Abstract
Spatial and temporal regulation of phosphoenolpyruvate carboxylase (PEPC) is critical to the function of C4 photosynthesis. The photosynthetic isoform of PEPC in the cytosol of mesophyll cells in Kranz-type C4 photosynthesis has distinctive kinetic and regulatory properties. Some species in the Chenopodiaceae family perform C4 photosynthesis without Kranz anatomy by spatial separation of initial fixation of atmospheric CO2 via PEPC from C4 acid decarboxylation and CO2 donation to Rubisco within individual chlorenchyma cells. We studied molecular and functional features of PEPC in two single-cell functioning C4 species (Bienertia sinuspersici, Suaeda aralocaspica) as compared to Kranz type (Haloxylon persicum, Salsola richteri, Suaeda eltonica) and C3 (Suaeda linifolia) chenopods. It was found that PEPC from both types of C4 chenopods displays higher specific activity than that of the C3 species and shows kinetic and regulatory characteristics similar to those of C4 species in other families in that they are subject to light/dark regulation by phosphorylation and display differential malate sensitivity. Also, the deduced amino acid sequence from leaf cDNA indicates that the single-cell functioning C4 species possesses a Kranz-type C4 isoform with a Ser in the amino terminal. A phylogeny of PEPC shows that isoforms in the two single-cell functioning C4 species are in a clade with the C3 and Kranz C4 Suaeda spp. with high sequence homology. Overall, this study indicates that B. sinuspersici and S. aralocaspica have a C4-type PEPC similar to that in Kranz C4 plants, which likely is required for effective function of C4 photosynthesis.
In C4 plants having Kranz anatomy, fully differentiated mesophyll cells (MCs) and bundle sheath cells (BSCs) cooperate to fix CO2 by the C4 pathway (Edwards and Walker, 1983). In these plants, atmospheric CO2 is first fixed into C4 acids in the MC by phosphoenolpyruvate (PEP) carboxylase (PEPC). These C4 acids are then transported to BSCs where they are decarboxylated and the CO2 is concentrated and donated to the C3 cycle.
Since the discovery of C4 photosynthesis, the spatial compartmentation of terrestrial C4 plants was consistently linked to the occurrence of Kranz-type anatomy. Recently, three succulent species in the Chenopodiaceae family, Bienertia cycloptera Bunge ex Boiss., Bienertia sinuspersici Akhani, and Suaeda aralocaspica (Bunge) Freitag and Schütze (formerly classified as Borszczowia), were found to have a unique mechanism of C4 photosynthesis, which occurs within individual photosynthetic cells by intracellular partitioning of enzymes and organelles (including dimorphic chloroplasts) into two compartments (Voznesenskaya et al., 2003; Edwards et al., 2004; Akhani et al., 2005). S. aralocaspica has a single layer of elongated, cylindrical chlorenchyma cells in which functions of C4 photosynthesis are spatially separated between opposite ends of the cells. The model for photosynthesis from studies on cell structure and compartmentation of photosynthetic enzymes is that atmospheric CO2 enters the chlorenchyma cell at the distal end and carbon is assimilated into C4 acids by the action of PEPC located in the cytosol. This is accomplished through use of the PEP generated in chloroplasts by pyruvate orthophosphate dikinase (PPDK) in this part of the cell. C4 acids diffuse from the distal to the proximal part of the cell through a thin, peripheral cytoplasmic space in the middle of the cell. There, C4 acids are decarboxylated by the NAD-malic enzyme (NAD-ME) in mitochondria, which are localized in the proximal end, and the CO2 captured by Rubisco in the chloroplasts (Voznesenskaya et al., 2003). B. cycloptera and B. sinuspersici have an unusual development of two cytoplasmic compartments in chlorenchyma cells, which consist of a large central cytoplasmic compartment packed with chloroplasts and mitochondria and a peripheral layer of cytoplasm with chloroplasts (Voznesenskaya et al., 2002, 2005; Akhani et al., 2005). The central compartment, which is surrounded by vacuoles, is connected to the peripheral cytoplasm by cytoplasmic channels. From our studies, the model for C4 photosynthesis in Bienertia spp. is that atmospheric CO2 enters the cell around the periphery and is incorporated into C4 acids by PEPC. C4 acids then diffuse to the central compartment through cytoplasmic channels and are decarboxylated by NAD-ME in the mitochondria specifically located there. Rubisco in the chloroplasts in the central compartment fixes the released CO2. A three-carbon product from this decarboxylation diffuses to the peripheral chloroplasts, where PPDK generates PEP from pyruvate for the PEPC reaction (Voznesenskaya et al., 2002).
The specialized organelle and enzyme compartmentation in these single-cell C4 functioning chenopods, and the particular features of the connecting cytoplasmic compartments, mimic the organization of BSCs and MCs, where there is intercellular spatial separation (Edwards et al., 2004). However, in the single-cell functioning C4 species, the cytosolic enzyme PEPC is not exclusively confined to the site of entry of atmospheric CO2. In the chlorenchyma cells of S. aralocaspica, PEPC is found throughout the cytoplasm (Voznesenskaya et al., 2001). In Bienertia, immunolabeling for PEPC shows the most intensive labeling in the peripheral cytoplasm, with some labeling in the central cytoplasmic compartment (Voznesenskaya et al., 2002). Selective function of the enzyme is necessary to prevent a futile cycle in the Rubisco-containing cellular compartment. In Bienertia and S. aralocaspica, PEPC catalysis in the central and proximal compartments, respectively, may be limited by (1) less PEPC protein because there is lower cytosolic space due to the high density of mitochondria and chloroplasts in these compartments; (2) selective control of PEPC activity by metabolites or covalent modification (Edwards et al., 2004); or (3) unavailability of PEP because PPDK is selectively localized in the peripheral and distal chloroplasts, respectively.
PEPC catalyzes the β-carboxylation of PEP by HCO3− in the presence of a divalent cation to yield oxaloacetate and inorganic phosphate (O'Leary, 1982). It plays a cardinal role in the initial fixation of atmospheric CO2 in leaf tissue of plants performing C4 photosynthesis and Crassulacean acid metabolism (CAM). PEPC also participates in diverse anaplerotic functions in basic plant metabolism, such as gluconeogenesis and nonautotrophic CO2 fixation in C3 leaves and nonphotosynthetic tissues (Andreo et al., 1987; Chollet et al., 1996). In Kranz-type C4 and CAM plants, PEPC is highly regulated in vivo with l-malate and Glc-6-P acting as negative and positive allosteric effectors, respectively. In addition, the enzyme is regulated by phosphorylation/dephosphorylation of a single Ser residue, located in its N terminus, resulting in an increase in catalytic activity by phosphorylation and, more notably, a decrease in l-malate sensitivity. PEPC is phosphorylated by a specific Ca2+-independent protein kinase closely related to plant calcium-dependent protein kinases and dephosphorylated by protein phosphatase 2A. The phosphorylation state of PEPC is highly controlled by the PEPC kinase, but not through changes in phosphatase activity (Nimmo et al., 1987; Carter et al., 1991; Chollet et al., 1996; Vidal and Chollet, 1997). In C4 plants, PEPC phosphorylation is triggered by illumination (Nimmo et al., 1987; Jiao and Chollet, 1991; Duff et al., 1995; Vidal and Chollet, 1997), whereas in CAM plants, enzyme phosphorylation occurs during the dark period according to circadian rhythm (Hartwell et al., 1996). Additionally, regulatory phosphorylation of a C4 form of PEPC in mature maize (Zea mays) plants is controlled not only by a light signal, but also by some other metabolic signals such as nitrogen status (Ueno et al., 2000; Echeverria and Vidal, 2003; Nimmo, 2003). PEPC kinase transcript levels from CAM plants are regulated in response to a circadian oscillator (Hartwell et al., 1999; Taybi et al., 2000), and those in C3 and C4 plants have been shown to be induced by light (Tsuchida et al., 2001; Fontaine et al., 2002).
Isoforms of enzymes of the C4 pathway, including PEPC, are present in C3 plants. The evolution of C4 plants was facilitated by a set of genes that already existed in ancestral C3 species. New expression patterns and regulatory elements of the genes were acquired to make them more efficient and spatially regulated, and genes modified in the region were transcribed to give forms with kinetic properties different from those in C3 (Lepiniec et al., 1994; Svensson et al., 2003).
