Abstract
Auxin and gibberellins (GAs) overlap in the regulation of multiple aspects of plant development, such as root growth and organ expansion. This coincidence raises questions about whether these two hormones interact to regulate common targets and what type of interaction occurs in each case. Auxins induce GA biosynthesis in a range of plant species. We have undertaken a detailed analysis of the auxin regulation of expression of Arabidopsis (Arabidopsis thaliana) genes encoding GA 20-oxidases and GA 3-oxidases involved in GA biosynthesis, and GA 2-oxidases involved in GA inactivation. Our results show that auxin differentially up-regulates the expression of various genes involved in GA metabolism, in particular several AtGA20ox and AtGA2ox genes. Up-regulation occurred very quickly after auxin application; the response was mimicked by incubations with the protein synthesis inhibitor cycloheximide and was blocked by treatments with the proteasome inhibitor MG132. The effects of auxin treatment reflect endogenous regulation because equivalent changes in gene expression were observed in the auxin overproducer mutant yucca. The results suggest direct regulation of the expression of GA metabolism genes by Aux/IAA and ARF proteins. The physiological relevance of this regulation is supported by the observation that the phenotype of certain gain-of-function Aux/IAA alleles could be alleviated by GA application, which suggests that changes in GA metabolism mediate part of auxin action during development.
The plant hormones GA and auxin overlap in the regulation of many developmental programs, most of them related to cell expansion and differentiation. For instance, both hormones seem to participate in the signals that trigger early fruit development in response to fertilization in many different species, including pea (Pisum sativum) and Arabidopsis (Arabidopsis thaliana; García-Martínez et al., 1991; Reinecke et al., 1995; Rodrigo et al., 1997; Vivian-Smith and Koltunow, 1999), as well as in the promotion of organ expansion, such as the Arabidopsis hypocotyl (Smalle et al., 1997; Cowling and Harberd, 1999; Collett et al., 2000) and pea stems (Ross and O'Neill, 2001), or in the promotion of Arabidopsis root growth (Fu and Harberd, 2003). Although it is possible that auxin and GAs exert their action through independent mechanisms, an interaction between these two hormones (and other convergent signals) has been demonstrated. This is the case of pericarp growth in pea fruits, which is synergistically induced by GAs and auxin (Ozga and Reinecke, 1999), or hypocotyl and stomata development, for which GAs are absolutely required, but their action is modulated by auxins and ethylene (Saibo et al., 2003).
The interaction between GAs and auxin has been intensively investigated recently, revealing at least two mechanisms. In roots, auxin signaling has been shown to induce the degradation of the negative GA-signaling element RGA, thereby promoting GA signaling and root growth (Fu and Harberd, 2003). In addition, there are numerous reports of the induction of GA biosynthesis by auxin in different systems. For example, in pea and tobacco (Nicotiana tabacum), the concentration of active GAs is lower in stems of plants that have been decapitated (and thus deprived of the apical source of auxins) compared with intact plants, whereas application of auxin to the apex restored wild-type levels of GAs (Ross et al., 2000; Wolbang and Ross, 2001). Similarly, auxin 4-chloroindole-3-acetic acid stimulated GA biosynthesis in pea pericarp (van Huizen, 1995), and auxin induction was proposed to be necessary for pericarp expansion (Ozga et al., 2003).
The regulation of GA biosynthesis by auxin has been attributed to changes in the expression level of the genes encoding GA biosynthetic or deactivating enzymes. The most important regulatory steps that determine the concentration of the active GA species (GA1 or GA4) are the final biosynthetic reactions, catalyzed by GA 20-oxidases (GA20ox) and GA 3-oxidases (GA3ox), and deactivation reactions catalyzed by GA 2-oxidases (GA2ox; Hedden and Phillips, 2000). The balance between biosynthesis and deactivation is also regulated by a homeostatic mechanism involving feedback inhibition by GAs on the expression of GA20ox and GA3ox genes and feed-forward promotion of GA2ox expression (Hedden and Phillips, 2000). Although stimulation of GA biosynthesis by auxin has been correlated with up-regulation of GA biosynthesis genes in all systems studied, the particular target genes for this regulation are different. Whereas decapitation in pea dramatically reduces the expression of a GA3ox gene in expanding internodes (Ross et al., 2000) and exogenous auxin treatments affect the expression of GA3ox and GA2ox genes in a dose-dependent manner (O'Neill and Ross, 2002), auxin induces both GA20ox and GA3ox expression in pea fruit pericarp (van Huizen et al., 1997; Ngo et al., 2002; Ozga et al., 2003), or only GA20ox in tobacco stems (Wolbang and Ross, 2001). Although the precise targets seem to vary between species and tissues, auxin regulation of the expression of genes encoding GA biosynthetic enzymes is ancient in evolutionary terms, as shown by the fact that auxin also stimulates GA production in the monocot barley (Hordeum vulgare) through the up-regulation of a GA3ox gene and the down-regulation of a GA2ox gene (Wolbang et al., 2004).
The mechanism of the auxin-signaling pathway allows the plant cell to transform very rapidly changes in auxin concentration into changes in gene expression. Upon auxin binding, the F-box-containing auxin receptors accelerate the 26S proteasome-dependent degradation of short-lived transcriptional repressors, Aux/IAA, which in turn allows ARF transcription factors to modulate gene expression by binding to the promoters of auxin-regulated genes (Dharmasiri et al., 2005a, 2005b; Kepinski and Leyser, 2005). Aux/IAA proteins exert their action by binding and sequestering the ARF transcription factors into nonactive complexes. In Arabidopsis, the Aux/IAA and ARF families contain 29 and 22 members, respectively (Hagen and Guilfoyle, 2002; Remington et al., 2004). The different combinations of these two classes of proteins in different tissues and organs and under different physiological contexts contribute to diversify the pervasive auxin control of gene expression.
A clearer picture of the molecular mechanism by which auxin regulates GA biosynthesis and inactivation may be obtained by answering the following questions. (1) Given that the genes encoding GA biosynthesis are regulated by both auxin and GA, is there a functional connection between these two processes? (2) Is auxin regulation of GA metabolism genes mediated directly by Aux/IAA-ARF auxin signaling elements? We have addressed these issues by systematically analyzing the expression of GA metabolism genes in Arabidopsis seedlings in response to auxin. We found that the magnitude and kinetics of gene activation by auxin are different, depending on the particular GA metabolism gene under study, and that activation by auxin also occurs in the absence of GA-signaling elements that modify the expression of these genes in the GA homeostatic mechanism. These results indicate that regulation of GA metabolism by auxin and GAs occurs through independent pathways. The fast induction by auxin occurs in the absence of de novo protein synthesis and is reduced by the proteasome inhibitor MG132, suggesting that it is dependent on the rapid turnover of short-lived Aux/IAA repressors of auxin signaling. In fact, several mutants harboring dominant versions of these repressors are defective in the auxin up-regulation of the GA metabolism genes, indicating a direct involvement of these signaling elements in this control. Furthermore, certain phenotypes of these mutants could be rescued by GA treatment.
