plant Physiol 130: 1983–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.