The aim of this work was to determine whether PEPC from B. sinuspersici and S. aralocaspica are regulated in such a way as to control day/night activity and to analyze whether these single-cell functioning C4 species possess a C4 PEPC isoform, as occurs in Kranz-type C4 plants. For comparison, we also studied PEPC from Suaeda linifolia (C3)—phylogenetically very close to S. aralocaspica—and Haloxylon persicum, Suaeda eltonica, and Salsola richteri (C4 species), also succulent species belonging to the Chenopodiaceae family. We biochemically characterized PEPC in crude extract from leaves and isolated and sequenced cDNAs of some members of the PEPC family. The sequence characteristics of each were studied and analyzed in an evolutionary context.
RESULTS
Western Blots for Enzymes Related to Carbon Fixation
To study PEPC from the single-cell functioning C4 plants B. sinuspersici and S. aralocaspica, and to compare the enzyme's molecular and biochemical features with those of other C3 and C4 species belonging to the Chenopodiaceae family, we analyzed leaf extracts from S. linifolia, H. persicum, S. richteri, and S. eltonica.
S. linifolia has been described as a C3 species according to its carbon isotope composition and leaf anatomy (Akhani et al., 1997; Kapralov et al., 2006); however, there are no biochemical data regarding the carbon fixation enzymes. Western-blot analysis carried out with total protein from this species (Fig. 1, lane 7) indicates that C3-type photosynthetic metabolism is operating in this plant, as shown by high levels of Rubisco (Fig. 1D, lane 7), very low levels of PEPC—consistent with it having a housekeeping function (Fig. 1C, lane 7)—and no reaction with antibodies against the malate decarboxylating enzymes NAD-ME and NADP-ME (Fig. 1, A and B, respectively).
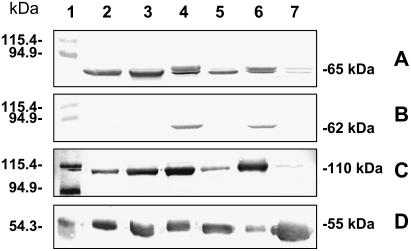
Figure 1.
Western blots of photosynthetic enzymes in the single-cell functioning C4 species B. sinuspersici and S. aralocaspica compared to Kranz C4 and C3 chenopod species. Western blots were revealed with antibodies against α-NAD-ME (A), NADP-ME (B), PEPC (C), and Rubisco (D). Twenty micrograms (A–C) or 5 μg (D) of total protein from B. sinuspersici (2), S. aralocaspica (3), H. persicum (4), S. eltonica (5), S. richteri (6), and S. linifolia (7) were loaded. Molecular masses of the Mr markers loaded in lane 1 are shown on the left. Molecular masses of the immunoreactive bands are shown on the right.
With respect to the other species, the occurrence of C4-type metabolism is indicated by the presence of high levels of a 110-kD immunoreactive PEPC (Fig. 1C, lanes 2–6) as compared to the C3 species S. linifolia (Fig. 1C, lane 7), by the presence of lower levels of the large subunit of Rubisco (rbcL; Fig. 1D, lanes 2–6) than in the C3 species (Fig. 1D, lane 7), and by high levels of the decarboxylating enzymes NAD-ME or NADP-ME (Fig. 1, A and B, lanes 2–6).
Both of the non-Kranz, single-cell functioning C4 species, B. sinuspersici and S. aralocaspica, are NAD-ME type, displaying an immunoreactive band of 65 kD with antibodies against NAD-ME (Fig. 1A, lanes 2 and 3) and no reaction with antibodies against NADP-ME (Fig. 1B, lanes 2 and 3). This subclassification has been previously reported for S. aralocaspica (Voznesenskaya et al., 2001), but not for B. sinuspersici, which was more recently classified as a single-cell functioning C4 plant (Akhani et al., 2005).
S. eltonica is classified as an NAD-ME-type C4 species as evidenced by the presence of the α-NAD-ME subunit of 65 kD (Fig. 1A, lane 5) and lack of NADP-ME. In agreement with previous work (Casati et al., 1999; Pyankov et al., 2000), H. persicum and S. richteri are classified as NADP-ME-type C4 species, as shown by the presence of an immunoreactive band with antibodies against maize NADP-ME (Fig. 1B, lanes 4 and 6). In addition, both species show strong immunoreactive bands in western blots with anti-α-NAD-ME from Amaranthus hypochondriacus (Fig. 1A, lanes 4 and 6). For S. richteri and H. persicum, the presence of both NADP and NAD decarboxylating enzymes is not surprising because in several species within the Chenopodiaceae family coexistence of both enzymes has been previously reported (Pyankov et al., 2000).
Biochemical Characterization of PEPC
PEPC Kinetic Parameters
Prior to measurements of PEPC activity, native gels were run followed by activity staining to check the oligomerization state of the enzyme. It was found that inclusion of 20% (v/v) glycerol in the extraction buffer and in the gels was necessary to maintain the enzyme in the tetrameric form and to obtain maximal activity. Glycerol and other compatible solutes are known to stabilize PEPC and they may affect the kinetic properties by protecting against deoligomerization and/or as osmoprotectants (Krall and Edwards, 1993; Ogawa et al., 1997). With glycerol, PEPC proteins from the different species displayed similar mobility, corresponding to that of a molecular mass of 440 kD. Because a similar PEPC subunit size (110 kD) was present in all samples (see western-blot analysis; Fig. 1C), the aggregation state in native gels indicates that the enzyme in single-cell functioning C4 and other species is a tetramer. When glycerol was omitted, some additional bands, corresponding to dimer forms, were observed (data not shown). To prevent dissociation of the enzyme during kinetic analysis, 20% (v/v) glycerol was included in extraction and activity media assay.
To biochemically characterize PEPC from different Chenopodiaceae species, we determined the kinetic parameters of the enzyme [apparent maximal specific activity of PEPC at saturating substrates (apparent Vmax), Km (PEP), and I50 (malate)] using desalted soluble protein from leaf extracts collected in the light period and the dark period (Table I). In all cases, when assayed at the optimal pH (8.0), the curve linking initial velocity to substrate (PEP) concentration was a rectangular hyperbola.
Table I.
Kinetic parameters of PEPC of the single-cell functioning C4 species B. sinuspersici and S. aralocaspica and related chenopod species
Leaves were collected after 5 h into the light and 5 h into the dark period. Activity was assayed at pH 8.0. Values represent the mean of at least three independent determinations (ses are shown).
| Species | Photosynthetic Type | Parameter | Light | Dark |
|---|---|---|---|---|
| B. sinuspersici | Single-cell C4 | Apparent Vmax (units/mg protein) | 1.96 ± 0.01 | 1.78 ± 0.04 |
| Km (PEP) (mm) | 0.06 ± 0.01 | 0.06 ± 0.01 | ||
| S. aralocaspica | Single-cell C4 | Apparent Vmax (units/mg protein) | 1.67 ± 0.03 | 1.06 ± 0.04 |
| Km (PEP) (mm) | 0.10 ± 0.01 | 0.12 ± 0.02 | ||
| H. persicum | Kranz C4 | Apparent Vmax (units/mg protein) | 1.27 ± 0.04 | 0.87 ± 0.04 |
| Km (PEP) (mm) | 0.12 ± 0.03 | 0.17 ± 0.03 | ||
| S. eltonica | Kranz C4 | Apparent Vmax (units/mg protein) | 0.83 ± 0.03 | 0.67 ± 0.08 |
| Km (PEP) (mm) | 0.09 ± 0.02 | 0.10 ± 0.03 | ||
| S. richteri | Kranz C4 | Apparent Vmax (units/mg protein) | 0.93 ± 0.03 | 1.00 ± 0.04 |
| Km (PEP) (mm) | 0.21 ± 0.04 | 0.22 ± 0.04 | ||
| S. linifolia | C3 | Apparent Vmax (units/mg protein) | 0.09 ± 0.01 | 0.08 ± 0.01 |
| Km (PEP) (mm) | 0.05 ± 0.01 | 0.05 ± 0.01 |
As anticipated for a C3-type PEPC, the enzyme from S. linifolia had much lower apparent Vmax values and 2- to 4-fold lower Km (PEP) than those of PEPC from the Kranz C4 species H. persicum, S. eltonica, and S. richteri. Interestingly, the highest values for apparent maximal specific activity of PEPC were from the non-Kranz, single-cell functioning C4 species. With respect to Km (PEP) for PEPC, in the case of S. aralocaspica, the values were 2-fold higher than C3 S. linifolia and comparable to that of the C4 plants H. persicum and S. eltonica. In contrast, Km (PEP) values of PEPC from B. sinuspersici were intermediate between those of the enzyme from the C3 S. linifolia and from the C4 species.