RESULTS
Auxin Up-Regulates GA Metabolism Genes
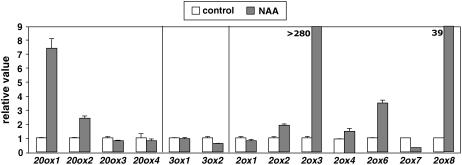
Control of expression of GA metabolism genes by auxin has been described for several species. To investigate whether this control also occurs in Arabidopsis, we analyzed changes in transcript levels of GA metabolism genes by quantitative reverse transcription (RT)-PCR in light-grown seedlings subject to exogenous auxin treatment. In Arabidopsis, the later regulatory enzymes in the GA pathway are encoded by small gene families: GA20ox are encoded by at least five genes (AtGA20ox1–AtGA20ox5); GA3ox are encoded by at least four genes (AtGA3ox1–AtGA3ox4); and GA2ox are encoded by at least seven genes (AtGA2ox1–AtGA2ox8, with AtGA2ox5 a pseudogene; Hedden et al., 2002; Schomburg et al., 2003). Among these genes, we were able to detect expression of 13 of them in seedlings (Fig. 1; see Supplemental Fig. S1 for comparison of gene expression levels between genes). Interestingly, we found contrasting effects of auxin application on the expression of these genes. For example, exogenous 1-naphthalene acetic acid (NAA) caused an increase in transcript levels for AtGA20ox1 (GA5) and AtGA20ox2 genes after 24 h, whereas it had no effect on the expression of the other members of the same family, AtGA20ox3 and AtGA20ox4, or on the expression of AtGA3ox1 (GA4) and AtGA3ox2. Furthermore, Arabidopsis AtGA2ox2, AtGA2ox3, AtGA2ox6, and AtGA2ox8 were up-regulated by NAA treatments after 24 h. This survey suggests that auxin differentially regulates the expression of several genes involved in GA metabolism in Arabidopsis light-grown seedlings.
Figure 1.
Transcriptional regulation of GA metabolism genes by auxin. Six-day-old wild-type Col-0 Arabidopsis seedlings were treated with 50 μm NAA or mock treated for 24 h. mRNA levels of GA metabolism genes were determined by quantitative RT-PCR as described in “Materials and Methods” and normalized against transcript levels of EF1-α. Transcript level in the control sample for each gene was set to 1, and level in the treated sample was calculated relative to the corresponding control value. Error bars represent sd.
Auxin Effect on GA Biosynthesis Genes Does Not Require the DELLA Proteins GAI and RGA
GA metabolism is tightly controlled by GA through negative feedback regulation of GA20ox and GA3ox expression and positive feed-forward regulation of GA2ox expression (Hedden and Phillips, 2000). For instance, it has been reported that reduced GA levels or signaling results in increased transcript levels of the Arabidopsis AtGA3ox1 gene (Dill et al., 2001), whereas increased GA levels or signaling causes the opposite effect. The feedback mechanism operates to control expression of all members of the GA20ox and GA3ox families analyzed in light-grown seedlings (Supplemental Fig. S2). Continuous exposure to 0.5 μm of the GA biosynthesis inhibitor paclobutrazol, which resulted in small, dark-green seedlings (data not shown), caused an increase in transcript levels for GA20ox and GA3ox genes. It reduced transcript levels of AtGA2ox4, AtGA2ox6, AtGA2ox7, and AtGA2ox8, but, unexpectedly, it also resulted in an increase in mRNA levels for AtGA2ox2 and AtGA2ox3. In addition, treatment of control seedlings with 100 μm GA3 for 1 h resulted in a strong reduction of mRNA levels of genes encoding GA20ox. The effect on transcript levels of genes encoding GA3ox and GA2ox was mild, resulting in a slight reduction and increase, respectively.
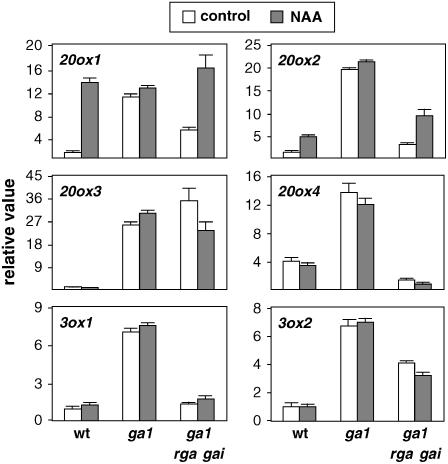
Significantly, expression of the GA biosynthesis genes AtGA20ox1 and AtGA20ox2 is up-regulated in response to at least two inputs: reduced GA levels and increased auxin levels. This raises the question as to whether these two inputs use the same or independent mechanisms to control the expression of these two target genes. Although the lack of correlation between up-regulation by auxin and feedback regulation by GAs for certain GA metabolism genes suggests that these mechanisms are independent, interaction may still occur specifically for those two genes that are up-regulated by both signals. This model for GA signaling proposes that the feedback mechanism operates through the activity of DELLA proteins (Fleet and Sun, 2005), such that stabilization of these proteins by reduced GA levels or signaling results in the increase in the mRNA level of the GA biosynthetic genes. For example, the high level of expression of the AtGA3ox1 gene observed in the Arabidopsis GA-deficient mutant ga1-3 is completely reversed by loss-of-function mutations in the DELLA genes GAI and RGA (Dill and Sun, 2001).
Our expression analysis shows that GAI and RGA mediate, to different extents, feedback regulation of AtGA20ox1, AtGA20ox2, AtGA20ox4, and AtGA3ox1 because the elevated mRNA levels of these genes in the GA-deficient ga1-3 were reduced in ga1-3 gai-t6 rga-24 triple-mutant seedlings (Fig. 2), which lack GAI and RGA activity (Dill and Sun, 2001; King et al., 2001). However, other members of the DELLA family may also have a role together with GAI and/or RGA in the feedback control of AtGA20ox3 and AtGA3ox2. More importantly, induction of AtGA20ox1 and AtGA20ox2 by auxin was as efficient in the absence of DELLA proteins GAI and RGA as in control seedlings (Fig. 2), which indicates that regulation by auxin can be separated from feedback regulation by GAs and thus occurs through an independent mechanism. It is not clear why auxin treatments were not efficient in ga1-3 seedlings; one likely explanation might be masking of the auxin induction by an already saturated response imposed by the feedback regulation.
Figure 2.
Transcriptional regulation of GA metabolism genes by auxin in GA-signaling mutants. Six-day-old wild-type Ler Arabidopsis seedlings and the GA mutants ga1-3 and ga1-3 rga-24 gai-t6 were treated with 50 μm NAA or mock treated for 24 h. mRNA levels were analyzed as explained in the Figure 1 legend. Transcript level in wild-type control sample was set to 1 for each gene, and the levels in all other samples were calculated relative to this value. Error bars represent sd.
Auxin Signaling Directly Controls Expression of Several GA Metabolism Genes
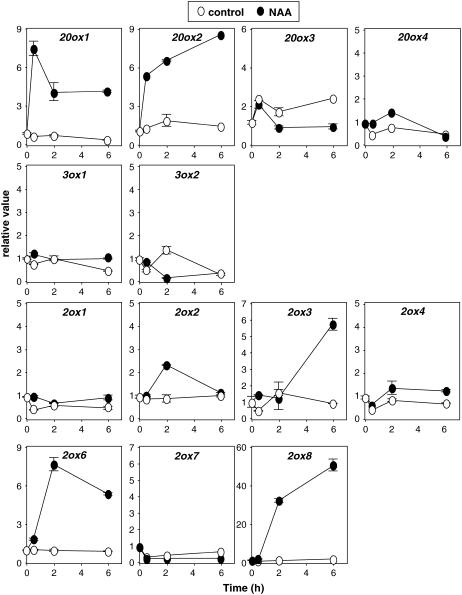
Auxin has a broad effect on gene expression (Goda et al., 2004; Nemhauser et al., 2004), and time-course analyses show that induction of primary targets can occur within minutes (Hagen and Guilfoyle, 1985; Goda et al., 2004). This raises the possibility that our 24-h treatment would have missed early transient inductions. Hence, we carried out a time-course experiment to study the kinetics of the induction of GA metabolism genes by NAA treatment during a 6-h period (Fig. 3). As expected, the magnitude of the response and the kinetics of induction varied from gene to gene, but short-term treatments with 50 μm NAA broadly corroborated results obtained with the 24-h treatment. AtGA20ox1 and AtGA20ox2, as well as AtGA2ox2, AtGA2ox6, and AtGA2ox8, were quickly up-regulated by NAA, some of them within the first 30 min. AtGA2ox3 transcript levels increased after a lag period of at least 2 h.
Figure 3.
Time course of auxin up-regulation of GA metabolism genes. Six-day-old wild-type Col-0 Arabidopsis seedlings were treated with 50 μm NAA or mock treated for the indicated times. Transcript levels of GA metabolism genes were analyzed as explained in the Figure 1 legend. Transcript level at time point 0 was set to 1 for each gene, and all other values were calculated relative to this value. Error bars represent sd.
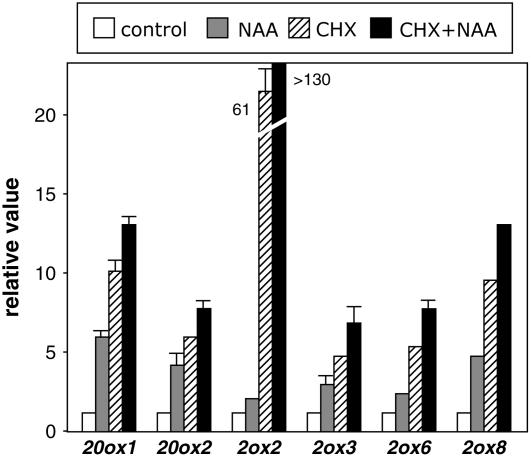
The fast up-regulation of these genes resembles the behavior of direct targets of auxin signaling. Based on this model for auxin signaling, we would predict that one or more of the short-lived Aux/IAA proteins would be involved in repressing, through physical interaction with ARF transcription factors, the ARF-mediated auxin induction of these genes (Dharmasiri et al., 2005a; Kepinski and Leyser, 2005). Preventing de novo protein synthesis must cause rapid depletion of Aux/IAA repressors in the cell, thus activating ARF-regulated genes. As shown in Figure 4, cycloheximide treatment resulted in high mRNA levels of the GA metabolism genes regulated by auxin. This result suggests that Aux/IAA proteins present in the cell before treatment might mediate the regulation of GA metabolism genes by auxin, and this is supported by the observation that NAA potentiated the effect of cycloheximide (Fig. 4). The transcript levels of the non-auxin-regulated genes AtGA3ox1, AtGA3ox2, and AtGA2ox1 slightly increased in response to cycloheximide treatment (data not shown), suggesting the activity of a labile repressor in the control of their basal expression, which may also contribute to the cycloheximide effect seen on GA metabolism genes regulated by auxin. In addition, impairment of Aux/IAA degradation by inhibition of the 26S proteasome with MG132 (Gray et al., 2001; Ouellet et al., 2001) prevented induction of GA metabolism genes by auxin (Supplemental Fig. S3). Treatment with MG132 in the absence of exogenous auxin did not alter expression levels of AtGA20ox1, AtGA20ox2, AtGA2ox2, and AtGA2ox6, suggesting that any effect on DELLA stability, which is also controlled by the proteasome pathway (Fu et al., 2002), is not sufficient to alter basal expression levels of these genes, and therefore the effect of the inhibitor on expression of GA metabolism genes is likely to be via the auxin pathway. Only in the case of AtGA2ox3 could the increase in expression after MG132 treatment be caused by stabilization of DELLA proteins because this gene was also up-regulated by paclobutrazol (Supplemental Fig. S2). The relatively different magnitude of the effect caused by MG132 on the induction of these genes may reflect different responsiveness of the promoters to the activity of the Aux/IAA proteins. Taken together, these results strongly suggest that some GA metabolism genes may represent primary targets in response to auxin, and that this induction may be regulated by Aux/IAA-ARF dimers.
Figure 4.
Effect of inhibitors of protein synthesis on the up-regulation of GA metabolism genes by auxin. Six-day-old wild-type Col-0 Arabidopsis seedlings were treated for 1 h with either 50 μm NAA, 10 μm cycloheximide (CHX), 50 μm NAA + 10 μm CHX, or mock treated. Transcript levels of GA metabolism genes were analyzed as explained in the Figure 1 legend. Transcript level in the control sample for each gene was set to 1, and level in the treated sample was calculated relative to the control value. Error bars represent sd.