When comparing the kinetic parameters of samples collected during the day and night, it is clear that PEPC from H. persicum is differentially regulated during the photoperiod, showing higher apparent maximal activity (1.5 times) and higher affinity for the substrate PEP during the day than at night. In crude extracts of S. eltonica (C4), although apparent Vmax estimated during the day was higher than that at night, PEP affinity remained almost unchanged. In the case of PEPC from S. aralocaspica, the enzyme parameters were affected in the same manner as those of PEPC from the C4 species S. eltonica, showing a 1.6 times increase in apparent Vmax and a 0.9 times decrease in Km (PEP) during the day versus night. In contrast to the other species, PEPC kinetic parameters from the Kranz C4 S. richteri, C3 S. linifolia, and single-cell functioning C4 B. sinuspersici remained almost unchanged.
Varying the l-malate concentration in the reaction mixture at suboptimal PEP concentration and pH 7.2 resulted in a progressive decrease of enzyme activity in all cases. The concentration of malate causing 50% inhibition of initial activity of PEPC [I50 (malate)] was measured in desalted crude extracts from samples collected during the day and night (Table II). Measurements were performed at a PEP concentration (0.1 mm) that was similar to estimated Km (PEP) values for most species (Table I). PEPC from S. linifolia did not show differences in malate sensitivity during the day and night periods, as revealed by essentially similar I50 (malate) values, which is expected for a C3-type PEPC (Gupta et al., 1994; Svensson et al., 2003).
Table II.
Inhibition of PEPC by malate [I50 (malate)] in soluble protein extracts from leaves of the single-cell functioning C4 species B. sinuspersici and S. aralocaspica and related chenopod species
Samples were taken after 5 h into the light and dark periods. PEPC from crude extracts was assayed at pH 7.2, 0.1 mm PEP. Values represent the mean of at least three independent determinations (ses are shown).
| Species | Photosynthetic Type |
I50 (malate)
|
I50 (malate)Light/Dark | |
|---|---|---|---|---|
| Light | Dark | |||
| mm | ||||
| B. sinuspersici | Single-cell C4 | 2.22 ± 0.03 | 2.00 ± 0.25 | 1.11 |
| 0.60 ± 0.02a | 0.23 ± 0.02a | 2.61a | ||
| S. aralocaspica | Single-cell C4 | 0.69 ± 0.07 | 0.33 ± 0.06 | 2.09 |
| H. persicum | Kranz C4 | 1.91 ± 0.04 | 0.47 ± 0.05 | 4.06 |
| S. eltonica | Kranz C4 | 0.89 ± 0.05 | 0.26 ± 0.01 | 3.42 |
| S. richteri | Kranz C4 | 2.17 ± 0.11 | 1.25 ± 0.14 | 1.74 |
| S. linifolia | C3 | 3.93 ± 0.06 | 3.90 ± 0.11 | 1.01 |
With B. sinuspersici, PEPC was also assayed at pH 7.2 with 0.055 mm PEP.
In B. sinuspersici and S. aralocaspica, malate sensitivity of PEPC was similar to that from Kranz-type C4 species. When comparing samples collected during the light and dark periods, in all PEPCs from Kranz-type C4 plants analyzed during the day, malate sensitivity was lower compared to the night, as shown by an increase in the I50 (malate) value ranging from 1.7 (S. richteri) to 4 times (H. persicum). In the same manner, the I50 (malate) value of PEPC from the single-cell functioning C4 species S. aralocaspica collected during the day is twice that measured during the night, indicating that the enzyme is more sensitive to malate at night. However, in the case of B. sinuspersici, only a slight increase (1.1 times) in the I50 (malate) was obtained for samples collected during the day. Nevertheless, when malate sensitivity was evaluated at a lower level of PEP (0.55 mm), which is the Km (PEP) value estimated for the enzyme in this species (Table I), there was a 2.6 times increase in the I50 (malate) values for samples collected in the light compared to those collected in the dark. In addition, the I50 (malate) values for PEPC for B. sinuspersici and S. aralocaspica were similar when determined around the respective Km (PEP) values for the two species.
PEPC Isoelectric Point
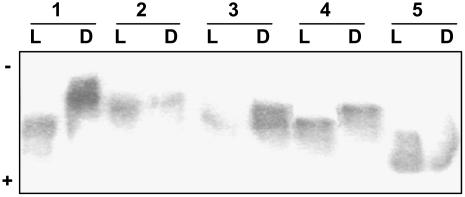
Data obtained from kinetic and malate inhibition studies of PEPC (on day versus night isolations) from B. sinuspersici and S. aralocaspica indicate there may be posttranslational modification of the enzyme. To evaluate this possibility, native isoelectric focusing (IEF) was conducted followed by activity staining, using samples taken during the night versus the day. In both species, PEPC from samples collected during the night showed a more alkaline pI than from samples harvested during the day. Representative results are shown in Figure 2, with two single-cell C4 species in lanes 1 (B. sinuspersici) and 2 (S. aralocaspica), two Kranz-type species in lanes 3 (H. persicum) and 4 (S. eltonica), and the C3 species (S. linifolia) in lane 5. The average pI values (from three replications) for samples collected from the day versus night, respectively, were 5.41 (±0.05) versus 5.55 (±0.05) for B. sinuspersici, and 5.47 (±0.03) versus 5.62 (±0.05) for S. aralocaspica. Similar results were obtained for the Kranz-type C4 chenopod species S. richteri (data not shown), H. persicum, and S. eltonica with average day versus night values for the three species of 5.52 (±0.04) and 5.63 (±0.03), respectively. In contrast, no light versus dark changes in the pI values (5.14 ± 0.02 versus 5.11 ± 0.05) were observed for PEPC from C3 S. linifolia. The light versus dark changes in pI in the single-cell C4 species provides additional evidence that their PEPC undergoes regulatory modifications during the day/night periods.
Figure 2.
Native IEF protein gel revealed by activity staining for PEPC in the single-cell functioning C4 species B. sinuspersici and S. aralocaspica and related chenopod species. Leaf samples were taken after 5 h into the light (L) and 5 h into the dark (D) period. Approximately 4 milliunits of PEPC from leaf extracts of B. sinuspersici (1), S. aralocaspica (2), H. persicum (3), S. eltonica (4), and S. linifolia (5) were loaded.
PEPC Phosphorylation Studies
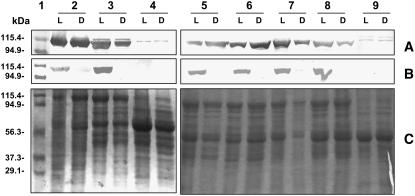
Based on changes in the kinetic properties and pI of PEPC during the day/night periods, B. sinuspersici and S. aralocaspica leaves were investigated to determine whether the enzyme undergoes a diel pattern of phosphorylation/dephosphorylation. For this purpose, we used samples collected during the day and night periods and performed western-blot analysis with antibodies raised against phosphorylated PEPC (Fig. 3B), which cross-reacts with PEPC from a number of C4 species (T. Furumoto and K. Izui, personal communication). As controls, crude extracts from the previously characterized species, Flaveria trinervia (C4) and Flaveria pringlei (C3), were included (Fig. 3, lanes 3 and 4). In F. trinervia leaves, the ppcA gene encoding the C4 isoform is the PEPC gene having the highest level of expression (Westhoff et al., 1997). Thus, the immunoreactive band observed in crude extracts of F. trinervia, which undergoes phosphorylation in the light, is considered the C4-type isoform. In F. pringlei, all the genes encoding PEPCs are expressed in low levels in leaves, and the antiphosphorylated PEPC antibody did not detect light/dark changes in the phosphorylation state, although F. pringlei PEPC-A has a phosphorylatable Ser near the N terminus (Westhoff et al., 1997; Bläsing et al., 2000). Figure 3, A and C, shows western blots with anti-PEPC from Amaranthus viridis and the corresponding Coomassie Blue SDS-PAGE carried out with the same samples with the same amount of protein loaded in each lane. The results indicate that the same levels of PEPC protein are present in samples collected during the day and night for each plant.
Figure 3.
Western blots for PEPC protein and for the phosphorylated form of PEPC from leaves harvested in the light versus dark. Samples were taken after 5 h into the light (L) and 5 h into the dark (D) period. Fifteen micrograms of total protein from S. richteri (2), F. trinervia (3), F. pringlei (4), B. sinuspersici (5), S. aralocaspica (6), H. persicum (7), S. eltonica (8), and S. linifolia (9) were loaded for western-blot analysis with antitotal PEPC (A) or with antiphosphorylated PEPC (B). Molecular masses of the Mr markers loaded in lane 1 are shown on the left. Twenty micrograms of total protein were loaded in each lane (2–9) for Coomassie Blue staining (C).