Expression of GA Metabolism Genes Is Regulated by Aux/IAA-ARF Auxin-Signaling Elements
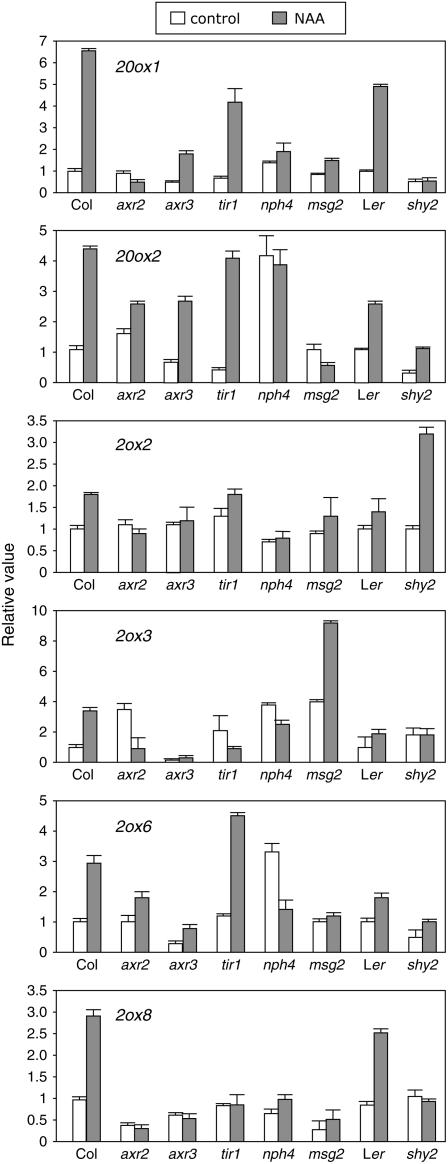
To confirm the involvement of Aux/IAA proteins in the control of GA metabolism genes by auxin, we analyzed the response of these genes to NAA treatments in mutant lines harboring gain-of-function alleles of some Aux/IAA genes: axr2-1/iaa7 (Nagpal et al., 2000), axr3-1/iaa17 (Leyser et al., 1996), msg2-1/iaa19 (Tatematsu et al., 2004), and shy2-2/iaa3 (Tian and Reed, 1999). In general, all gain-of-function alleles of IAA genes tested showed defects in the induction by auxin of at least one of the GA metabolism genes analyzed (Fig. 5). Among these genes, AtGA20ox1 and AtGA2ox6 were clearly affected by all mutants tested. However, certain mutants showed the opposite phenotype. For instance, shy2-2 rendered AtGA2ox2 hyperresponsive to auxin and msg2-1 showed increased basal levels of AtGA2ox3. This behavior is not unexpected because it has been reported that gain-of-function alleles of Aux/IAA genes cause hypersensitivity and reduced sensitivity depending on the phenotype tested (Leyser et al., 1996; Collett et al., 2000).
Figure 5.
Involvement of Aux/IAA-ARF signaling elements in the regulation of GA metabolism genes by auxin. Six-day-old wild-type Col-0 and Ler Arabidopsis seedlings and several auxin-related mutants were treated with 50 μm NAA or mock treated for 1 h. mRNA levels were analyzed as explained in the Figure 1 legend. Transcript levels in the wild-type control samples were set to 1 for each gene, and the levels in all other samples were calculated relative to this value. Error bars represent sd.
The involvement of ARF transcription factors was tested for NPH4/ARF7 by analyzing NAA induction in nph4-1 null mutant seedlings (Harper et al., 2000; Fig. 5). NPH4/ARF7 seems to be involved in the regulation of all the auxin-inducible genes, in some cases controlling NAA induction and in other cases regulating basal transcript levels. It has been shown that nph4-1 null mutants resemble msg2-1 dominant mutants regarding auxin regulation of differential hypocotyl growth; moreover, both proteins interact physically, indicating that they act together in this control (Tatematsu et al., 2004). Although the behavior of AtGA20ox1 and AtGA2ox2 was similar in nph4-1 and msg2-1 seedlings—indicating that MSG2/IAA19 and NPH4/ARF7 may act together in the regulation of these two genes by auxin—their effects on the expression of the other genes were both qualitatively and quantitatively different, suggesting that other Aux/IAA and ARFs must also be involved.
Consistent with the hypothesis that Aux/IAA-ARF pairs direct the regulation of GA metabolism genes by auxin, loss of function of TIR1, a member of the small family of F-box proteins that act as auxin receptors (Dharmasiri et al., 2005a, 2005b; Kepinski and Leyser, 2005), caused mild and strong insensitivity toward auxin induction of AtGA20ox1 and AtGA2ox3/AtGA2ox8, respectively (Fig. 5). The response of the other genes in the mutant was very similar to the wild type, most probably because of redundancy with other members of the auxin receptors family (Dharmasiri et al., 2005b).
In summary, these results suggest that auxin controls GA metabolism gene expression through a highly branched signaling network involving different Aux/IAA-ARF elements.
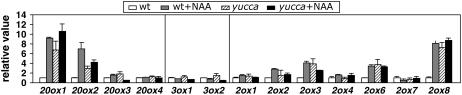
Endogenous Auxins Up-Regulate GA Metabolism Genes
A connection has been clearly established between auxin and GA metabolism genes in the context of transient, exogenous auxin applications. To investigate whether this regulation reflects a mechanism that operates in response to changes in endogenous auxin concentration, we studied steady-state mRNA levels of GA metabolism genes in the auxin-overproducing mutant yucca (Zhao et al., 2001). As shown in Figure 6, only the genes that responded to transient NAA treatments showed increased transcript levels in yucca seedlings compared to the wild type. Besides, exogenous NAA produced little or no further increase in expression of these genes in the yucca mutant, suggesting that exogenous auxin treatments and endogenous auxin act through the same mechanism.
Figure 6.
Expression of GA metabolism genes in plants adapted to auxin overproduction. Six-day-old Arabidopsis seedlings of wild-type Col-0 and the auxin-overproducing mutant yucca were treated with 50 μm NAA or mock treated for 1 h. mRNA levels were analyzed as explained in the Figure 1 legend. Transcript levels in the wild-type control samples were set to 1 for each gene, and the levels in all other samples were calculated relative to this value. Error bars represent sd.