Using the antibody with specificity to the phosphorylated form of PEPC, in the Kranz-type C4 species F. trinervia (used as a control; Fig. 3A, lane 3L), PEPC is phosphorylated during the day and dephosphorylated during the night. Likewise, PEPC in the single-cell functioning C4 species B. sinuspersici (lane 5L) and S. aralocaspica (lane 6L), and in all C4 Kranz species studied from Chenopodiaceae (lanes 2L, 7L, and 8L), is phosphorylated during the day and dephosphorylated during the night.
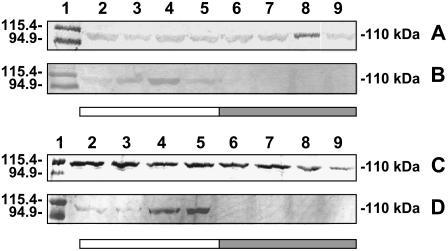
In addition, studies were done to establish whether the changes in the PEPC phosphorylation state in the single-cell functioning C4 species respond to light stimulus or to endogenous circadian rhythm. For these experiments, samples were collected at different times in the day/night cycle and also after 17 and 41 h of complete darkness (which includes time corresponding to that of the day period for the usual sampling). Western blots with anti-A. viridis PEPC showed that similar levels of PEPC protein were loaded in each lane (Fig. 4, A and C). On the other hand, western blots with antiphosphorylated PEPC, using B. sinuspersici (Fig. 4B) and S. aralocaspica (Fig. 4D) crude extracts, indicate that PEPC is phosphorylated under light conditions (lanes 2–5), showing a major proportion of phosphorylated PEPC at 1 and 5 pm (Fig. 4D, lanes 4 and 5, respectively) in the case of S. aralocaspica and at 1 pm in the case of B. sinuspersici (Fig. 4B, lane 4). Immunoreaction with antiphosphorylated PEPC was not observed in samples collected from plants during the night period (lanes 6 and 7) or during the extended dark periods when it would usually be phosphorylated under illumination (lanes 8 and 9). These results indicate that, as in the case of C4-type PEPC from Kranz species, phosphorylation of the enzyme from B. sinuspersici and S. aralocaspica is light dependent.
Figure 4.
Western-blot analyses showing PEPC phosphorylation time course in the single-cell functioning C4 species B. sinuspersici and S. aralocaspica. B. sinuspersici (A and B) and S. aralocaspica (C and D) samples were taken at 7 am (2), 10:30 am (3), 1 pm (4), 5 pm (5), 8:30 pm (6), and 12:30 pm (7), or after 17 (8) or 41 (9) h of complete darkness. In the case of B. sinuspersici, lights were turned on at 6 am and turned off at 8 pm, whereas for S. aralocaspica the light period was 6 am to 6 pm. Fifteen micrograms of total protein were loaded in lanes 2 to 9 for western-blot analysis with anti-PEPC (A and C) or with antiphosphorylated PEPC (B and D). Molecular masses of the Mr markers loaded in lane 1 are shown on the left. Molecular masses of the immunoreactive bands are shown on the right.
Molecular Characterization of PEPC
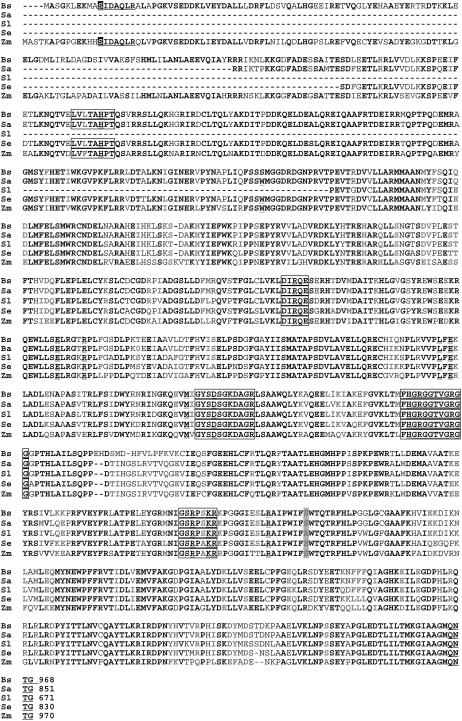
To gain insight into PEPC molecular characterization and evolution within the Chenopodiaceae in relation to the single-cell functioning C4 species, we isolated by reverse transcription (RT)-PCR the ppc transcripts expressed in major proportions in leaves of B. sinuspersici and S. aralocaspica, in the Kranz C4 plant S. eltonica, and in the C3 species S. linifolia. In the case of B. sinuspersici, the complete cDNA (2,909 bp; GenBank accession no. DQ538352) encoding PEPC was isolated. The nucleotide sequence obtained has 85% homology to the C4 PEPC from the dicot A. hypochondriacus (GenBank accession no. L4917). In the case of S. aralocaspica, S. eltonica, and S. linifolia, partial cDNAs were isolated (GenBank accession nos. DQ538353, DQ538354, DQ538355), where the nucleotides encoding for the first 116, 136, and 296 amino acid residues of the sequence were missing. Figure 5 shows the sequence alignment from the deduced amino acids from the PEPC sequences isolated in this work compared with that from maize, which was used for analysis of the three-dimensional structure of the enzyme (Kai et al., 2003).
Figure 5.
The deduced amino acid sequence of PEPC from B. sinuspersici (Bs), S. aralocaspica (Sa), S. linifolia (Sl), and S. eltonica (Se) aligned with the C4 PEPC from maize (Zm) using ClustalW analysis. Identical amino acid residues are highlighted in black, whereas highly conserved amino acids are highlighted in light gray. The strictly conserved, nonphosphorylatable Ala residue in PEPC near position 774, which when changed to Ser comprises a necessary, but not sufficient, determinant of C4-specific kinetics of C4 PEPCs (Svensson et al., 2003), and the corresponding Ser are shaded in a light background. The Ser residue located in the amino-terminal region subjected to day/night regulatory phosphorylation is shown in white and shaded in a dark background. Motifs are underlined, whereas conserved residues are double underlined. Boxes mark domains important for enzyme catalysis.
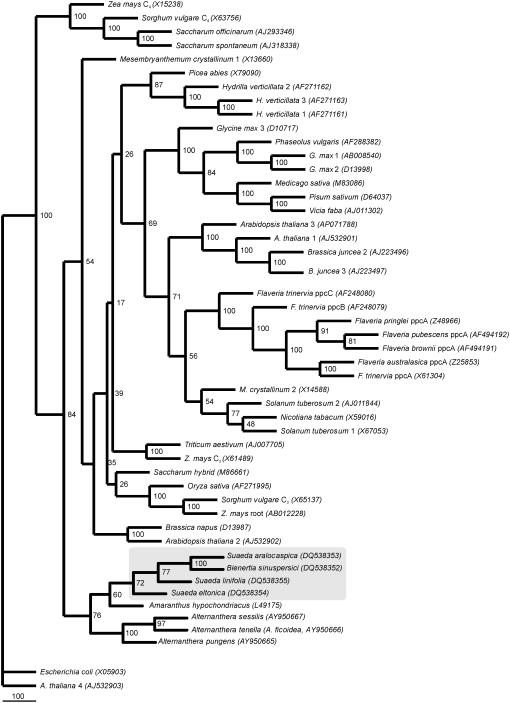
A phylogenetic tree constructed from amino acid sequences of PEPC, from both monocot and dicot species, shows that sequence information on the forms of PEPC isolated from Chenopodiaceae and Amaranthaceae species form a separate clade (Fig. 6). In our tree, the other C4 PEPC of grasses such as maize, Sorghum vulgare, sugar cane (Saccharum officinarum), and Sorghum spontaneum are basal. Surprisingly, both single-cell functioning C4 species, B. sinuspersici and S. aralocaspica, are grouped together in a subclade with C3 S. linifolia, C4 S. eltonica, and C4 A. hypochondriacus.
Figure 6.