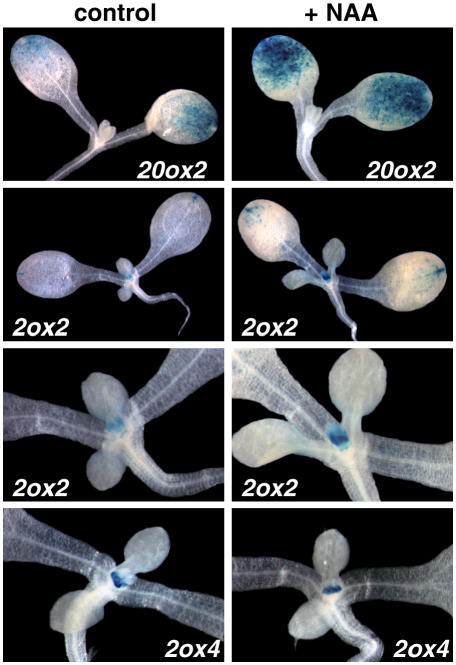
Auxin Regulation of GA Metabolism Genes Is Tissue Specific
To gain insight into the actual localization of the auxin regulation of GA metabolism gene expression, we investigated the spatial regulation of some GA metabolism genes in response to auxin treatment by using reporter-gene fusions (Fig. 7). AtGA20ox2 and AtGA2ox2 reporter genes showed complementary expression patterns in cotyledons of control seedlings; AtGA2ox2 was also expressed in the shoot apical region (Jasinsky et al., 2005). More importantly, enhanced expression of these reporter gene fusions after auxin treatment was confined basically to the same tissues. This suggests that the effect of auxin on GA metabolism may be different, enhancing GA biosynthesis or inactivation, depending on the tissue. As expected, expression of the AtGA2ox4 reporter gene, which is also expressed at the shoot apical region (Jasinsky et al., 2005), did not change after auxin treatment.
Figure 7.
Expression of promoter-GUS fusions for several genes involved in GA metabolism. Seedlings were grown in the light for 7 d and then treated with 50 μm NAA or mock treated for 1 h and then fixed and stained for GUS activity as described in “Materials and Methods.”
GAs Partially Mediate the Action of Auxin in Seedlings
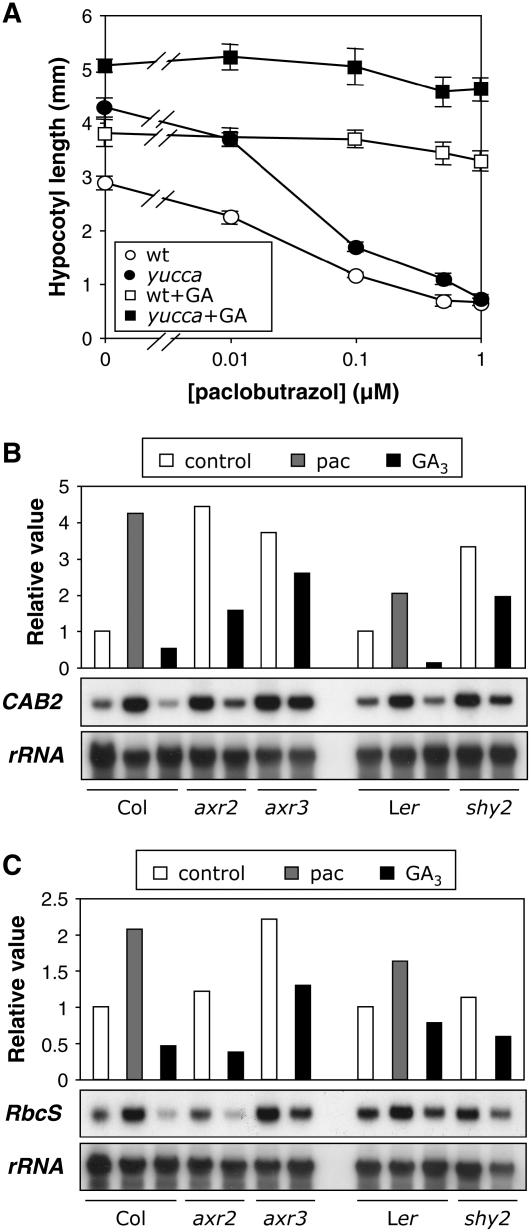
The observation that the expression of GA metabolism genes responds to changes in endogenous auxin concentration and that the regulation is tissue specific prompted the question of whether this regulation is physiologically relevant. Indeed, the exaggerated hypocotyl elongation provoked by auxin overproduction in the yucca mutant was completely dependent on GA biosynthesis, as observed in the dose-response curve of hypocotyl growth relative to increasing concentrations of paclobutrazol (Fig. 8A); the paclobutrazol effect was completely reverted by simultaneous application of GA3. In a similar way, the enhanced hypocotyl growth favored by the stimulation of auxin synthesis at higher temperatures (Gray et al., 1998) was prevented when GA biosynthesis was impaired (data not shown).
Figure 8.
Effect of GAs on the phenotype of auxin-related mutants. A, Long hypocotyl of light-grown yucca mutants was reduced by paclobutrazol treatment. Wild-type and yucca seedlings were grown in different concentrations of paclobutrazol for 7 d. Measurements are average of 10 to 15 seedlings ± sd per treatment. B, Expression level of CAB2 in 7-d-old seedlings grown in darkness. C, Expression level of RbcS in 7-d-old seedlings grown in darkness.
On the other hand, the short hypocotyl and the de-etiolation phenotypes shown by several gain-of-function mutants in Aux/IAA genes when grown in darkness (Leyser et al., 1996; Kim et al., 1998; Nagpal et al., 2000) are similar to the phenotype of dark-grown GA-deficient mutants (Alabadí et al., 2004). Exogenous application of GA3 to dark-grown axr2-1, axr3-1, and shy2-2 mutants partially compensated for the defect in hypocotyl elongation, but did not compensate for cotyledon opening (data not shown). Additionally, GA3 treatment partially rescued the molecular phenotype of CAB2 and RbcS observed in these mutants when grown in darkness (Fig. 8, B and C). These results suggest that at least part of the phenotype caused by auxin insensitivity in these mutants is mediated by a deficiency in GA biosynthesis.
To identify other potential targets for auxin regulation via GA, we reviewed microarray expression data available in Genevestigator (https://www.genevestigator.ethz.ch/at) and searched for genes up-regulated by auxin and GA. Among the genes selected, we examined by quantitative RT-PCR the expression of one of them (At4g37770) and, as shown in Supplemental Figure S4, auxin-induced up-regulation was dramatically reduced in the presence of paclobutrazol. In contrast, the expression of IAA32 (At2g01200), which is only induced by auxin, showed no alteration in the response to auxin even in the presence of paclobutrazol. This result confirms that auxin can indeed exert its action through GA or in an autonomous manner, depending on the particular target examined.
DISCUSSION
Interaction between hormone pathways appears as an important strategy used by plants to integrate environmental and developmental cues. These interactions may occur through shared signaling elements, as illustrated by the SPINDLY protein, which participates in the transduction of signals triggered by GAs and cytokinins in Arabidopsis (Greenboim-Wainberg et al., 2005), or the DELLA protein RGA that integrates signals from ethylene, auxin, and GAs in growth processes (Achard et al., 2003; Fu and Harberd, 2003; Vriezen et al., 2004). An additional strategy developed by plants is to modulate metabolism of one hormone by a second hormone. This is the case, for example, for the homeostatic control between auxin and cytokinin in Arabidopsis, in which auxin exerts negative control on cytokinin pools by acting on its biosynthetic pathway (Nordström et al., 2004). In this work, we show that auxins control the expression of genes encoding enzymes involved in GA metabolism in Arabidopsis seedlings. Auxins act on the GA biosynthesis pathway by regulating specifically the expression of two genes encoding enzymes that participate in rate-limiting steps, AtGA20ox1 and AtGA20ox2, supporting the standing model that auxin generally promotes GA biosynthesis. This step in GA biosynthesis would then represent the key entry point for auxin control of GA biosynthesis in Arabidopsis seedlings. This situation is comparable to the auxin promotion of GA biosynthesis in tobacco (Wolbang and Ross, 2001), whereas auxin induces GA biosynthesis in pea through the control of a different step, the one catalyzed by PsGA3ox1 (O'Neill and Ross, 2002). Nevertheless, PsGA3ox1 induction by auxin seems to proceed through a different mechanism, given that cycloheximide is able to block this induction, contrary to the case for AtGA20ox1 and AtGA20ox2 genes in Arabidopsis. Another important difference is that, unlike pea and tobacco, certain GA2ox genes are also up-regulated by auxin treatments in Arabidopsis seedlings. Moreover, this induction is likely direct (see below) and not the consequence of positive feed-forward regulation by GAs, as proposed for PsGA2ox2 in pea (O'Neill and Ross, 2002). AtGA20ox1 expression has also been shown to be up-regulated in response to auxin transport inhibitors and in the tir3-1 mutant, which also alters auxin transport, and in both cases this up-regulation has been related to reduced growth caused by the altered auxin distribution (Desgagné-Penix et al., 2005).
To interpret the physiological consequences of the regulation by auxin of GA metabolism genes, we have to consider at least one additional level of regulation: tissue specificity for the expression of the different GA metabolism genes. Our analysis resolves the actual localization of the regulation by auxin in seedlings for AtGA20ox2 and AtGA2ox2. The expression patterns of these genes do not overlap so that localized auxin accumulation would promote either GA biosynthesis or deactivation, depending on the tissue, leading to different responses. For example, auxin might participate in maintaining shoot apical meristem activity by enhancing GA deactivation through specific regulation of AtGA2ox2 gene expression at the boundary of the meristem (Jasinsky et al., 2005). On the other hand, it might promote cotyledon growth by supporting GA biosynthesis through regulation of AtGA20ox2 gene. However, we cannot rule out a different scenario, with GA20ox and GA2ox expressed in the same cell types, where auxin up-regulation of both classes of genes might result in a narrow, transient accumulation of active GAs, representing a very effective mechanism to tightly fine tune developmental and growth processes. This latter possibility is supported by the faster kinetics of induction of AtGA20ox1 and AtGA20ox2 genes when compared to that of genes encoding deactivating enzymes (Fig. 3). Although all parts of Arabidopsis seedlings have the capacity to synthesize auxin, there exist mechanisms that regulate the accumulation of auxin in the different tissues (Ljung et al., 2001), suggesting that they may also affect auxin control on GA metabolism gene expression in a tissue- or organ-specific manner. Moreover, auxin treatments seem to reflect endogenous regulation of GA metabolism genes by auxin, as indicated by the results obtained with the auxin-overproducing mutant yucca (Fig. 6).
Auxin-induced degradation of the DELLA proteins GAI and RGA mediates several growth responses in Arabidopsis (Achard et al., 2003; Fu and Harberd, 2003; Vriezen et al., 2004). Conversely, stabilization of these proteins promotes increased expression of GA biosynthesis genes (Dill and Sun, 2001; King et al., 2001). This suggests that the GA biosynthesis genes and GAI/RGA stability may have different sensitivity to auxin. Consistent with this view, auxin up-regulation of GA biosynthesis genes occurs through a mechanism that is independent of the DELLA proteins GAI and RGA (Fig. 2). Rather, our results show that several GA metabolism genes would be primary targets of auxin signaling (Figs. 3–5). Although there is no biochemical evidence available to confirm this point, several pieces of evidence suggest direct regulation: (1) induction of some of the GA metabolism genes occurs rapidly after exposure to auxin; (2) up-regulation does not require de novo protein synthesis; (3) induction is blocked with proteasome inhibitors; and (4) induction is altered in auxin-signaling mutant seedlings. Classical studies have shown that auxin modulates some morphological phenotypes very rapidly, for example, induction of cell elongation (Cleland, 2004). In addition, some plant responses to environmental cues, such as changing gravitropic or phototropic vectors, are mediated by fast changes in auxin transport and distribution (Friml and Palme, 2002; Swarup et al., 2002). This indicates that the auxin signal is transformed quickly into changes in gene expression that bring about physiological responses. Many genes are regulated by auxin within minutes (Hagen and Guilfoyle, 1985; Goda et al., 2004), and for many of them the regulation has been shown to be direct (Hagen and Guilfoyle, 2002). Among these, there are genes that encode auxin-signaling elements, such as Aux/IAA genes, as well as others of unknown functions. Our results suggest that auxin action in Arabidopsis seedlings might be executed, at least in part, by GAs, similarly to the effect of auxin on pea stem growth (O'Neill and Ross, 2002). Mediation of auxin action by GAs could represent a strategy to amplify and potentiate certain responses. Alternatively, auxin could delegate GAs to control certain responses without overlapping in the control of downstream genes. For instance, some phenotypes caused by high endogenous auxin levels, such as long hypocotyls in light-grown Arabidopsis seedlings, are highly dependent on GA biosynthesis (Fig. 8A; data not shown).
Although induction by auxin of GA metabolism genes fits this model of auxin signaling, it is difficult to assign roles to individual Aux/IAA proteins in this particular auxin response based solely on the analysis of the effects of aux/iaa-dominant mutations (Fig. 5), given both the nature of the mutations and also that, in many instances, gain of function of one particular Aux/IAA gene alters the expression of other members in the family (Tian et al., 2002; Tatematsu et al., 2004; Yang et al., 2004). Thus, we can only conclude that one or more Aux/IAA transcriptional repressors are involved in maintaining low auxin-regulated activity at the promoters of the GA metabolism genes, most likely through inhibition of the activity of at least one ARF transcription factor, NPH4/ARF7 (Fig. 5).