Phylogenetic analysis of various plant PEPC isoforms (based on the last 70 amino acid residues of the 3′ side of the coding region) using a neighbor-joining consensus tree including the single-cell functioning C4 species B. sinuspersici and S. aralocaspica. The tree was oriented with PEPC amino acid sequence from Escherichia coli and Arabidopsis bacterial-type PEPC as functional outgroups. The gray box indicates the distribution of members of the Chenopodiaceae family whose sequences have been obtained in this work. The GenBank accession numbers of the sequences used to construct the phylogenetic tree are shown in the figure. Bootstrap analysis was carried out with 1,000 replicates. Bootstrap values of the branches are indicated in a 100 basis.
DISCUSSION
In this study, we have characterized PEPC from leaves of different Chenopodiaceae species, including a photosynthetic PEPC from single-cell functioning C4 species. Although more than one isoform may be present in leaves of the species under study, as previously seen in Arabidopsis (Arabidopsis thaliana), Flaveria spp., Alternanthera spp., and maize, among others (Westhoff et al., 1997; Dong et al., 1999; Sánchez and Cejudo, 2003; Gowik et al., 2006), activity on native IEF gels shows only one activity band for each species (Fig. 2). Even if there are multiple isoforms in leaves, the major activity band observed indicates that either there is one isoform that is expressed at higher levels or there is an isoform that displays higher specific activity.
Previously, it was shown with PEPC from Kranz-type C4 plants that the photosynthetic isoform is predominant in quantity and has a higher specific activity than the C3 isoform (O'Leary, 1982). In the case of S. linifolia, a C3 plant, the form of PEPC present in leaves should have a housekeeping, anaplerotic function. As is typical for C3-type PEPC, the enzyme from S. linifolia exhibited lower Km (PEP) and apparent Vmax (Table I) and did not show differential day-night sensitivity to the malate inhibitor (Table II). Even though we were not able to isolate the 5′ end of this ppc and evaluate whether a Ser or Ala residue is present in the plant enzyme conserved motif (S/A) IDAQLR, no regulatory changes in the protein were observed by using antibodies against phosphorylated PEPC (Fig. 3) or IEF analysis (Fig. 2).
With respect to H. persicum, S. eltonica, and S. richteri, the higher apparent Vmax and Km (PEP) values for the enzyme compared to S. linifolia indicate that a C4-type isoform is responsible for these differences (Table I). Using purified proteins, considerable species-to-species variability has been reported in these parameters, but C3 PEPCs always exhibited a lower Km (PEP) than their C4 counterparts (Svensson et al., 2003). Changes in apparent Vmax and Km (PEP) during the day versus night periods, as well as a decrease in malate sensitivity of up to 4 times during the day (Table II), are a consequence of regulatory phosphorylation occurring during the day. Native IEF (Fig. 2) and western blot with antibodies against phosphorylated PEPC (Fig. 3) support this conclusion.
The single-cell functioning C4 species B. sinuspersici and S. aralocaspica have the highest apparent Vmax values (Table I) among the species tested, whereas their Km (PEP) values and changes in malate sensitivity during the day and night periods (Table II) are similar to those of the PEPCs from the Kranz-type C4 species (in the case of B. sinuspersici, PEP affinity is only slightly lower than that of C3 S. linifolia). Together with native IEF (revealed by activity; Fig. 2), the use of antibodies that react with the phosphorylated amino terminal of the protein shows that PEPC from both species is subject to regulatory phosphorylation (Fig. 3), which occurs during the day and peaks around noon (Fig. 4). Future studies are needed with purified or recombinant PEPC from single-cell C4 species and related photosynthetic chenopods to provide detailed analyses of their properties.
In general, the PEPC sequences from the chenopods have the typical conserved features of plant PEPCs for residues involved in catalysis, mobile loops, and regulation. This is the case for residues from maize, R456, R759, and R773, which have been shown to be catalytically essential; residues W288, L564, and M598 in the hydrophobic pocket around PEP; E566 and D603, which bind Me2+; and the basic residues K762 and R763, involved in the binding of bicarbonate, which are all also conserved in the sequences analyzed. As in maize PEPCs, residues R498 and E493, which are responsible for the formation of the dimer-of-dimer homotetrameric structure, are also conserved in the chenopod sequences presented here. The functionally essential R647 residue in the 640GRGGTVGRGG649 motif and the H177 residue involved in protein movements directly participating in the carboxylation reaction and in Asp inhibition, as well as the H639 residue of central importance found in maize and other plant PEPCs, are also present in the chenopod PEPC sequences we studied. In addition, the conserved C-terminal QNTG motif, indispensable for maximal catalytic activity of PEPC (Dong et al., 1999), is also found (Fig. 5).
Sequence comparison of plant PEPCs revealed that C4 enzymes investigated to date, of both monocot and dicot origin, harbor a Ser residue at a position corresponding to 774 of F. trinervia PEPC, 775 of Alternanthera pungens, or 780 in maize C4-PEPC (Blässing et al., 2000; Gowik et al., 2006), whereas this position is occupied by an Ala in all nonphotosynthetic and CAM PEPCs. As expected for C4-type PEPC, the deduced amino acid sequence obtained from S. eltonica displays a Ser residue in the equivalent position, whereas the equivalent position in C3 S. linifolia PEPC has an Ala. In agreement with kinetic assays performed in this work, a Ser residue is found in the deduced sequences from B. sinuspersici and S. aralocaspica (Fig. 5). It has been proposed that the substitution of Ser for Ala at position 774 in Flaveria PEPCs or in the equivalent position in other C4 PEPCs was essential to create a fully functional C4 isoform, and that this occurred as one of the last steps in the evolutionary transformation of a C3 PEPC to the C4 isoform. The comparison of ppcA PEPCs from various Flaveria spp. indicates that, in addition to the C3 species, both the C3-C4 intermediate and the C4-like species still have an Ala at position 774 (Svensson et al., 2003).
In the case of B. sinuspersici, the deduced amino acid sequence from the cDNA shows a Ser residue in the motif of 11SIDAQLR17 of the amino-terminal region of the enzyme, which is only found in plant proteins. It is involved in the regulation of activity by light or darkness in C4 and CAM PEPCs (Lepiniec et al., 1994) through a phosphorylation/dephosphorylation process. Even though the presence of Ser in the SIDAQLR motif has been described in some PEPCs from C3 species, such as Arabidopsis (Atppc1-3; Sánchez and Cejudo, 2003), the occurrence of an Ala residue instead of a Ser residue in a position equivalent to 774 from Flaveria PEPCs seems to be a constant. Thus, molecular analysis of PEPC from the single-cell functioning C4 species indicates that these isoforms are really highly evolved C4-type proteins, supporting the biochemical data obtained. The day/night modification of the enzyme by phosphorylation/dephosphorylation selectively maintains a more active state in the light during photosynthesis, and the C4-type kinetic properties, with respect to apparent Vmax (even higher than related Kranz-type C4 species) and Km (PEP), may provide optimal function in the C4 pathway.
The clustering of PEPC sequences confirms the hypothesis that the C4 isoform of grasses could have evolved earlier than that of dicots (Fig. 6; Lepiniec et al., 1994; Gehring et al., 1998). Among the dicot families having C4 species, the Chenopodiaceae seems to be the earliest C4 lineage in accordance with the phylogenetic relationships in angiosperms (Angiosperm Phylogeny Group, 2003). The eight species of Amaranthaceae and Chenopodiaceae in the phylogeny in Figure 6 include single-cell and Kranz-type C4 and C3 species. They form a monophyletic clade, which is consistent with previous phylogenetic studies on these families (Kadereit et al., 2003; Müller and Borsch, 2005). The C4 and non-C4 PEPCs of grasses have been shown to be well differentiated, whereas it appears that, in dicotyledonous plants, C4 PEPCs are more related to non-C4 PEPCs. The phylogenetic tree constructed here is consistent with this hypothesis, as one can see well-defined branches of the C4 and C3 PEPC isoforms of the monocots maize and sorghum in the Poaceae family, whereas all PEPCs from the dicots Flaveria in the Asteraceae and Amaranthaceae s. l. family are grouped together (Fig. 6), consistent with phylogenetic studies in these families (Angiosperm Phylogeny Group, 2003).