Studies on the auxin control of primary response genes led to the discovery of a sequence element in their promoters, TGTCTC, important for this response and that allowed the identification of ARF1, an Arabidopsis transcription factor that binds to this sequence element (Ulmasov et al., 1995, 1997). This sequence motif has been found in other auxin primary response genes and is thought to be the target point for auxin signaling at their promoters (Hagen and Guilfoyle, 2002). However, recent genome-wide analysis of auxin-regulated genes in Arabidopsis has shown that many targets lack this sequence element in their promoters (Nemhauser et al., 2004). Instead, the presence of two repeats of the core sequence, TGTC, within a window of 50 bp in the promoter is associated with induction by auxin. In a survey of the promoter region of the GA metabolism genes that are regulated by auxin, the TGTCTC element was only found in the promoter of the AtGA2ox8 gene; however, every auxin-regulated GA metabolism gene contains between one and six copies of the core sequence in its promoter, and in three of them, AtGA20ox2, AtGA2ox6, and AtGA2ox8, the core sequences are within a 50-bp window (http://bbc.botany.utoronto.ca/ntools/cgi-bin/BAR_Promomer.cgi; Toufighi et al., 2005). This suggests that auxin signaling may target these promoters at other sequence elements, alone or in combination with the TGTC sequences found.
In conclusion, our results indicate that auxin signaling affects GA metabolism mainly via direct up-regulation of expression of several AtGA20ox and AtGA2ox genes. This regulation operates through a mechanism that is independent of the feedback regulation mediated by the DELLA proteins GAI and RGA, and involves one or more Aux/IAA and ARF-signaling elements.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) accessions Columbia-0 (Col-0) and Landsberg erecta (Ler) were used as wild type. Seeds of the GA-deficient ga1-3 mutant were obtained from the Arabidopsis Biological Resource Center (ABRC), and ga1-3 rga-24 gai-t6 were provided by Dr. Tai-ping Sun (Duke University), both in Ler background. Seeds of the auxin-signaling mutants tir1-1, axr2-1, and axr3-1 were obtained from the ABRC; nph4-1 seeds were provided by Dr. Emmanuel Liscum (University of Missouri); msg2-1 by Dr. Kotaro Yamamoto (Hokkaido University); yucca by Dr. Yunde Zhao (University of California) and Dr. Joanne Chory (Salk Institute), all in Col-0 background; and shy2-2 was provided by Dr. Jason Reed (University of North Carolina) in Ler background.
All seeds were surface sterilized for 15 min in 70% (v/v) ethanol and 0.01% (v/v) Triton X-100, followed by 10 min in 96% (v/v) ethanol. Seeds were stratified in sterile water at 4°C for 4 d in darkness and then germinated in 50 mL of 0.5× Murashige and Skoog liquid medium, supplemented with 1% (w/v) Suc. Stratification of ga1-3 and ga1-3 rga-24 gai-t6 seeds was done in water containing 10 μm GA3 and the seeds were then rinsed several times with water before sowing. Seedlings were grown at 20°C under continuous fluorescent white light (fluence rate of 90–100 μmol m−2 s−1) for 6 d. For the experiments shown in Figure 8, seeds were sterilized as above, sown on 0.5× Murashige and Skoog agar plates supplemented with 1% (w/v) Suc, and stratified at 4°C for 4 d in the dark. For light-grown seedlings, plates were kept at 20°C under continuous fluorescent white light (fluence rate of 90–100 μmol m−2 s−1 for the first 2 d and 10–15 μmol m−2 s−1 for the next 5 d). For dark-grown seedlings, plates were placed in the light for 8 to 9 h, wrapped in several layers of aluminum foil, and kept at 20°C for a total of 7 d.
Seedlings grown in liquid media were treated with 50 μm NAA (Sigma), 100 μm GA3 (Duchefa), or mock treated with ethanol 0.14% (v/v, final concentration) for the indicated times. Paclobutrazol (Duchefa) was added to a final concentration of 0.5 μm, whereas control seedlings were mock treated with acetone 0.005% (v/v, final concentration). Inhibitors of protein synthesis (cycloheximide, 10 μm final concentration; Sigma) and 26S proteasome (MG132, 50 μm final concentration; Sigma) were added to the media for the indicated times; control seedlings for the MG132 treatment were mock treated with DMSO 0.1% (v/v, final concentration). In agar plates, GA3 (10 μm final concentration) and paclobutrazol (0.01–1 μm, final concentration) were added to the media after autoclaving. Control plates for GA3 and paclobutrazol treatments contained 0.014% ethanol (v/v, final concentration) and 0.01% acetone (v/v, final concentration), respectively.
RNA Extraction, cDNA Synthesis, Quantitative RT-PCR, and Northern Blotting
Total RNA was extracted from frozen, whole seedlings using the RNeasy plant mini kit (Qiagen). Genomic DNA was eliminated during RNA purification with the RNase-free DNase set (Qiagen). Two micrograms of total RNA were used to synthesize first-strand cDNA using the SuperScript first-strand synthesis system for RT-PCR (Invitrogen). cDNA samples were diluted in a total volume of 40 μL.
One microliter of cDNA was used for quantitative RT-PCR using SYBR Green PCR master mix (Applied Biosystems) following the manufacturer's recommendations and an ABI Prism 7000 sequence detection system (Applied Biosystems). Each sample was assayed in triplicate. Expression levels were calculated relative to EF1-α using the ΔΔthreshold cycle (Ct) method (Applied Biosystems). Primer sequences for amplification of GA metabolism and EF1-α genes and the corresponding AGI number appear in Supplemental Table S1 and in Curaba et al. (2004). Mean Ct values for each detected gene corresponding to the control sample in Figure 1 also appear in Supplemental Table S1. All quantitative RT-PCR determinations were done in at least two different biological replicates, obtaining very similar results, although only one experiment is shown in this article.
Two micrograms of total RNA were used for northern analysis. Northern blotting, hybridization with 32P-labeled CAB2 and RbcS probes, and washes were carried out as described previously (Alabadí et al., 2004). Membranes were reprobed, without previous stripping, with Arabidopsis 32P-labeled 18S rDNA as a loading control. For signal quantification, membranes were exposed to Fuji BAS-MS imaging plates and analyzed using a Fluor Imager FLA-5100 (Fujifilm). Northern analyses were done in two different biological replicates, obtaining very similar results, although only one experiment is shown in this article.