The cluster of Amaranthaceae/Chenopodiaceae is branched into two subclades, one including Alternanthera and another including Amaranthus, Suaeda, and Bienertia. All species of Chenopodiaceae are sister to A. hypochondriacus. Suaeda and Bienertia belong to the Suaedoideae subfamily, each representing monotypic tribes Suaedeae and Bienertieae (Schütze et al., 2003; Kapralov et al., 2006). However, Amaranthus belongs to the Amaranthoideae subfamily in Amaranthaceae s. str. and Alternanthera belongs to the Gomphrenoideae subfamily in the same family (Müller and Borsch, 2005). The two single-cell functioning C4 species, B. sinuspersici and S. aralocaspica, provide a monophyletic clade for PEPC phylogeny with 100% bootstrap support. This is interesting because phylogenetic results obtained from various chloroplast and nuclear markers, including Rubisco large subunit (rbcL), maturaseK(matK), the atpB/rbcL intergenic spacer (atpB-rbcL), psbB gene, psbB-psbT intergenic spacer, psbT gene, psbT-psbN intergenic spacer, psbN gene, psbN-psbH intergenic spacer (psbB-psbH), the trnL intron, the trnL exon 2, the trnL/trnF intergenic spacer (trnL-trnF), and the tRNA-Lys intron with matK inserted in the middle of the trnK intron (matK-trnK), indicate that two types of single-cell functioning C4 photosynthesis evolved independently (Schütze et al., 2003; Müller and Borsch, 2005; Kapralov et al., 2006). However, they have very similar C4 isoforms of PEPC, suggesting their ppc-C4 gene was either derived from a common ancestor or arose independently by convergence. Both species are NAD-ME-type C4 and both have remarkably similar PEPCs. Thus, their C4 biochemistry appears very conserved, whereas they evolved independent, novel means of spatial separation of functions in C4 photosynthesis. Future phylogenetic studies of the PEPC isoforms in various species in the Suaedoideae subfamily are needed to gain insight into evolution of the ppc-C4 gene in relation to the different structural forms of C4 photosynthesis.
C4 plants have evolved independently several times from ancestral C3 plants due to selective environmental conditions. This required adaptation of enzymes for effective functioning of the C4 cycle, which involved changes in the enzyme's kinetics and regulation. C4 photosynthesis evolved much earlier in monocots than in dicots (Cerling, 1999). In the case of PEPC, C4-type isoforms are derived from non-C4 isoforms. The C4 and non-C4 PEPCs of grasses have been shown to be well differentiated, whereas it appears that, in dicotyledonous plants, C4 PEPCs are more related to non-C4 PEPCs. Gehring et al. (1998) noted that PEPC isoforms are valuable molecular markers, which can be used to visualize trends in evolution and provide a useful tool to help answer taxonomic questions. Thus, further sequencing of other PEPC isoforms from B. sinuspersici and S. aralocaspica, as well as from other species from the family, may allow deciphering of the origin and evolutionary development of C4 photosynthesis. Overall, biochemical and molecular data obtained in this work suggest that PEPCs from the single-cell functioning C4 species, B. sinuspersici and S. aralocaspica, share the same characteristics as those from Kranz-type C4 plants. Analyses carried out with Flaveria PEPCs reveal that only small changes are required to convert a C3 PEPC into C4 (Blässing et al., 2000). In addition, C4 photosynthesis is of polyphyletic origin. Thus, perhaps it is not surprising that, in the single-cell species B. sinuspersici and S. aralocaspica, evolution of C4 photosynthesis was accomplished by the expression of C4-type PEPC isoforms with development of a completely different structural means of spatial compartmentation of the C4 process. The polyphyletic origin of C4 photosynthesis implies that, in genetic terms, it was comparatively simple for C4 plants to evolve from C3 ancestors (Westhoff and Gowik, 2004). However, genetic control of development of the spatial separation of functions of C4 photosynthesis in both the Kranz and single-cell C4 systems in terrestrial plants remains to be elucidated.
MATERIALS AND METHODS
Plant Material
The species used in the study were Bienertia sinuspersici Akhani, Suaeda aralocaspica (Bunge) Freitag and Schütze (=Borszczowia aralocaspica Bunge; Schütze et al., 2003), Haloxylon persicum Bunge, Suaeda eltonica Iljin, Salsola richteri (Moq.) Karel. ex Litv., and Suaeda linifolia Pall. With the exception of S. aralocaspica, plants were grown from seeds and kept in the greenhouse under natural sunlight (summer) plus supplemental light from sodium vapor lamps (supplying a photosynthetic photon flux density of 400 μmol m−2 s−1), with a 14-h light/10-h dark photoperiod and a 25°C day/15°C night regime. The S. aralocaspica plants were kept in a growth chamber under the same conditions, except the photoperiod was 12-h light/12-h dark. Samples were taken after 5 h into the light period (day samples) or after 5 h into the dark period (night samples). After collection, the leaves were immediately frozen in liquid N2 and stored at −80°C prior to analysis.
Protein Extraction
Total protein from the different samples was extracted using a buffer containing 100 mm Tris-HCl, pH 7.3, 1 mm EDTA, 10 mm MgCl2, 15 mm β-mercaptoethanol, 20% (v/v) glycerol, 1 mm NaF, 50 mm KH2PO4, 1 mm phenylmethylsulfonylfluoride, 10 μg mL−1 leupeptin, 10 μg mL−1 chymostatin, and 10 μL/mL of protease inhibitor cocktail (Sigma) extraction buffer. The samples were ground completely in a cold mortar and centrifuged at 10,000g for 10 min at 4°C. The supernatant of crude extracts was desalted in a cold Sephadex G-25 column preequilibrated with the above buffer according to Penefsky (1977). This extract was used for activity measurements or diluted in 0.25 m Tris-HCl, pH 7.5, 2% (w/v) SDS, 0.5% (v/v) β-mercaptoethanol, and 0.1% (v/v) bromphenol blue and boiled for 2 min for SDS-PAGE. For IEF gels, the extract was diluted in 0.25 m Tris-HCl, pH 6.8, 0.05% (v/v) bromphenol blue, and 50% (v/v) glycerol. Protein concentration was determined in desalted crude extracts by the method of Bradford (1976) using Bio-Rad protein assay reagent and bovine serum albumin as standard.
Enzyme Assay
PEPC activity was determined at 30°C in a coupled reaction with malate dehydrogenase by monitoring NADH oxidation at 340 nm. The standard assay medium contained 50 mm Tris-HCl, pH 8.0, 20% (v/v) glycerol, 10 mm MgCl2, 10 mm NaHCO3, 0.15 mm NADH, 10 units of malate dehydrogenase, and PEP in a final volume of 1 mL. The reaction was started by addition of PEPC. The assay with enzyme extract was also performed in the absence of PEP to correct for any oxidation of NADH independent of PEPC. One unit of enzyme activity is defined as the amount of enzyme resulting in the consumption of 1 μmol of NADH min−1. Initial velocity studies were performed by varying the concentration of the substrate PEP around its Km while keeping the other substrate concentrations at saturating levels. The Km values of the substrates were calculated by nonlinear least-squares regression using Sigma Plot.
To determine l-malate sensitivity of PEPC, the assay was performed at pH 7.2, 20% (v/v) glycerol, and 0.10 mm or 0.55 mm PEP. l-malate was added within a range where the reaction was linear, after enzyme addition, and to encompass the inhibitor concentration causing 50% inhibition of the initial PEPC activity (I50). I50 was estimated according to Brooks (1992).
Gel Electrophoresis
SDS-PAGE was performed in 8% or 10% (w/v) polyacrylamide gels according to Laemmli (1970). Proteins were visualized with Coomassie Blue or electroblotted onto a nitrocellulose membrane for immunoblotting according to Burnette (1981). Bound antibodies were located by linking to alkaline phosphatase-conjugated goat anti-rabbit IgG according to the manufacturer's instructions (Sigma). The antibodies used for detection were the following: 1:200 anti-Amaranthus viridis PEPC (Colombo et al., 1998); 1:1,000 antiphosphorylated PEPC (the antibody was prepared in the same way as described by Ueno et al.,2000, except for the use of a synthetic phosphonopeptide as an antigen; T. Furumoto and K. Izui, unpublished data); 1:200 anti-maize (Zea mays) 62-kD NADP-ME (Saigo et al., 2004); serum against the α-subunit of NAD-ME (diluted 1:1,000) from Amaranthus hypochondriacus (Long et al., 1994); 1:10,000 against spinach (Spinacia oleracea) rbcL (provided by Dr. A. Viale). The molecular masses of the polypeptides were estimated from a plot of the log of molecular mass of marker standards versus migration distance.
Native IEF was carried out on precast polyacrylamide slab gels (pH range 5–8). Gels were prerun for 30 min at 200 V with 20 mm NaOH in the cathode and 10 mm H3PO4 in the anode, and then run for 1.5 h at a constant voltage of 200 V followed by 1.5 h at 400 V and 6°C. After electrophoresis, PEPC was detected by incubating the gel in a solution containing 50 mm Tris-HCl, pH 8.0, 20% (v/v) glycerol, 10 mm NaHCO3, 10 mm MgCl2, and 2 mm PEP for 1 h at 25°C. The gel was washed with distilled water and incubated in the dark at room temperature with Fast Violet B salt (1 mg mL−1) until red bands developed where oxaloacetic acid was produced. Excess dye was removed by rinsing with distilled water (Vidal et al., 1976).
RNA Isolation
Total RNA from leaves of B. sinuspersici, S. aralocaspica, S. eltonica, and S. linifolia was isolated from 1 g of tissue using the lithium chloride method, according to the manufacturer's instructions (Ambion). Following extraction of total RNA, a DNase treatment (DNA-free; Ambion) was performed to eliminate contamination with genomic DNA. The integrity of the RNA was verified by agarose electrophoresis. The quantity and purity of RNA were determined spectrophotometrically according to the method described by Sambrook et al. (1989).
RACE and RT-PCR
One microgram of RNA was converted into first-strand cDNA using alfalfa mosaic virus reverse transcriptase following the manufacturer's instructions (Promega). A nested PCR was performed using the forward 296F primer (5′-GGATGGGTGGTGACCGTGATGGCA-3′) and reverse M3RT primer (5′-GGTAGATGAAACCTGGTTTGTGTCC-3′) and the cDNA template generated was employed as a template. For the 3′ terminal, a nested PCR was performed using the forward 35RT primer (5′-ACCCATCTTGCCATTTTGTCTCAACC-3′) and the reverse 33RT primer (5′-TAACCGGTGTTCTGCATTCC-3′) and the cDNA template generated before was employed as template. The 5′ end of PEPC was amplified with the 5′/3′-RACE system (Roche) according to the manufacturer's instructions using gene-specific primers RACE53 (5′-GCCATCATTCTGGCTAGCAAACT-3′) for the first-strand synthesis and 296R (5′-GCCATCACGGTCACCACCCATCCA-3′) for the PCR amplification of dA-tailed cDNA. The amplified products were purified using the QIA quick extraction kit (Qiagen) and both strands fully sequenced using the ABI Prism 3730 genetic analyzer and the ABI Prism big dye terminator cycle sequencing ready reaction kit (PE Biosystems) at Washington State University sequencing facilities.
DNA and Protein Sequence Comparison and Phylogenetic Analysis
DNA sequence data were analyzed using the DNA Star sequence analysis programs. Database searches were conducted using the National Center for Biotechnology Information network version of BLAST 2.2.13 (Altschul et al., 1997). Multiple alignment of complete amino acid sequences of PEPC from different sources accessible from public databases was performed with the ClustalW multiple alignment program (Thompson et al., 1994), removing gap-only columns. This alignment was used to construct the phylogenetic tree with the neighbor-joining method (protein distance after Kimura) using the Phylip software package (Felsenstein, 1989). Bootstrap analysis was computed with 1,000 replicates and excluding positions with gaps. Trees were orientated with PEPC amino acid sequences from Escherichia coli and Arabidopsis (Arabidopsis thaliana) bacterial-type PEPC as functional outgroups.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ538352 to DQ538355.
This work was supported by the National Science Foundation (grant no. IBN–0131098 to G.E.E. and grant no. IBN–0236959 to G.E.E. and S.D.X.C.), by the United Nations Educational, Scientific and Cultural Organization-L'Oréal (grant to M.V.L.), and by the University of Tehran (project no. 6104037/1/01 to H.A.). M.V.L. and C.S.A. are Researcher Career Members of the Consejo Nacional de Investigaciones Científicas y Técnicas, Argentina.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Gerald E. Edwards (edwardsg@wsu.edu).
References
- Akhani H, Barroca J, Koteeva N, Voznesenskaya EV, Franceschi VR, Edwards GE, Ghaffari SM, Ziegler H (2005) Bienertia sinuspersici (Chenopodiaceae): a new species from southwest Asia and discovery of a third terrestrial C4 plant without Kranz anatomy. Syst Bot 30: 290–301 [Google Scholar]
- Akhani H, Trimborn P, Ziegler H (1997) Photosynthetic pathways in Chenopodiaceae from Africa, Asia and Europe with their ecological, phytogeographical and taxonomical importance. Plant Syst Evol 206: 187–221 [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreo CS, González DH, Iglesias A (1987) Higher plant phosphoenolpyruvate carboxylase: structure and regulation. FEBS Lett 213: 1–8 [Google Scholar]
- Angiosperm Phylogeny Group (2003) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc 141: 399–435 [Google Scholar]
- Blässing OE, Westhoff P, Svensson P (2000) Evolution of C4 phosphoenolpyruvate carboxylase in Flaveria, a conserved serine residue in the carboxyl-terminal part of the enzyme is a major determinant for the C4-specific characteristics. J Biol Chem 275: 27917–27923 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) Rapid and quantitative method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–252 [DOI] [PubMed] [Google Scholar]
- Brooks SPJ (1992) A simple computer program with statistical tests for the analysis of enzyme kinetics. Biotechniques 13: 906–910 [PubMed] [Google Scholar]
- Burnette WN (1981) “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem 112: 195–203 [DOI] [PubMed] [Google Scholar]
- Carter PJ, Nimmo HG, Fewson CA, Wilkings MB (1991) Circadian rhythms in the activity of a plant protein kinase. EMBO J 10: 2063–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P, Andreo CS, Edwards GE (1999) Characterization of NADP-malic enzyme from two species of Chenopodiaceae: Haloxylon persicum (C4) and Chenopodium album (C3). Phytochemistry 52: 985–992 [Google Scholar]
- Cerling TE (1999) Paleorecords of C4 plants and ecosystems. In RF Sage, RK Monson, eds, C4 Plant Biology. Academic Press, London, pp 445–469
- Chollet R, Vidal J, O'Leary MH (1996) Phosphoenolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 273–298 [DOI] [PubMed] [Google Scholar]
- Colombo SL, Andreo CS, Chollet R (1998) The interaction of shikimic acid and protein phosphorylation with PEP carboxylase from the C4 dicot Amaranthus viridis. Phytochemistry 48: 55–59 [DOI] [PubMed] [Google Scholar]
- Dong L, Patil S, Condon SA, Has EJ, Chollet R (1999) The conserved C-terminal tetrapeptide of sorghum C4 phosphoenolpyruvate carboxylase is indispensable for maximal catalytic activity, but not for homotetramer formation. Arch Biochem Biophys 371: 124–128 [DOI] [PubMed] [Google Scholar]
- Duff SMG, Andreo CS, Pacquit V, Lepiniec L, Sarath G, Codon SA, Vidal J, Gadal P, Chollet R (1995) Kinetic analysis of the non-phosphorylated, in vitro phosphorylated, and phosphorylation-site-mutant (Asp8) forms of the intact recombinant C4 phosphoenolpyruvate carboxylase from sorghum. Eur J Biochem 228: 92–95 [PubMed] [Google Scholar]
- Echeverria C, Vidal J (2003) The unique phosphoenolpyruvate carboxylase kinase. Plant Physiol Biochem 41: 541–547 [DOI] [PubMed] [Google Scholar]
- Edwards GE, Franceschi VR, Voznesenskaya EV (2004) Single-cell C4 photosynthesis versus the dual-cell (Kranz) paradigm. Annu Rev Plant Biol 55: 173–196 [DOI] [PubMed] [Google Scholar]
- Edwards GE, Walker DA (1983) C3, C4: Mechanisms, and Cellular and Environmental Regulation of Photosynthesis. Blackwell Scientific, Oxford
- Felsenstein J (1989) PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 5: 164–166 [Google Scholar]
- Fontaine V, Hartwell J, Jenkins G, Nimmo HG (2002) Arabidopsis thaliana contains two phosphoenolpyruvate carboxylase kinase genes with different expression patterns. Plant Cell Environ 25: 115–122 [Google Scholar]
- Gehring HH, Heute V, Kluge M (1998) Toward a better knowledge of the molecular evolution of phosphoenolpyruvate carboxylase by comparison of partial cDNA sequences. J Mol Evol 46: 107–114 [DOI] [PubMed] [Google Scholar]
- Gowik U, Engelman S, Blässing OE, Raghavendra AS, Westhoff P (2006) Evolution of C4 phosphoenolpyruvate carboxylase in the genus Alternanthera: gene families and the enzymatic characteristics of the C4 isoenzyme and its orthologues in C3 and C3/C4 Alternanthera. Planta 223: 359–368 [DOI] [PubMed] [Google Scholar]
- Gupta SK, Ku MSB, Lin J-H, Zhang D, Edwards GE (1994) Light/dark modulation of phosphoenolpyruvate carboxylase in C3 and C4 species. Photosynth Res 42: 133–143 [DOI] [PubMed] [Google Scholar]
- Hartwell J, Aideen G, Nimmo GA, Wilkins MB, Jenkins GI, Nimmo HG (1999) Phosphoenolpyruvate carboxylase kinase is a novel protein kinase regulated at the level of expression. Plant J 20: 333–342 [DOI] [PubMed] [Google Scholar]
- Hartwell J, Smith LH, Wilkins MB, Jenkins GI, Nimmo HG (1996) Higher plant phosphoenolpyruvate carboxylase kinase is regulated at the level of translatable mRNA in response to light or a circadian rhythm. Plant J 10: 1071–1078 [Google Scholar]
- Jiao JA, Chollet R (1991) Posttranslation regulation of phosphoenolpyruvate carboxylase in C4 and Crassulacean acid metabolism plants. Plant Physiol 95: 981–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadereit G, Borsch T, Weising K, Freitag H (2003) Phylogeny of Amaranthaceae and Chenopodiaceae and the evolution of C4 photosynthesis. Int J Plant Sci 164: 959–986 [Google Scholar]
- Kai Y, Matsumura H, Izui K (2003) Phosphoenolpyruvate carboxylase: three-dimensional structure and molecular mechanisms. Arch Biochem Biophys 414: 170–179 [DOI] [PubMed] [Google Scholar]
- Kapralov MV, Akhani H, Voznesenskaya E, Edwards G, Franceschi VR, Roalson EH (2006) Phylogenetic relationships in the Salicornioideae/Suaedoideae/Salsoloideae s.l. (Chenopodiaceae) clade and a clarification of the phylogenetic position of Bienertia and Alexandra using multiple DNA sequence datasets. Syst Bot 31: 571–585 [Google Scholar]
- Krall JP, Edwards GE (1993) PEP carboxylases from two C4 species of Panicum with markedly different susceptibilities to cold inactivation. Plant Cell Physiol 34: 1–11 [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lepiniec L, Vidal J, Chollet R, Gadal P, Crétin C (1994) Phosphoenolpyruvate carboxylase: structure, regulation and evolution. Plant Sci 99: 111–124 [Google Scholar]
- Long JJ, Wang J-L, Berry JO (1994) Cloning and analysis of the C4 NAD-dependent malic enzyme of amaranth mitochondria. J Biol Chem 269: 2827–2833 [PubMed] [Google Scholar]
- Müller K, Borsch T (2005) Phylogenetics of Amaranthaceae using matK/trnK sequence data—evidence from parsimony, likelihood and Bayesian approaches. Ann Mo Bot Gard 92: 66–102 [Google Scholar]
- Nimmo GA, McNaughton GA, Fewson CA, Wilkins MB, Nimmo HG (1987) Changes in the kinetic properties and phosphorylation state of phosphoenolpyruvate carboxylase in Zea mays leaves in response to light and dark. FEBS Lett 213: 18–22 [Google Scholar]
- Nimmo HG (2003) Control of the phosphorylation of phosphoenolpyruvate carboxylase in higher plants. Arch Biochem Biophys 414: 189–196 [DOI] [PubMed] [Google Scholar]
- Ogawa N, Kai T, Yabuta N, Izui K (1997) Phosphoenolpyruvate carboxylase of maize leaves: an improved method for purification and reduction of the inhibitory effect of malate by ethylene glycol and bicarbonate. Plant Cell Physiol 38: 76–80 [Google Scholar]
- O'Leary MH (1982) Phosphoenolpyruvate carboxylase: an enzymologist's view. Annu Rev Plant Physiol 33: 297–315 [Google Scholar]
- Penefsky H (1977) The reversible binding of Pi by beef heart mitochondrial adenosine triphosphatase. J Biol Chem 252: 2891–2899 [PubMed] [Google Scholar]
- Pyankov VI, Voznesenskaya EV, Kuz'min AN, Ku MSB, Ganko E, Franceschi VR, Black CC Jr, Edwards GE (2000) Occurrence of C3 and C4 photosynthesis in cotyledons and leaves of Salsola species (Chenopodiaceae). Photosynth Res 63: 69–84 [DOI] [PubMed] [Google Scholar]
- Saigo M, Bologna F, Maurino VG, Detarsio E, Andreo CS, Drincovich MF (2004) Maize recombinant non-C4 NADP-malic enzyme: a novel dimeric malic enzyme with high specific activity. Plant Mol Biol 55: 97–107 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning. In C Nolan, ed, A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sánchez R, Cejudo FJ (2003) Identification and expression analysis of a gene encoding a bacterial-type phosphoenolpyruvate carboxylase from Arabidopsis and rice. Plant Physiol 132: 949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütze P, Freitag H, Weising K (2003) An integrated molecular and morphological study of the subfamily Suaedoideae Ulbr. (Chenopodiaceae). Plant Syst Evol 239: 257–286 [Google Scholar]
- Svensson P, Blässing OE, Westhoff P (2003) Evolution of C4 phosphoenolpyruvate carboxylase. Arch Biochem Biophys 414: 180–188 [DOI] [PubMed] [Google Scholar]
- Taybi T, Patil S, Chollet R, Cushman JC (2000) A minimal serine/threonine protein kinase circadianly regulates phosphoenolpyruvate carboxylase activity in Crassulacean acid metabolism-induced leaves of the common ice plant. Plant Physiol 123: 1471–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida Y, Furumoto T, Izumida A, Hata S, Izui K (2001) Phosphoenolpyruvate carboxylase kinase involved in C4 photosynthesis in Flaveria trinervia: cDNA cloning and characterization. FEBS Lett 507: 318–322 [DOI] [PubMed] [Google Scholar]
- Ueno Y, Imanari E, Emura J, Yoshizawa-Kumagaye K, Nakajima K, Inami K, Shiba T, Sakakibara H, Sugiyama T, Izui K (2000) Immunological analysis of the phosphorylation state of maize C4-form phosphoenolpyruvate carboxylase with specific antibodies raised against a synthetic phosphorylated peptide. Plant J 21: 17–26 [DOI] [PubMed] [Google Scholar]
- Vidal J, Cavalié GY, Gadal P (1976) Etude de la phosphoenolpyruvate carboxylase du Haricot et du Sorgho par électrophorèse sur gel de polyacrylamide. Plant Sci Lett 7: 265–270 [Google Scholar]
- Vidal J, Chollet R (1997) Regulatory phosphorylation of C4 PEP carboxylase. Trends Plant Sci 2: 230–237 [Google Scholar]
- Voznesenskaya EV, Edwards GE, Kiirats O, Artyusheva EG, Franceschi VR (2003) Development of biochemical specialization and organelle partitioning in the single celled C4 system in leaves of Borszczowia aralocaspica (Chenopodiaceae). Am J Bot 90: 1669–1680 [DOI] [PubMed] [Google Scholar]
- Voznesenskaya EV, Franceschi VR, Kiirats O, Artyusheva EG, Freitag H, Edwards GE (2002) Proof of C4 photosynthesis without Kranz anatomy in Bienertia cycloptera (Chenopodiaceae). Plant J 31: 649–662 [DOI] [PubMed] [Google Scholar]
- Voznesenskaya EV, Franceschi VR, Kiirats O, Freitag H, Edwards GE (2001) Kranz anatomy is not essential for terrestrial C4 plant photosynthesis. Nature 414: 543–546 [DOI] [PubMed] [Google Scholar]
- Voznesenskaya EV, Koteyeva NK, Choung SD, Akhani H, Edwards GE, Franceschi V (2005) Differentiation of cellular and biochemical features of the single-cell syndrome during leaf development in Bienertia cycloptera (Chenopodiaceae). Am J Bot 92: 1784–1795 [DOI] [PubMed] [Google Scholar]
- Westhoff P, Gowik U (2004) Evolution of C4 phosphoenolpyruvate carboxylase. Genes and proteins: a case study with the genus Flaveria. Ann Bot (Lond) 93: 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhoff P, Svensson P, Ernst K, Bläsing O, Burscheidt J, Stockhaus J (1997) Molecular evolution of C4 phosphoenolpyruvate carboxylase in the genus Flaveria. Aust J Plant Physiol 24: 429–436 [Google Scholar]