Construction of AtGA20ox2∷β-Glucuronidase Reporter Fusion and Plant Transformation
The AtGA20ox2∷β-glucuronidase (GUS) reporter gene was constructed as a translation fusion comprising the promoter and transcribed region of AtGA20ox2 (from −1,507 bp relative to the translational start to the seventh codon of the third exon), fused in-frame with the GUS reporter gene. The PCR primers used were At20ox2-PF (5′-AAAGGTACCTCACATATCATTCGTGGGT-3′; AtGA20ox2-specific sequence is underlined) and At20ox2-PR (5′-AAAGGGCCCGAATATCCCGTTCGATAGAG-3′), which amplified a 2,617-bp fragment from a λEMBL3 genomic clone containing the At20ox2 gene (J.P. Coles and A.L. Phillips, unpublished data). Construction of the AtGA20ox2 promoter∷GUS fusion and Agrobacterium-mediated transformation of Arabidopsis were carried out as described for AtGA20ox1∷GUS (Desgagné-Penix et al., 2005). More than 70 transgenic lines were generated, the vast majority with consistent GUS histochemical staining patterns; 10 lines containing single transgenic loci were taken forward to a homozygous state as determined by resistance to kanamycin.
GUS Staining
For histochemical analyses, whole seedlings were prefixed for 20 min in cold 90% (v/v) acetone at room temperature, washed once in cold staining buffer (50 mm sodium phosphate buffer, pH 7, 0.2% [v/v] Triton X-100, 2 mm potassium ferricyanide, 2 mm potassium ferrocyanide), and incubated in 2 mm X-GlcA (in staining buffer) first on ice for 1 h, then at 37°C for 8 h. Tissue was dehydrated in ethanol series (20% [v/v], 35% [v/v], 50% [v/v], 30 min each at room temperature), then postfixed 30 min at room temperature in 50% (v/v) ethanol, 5% (v/v) formaldehyde, 10% (v/v) acetic acid. Tissue was further dehydrated in 70% (v/v) ethanol and photographed with Nomarski optics.
Hypocotyl Length Measurements
Seedlings were placed on an acetate sheet and scanned at a resolution of 800 dots per inch. Hypocotyl lengths were measured as described previously (Alabadí et al., 2004).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Transcriptional regulation of GA metabolism genes by auxin.
Supplemental Figure S2. Feedback regulation of GA metabolism genes by GA.
Supplemental Figure S3. Effect of inhibitors of a proteasome inhibitor on the up-regulation of GA metabolism genes by auxin.
Supplemental Figure S4. Effect of paclobutrazol on the up-regulation of gene expression by auxin.
Supplemental Table S1. Primers used for gene expression studies.
Supplementary Material
Acknowledgments
We thank Dr. Joanne Chory (Salk Institute, La Jolla, CA), Dr. Emmanuel Liscum (University of Missouri, Columbia, MO), Dr. Jason Reed (University of North Carolina, Chapel Hill, NC), Dr. Tai-ping Sun (Duke University, Raleigh, NC), Dr. Kotaro Yamamoto (Hokkaido University, Sapporo, Japan), and Dr. Yunde Zhao (University of California, La Jolla, CA) for seeds; Dr. G. Vachon (Université Joseph Fourier, Grenoble, France) for advice on the quantitative RT-PCR determination of GA metabolism gene expression; and Dr. J. Carbonell, Dr. J.L. García-Martínez, Dr. O. Nilsson, and Dr. M.A. Pérez-Amador for discussions and useful comments on the manuscript.
This work was supported by the Spanish Ministry of Education and Science (MEC; grant nos. BIO2001–1558 and BIO2004–02355); the European Molecular Biology Organization Young Investigator Programme (grant to M.A.B.); a Ramón y Cajal contract with the Spanish MEC (to D.A.); a Spanish FPI fellowship (to M.F.); the European Union Research Training Network INTEGA (J.P.-G.); and a Core Strategic Grant from the Biotechnology and Biological Sciences Research Council to Rothamsted Research (to A.L.P. and P.H.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Miguel A. Blázquez (mblazquez@ibmcp.upv.es).
The online version of this article contains Web-only data.
References
- Achard P, Vriezen WH, Van Der Straeten D, Harberd NP (2003) Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15: 2816–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadí D, Gil J, Blázquez MA, García-Martínez JL (2004) Gibberellins repress photomorphogenesis in darkness. Plant Physiol 134: 1050–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland RE (2004) Auxin and cell elongation. In PJ Davies, ed, Plant Hormones: Biosynthesis, Signal Transduction, Action!, Ed 3. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 204–220
- Collett CE, Harberd NP, Leyser O (2000) Hormonal interactions in the control of Arabidopsis hypocotyl elongation. Plant Physiol 124: 553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling RJ, Harberd NP (1999) Gibberellins control Arabidopsis hypocotyl growth via regulation of cellular elongation. J Exp Bot 50: 1351–1357 [Google Scholar]
- Curaba J, Moritz T, Blervaque R, Parcy F, Raz V, Herzog M, Vachon G (2004) AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol 136: 3660–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desgagné-Penix I, Eakanunkul S, Coles JP, Phillips AL, Hedden P, Sponsel VM (2005) The auxin transport inhibitor response 3 (tir3) allele of BIG and auxin transport inhibitors affect the gibberellin status of Arabidopsis. Plant J 41: 231–242 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005. a) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Juürgens G, Estelle M (2005. b) Plant development is regulated by a family of auxin receptor F-box proteins. Dev Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Dill A, Jung H-S, Sun T-p (2001) The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc Natl Acad Sci USA 98: 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Sun T-p (2001) Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159: 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleet CM, Sun T-p (2005) A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr Opin Plant Biol 8: 1–9 [DOI] [PubMed] [Google Scholar]
- Friml J, Palme K (2002) Polar auxin transport—old questions and new concepts? Plant Mol Biol 49: 273–284 [PubMed] [Google Scholar]
- Fu X, Harberd NP (2003) Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421: 740–743 [DOI] [PubMed] [Google Scholar]
- Fu X, Richards DE, Ait-ali T, Hynes LW, Ougham H, Peng J, Harberd NP (2002) Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor. Plant Cell 14: 3191–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Martínez JL, Santes C, Croker SJ, Hedden P (1991) Identification, quantitation and distribution of gibberellins in fruits of Pisum sativum L. cv Alaska during pod development. Planta 184: 53–60 [DOI] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S (2004) Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 134: 1555–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Östin A, Sandberg G, Romano CP, Estelle M (1998) High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA 95: 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Greenboim-Wainberg Y, Maymon T, Borochov R, Alvarez J, Olszewski N, Ori N, Eshed Y, Weiss D (2005) Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17: 92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle TJ (1985) Rapid induction of selective transcription by auxin. Mol Cell Biol 5: 1197–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle TJ (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49: 373–385 [PubMed] [Google Scholar]
- Harper RM, Stowe-Evans EL, Luesse DR, Muto H, Tatematsu K, Watahiki MK, Yamamoto K, Liscum E (2000) The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12: 757–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Phillips AL (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5: 523–530 [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL, Rojas MC, Carrera E, Tudzynski B (2002) Gibberellin biosynthesis in plants and fungi: a case of convergent evolution? J Plant Growth Regul 20: 319–331 [DOI] [PubMed] [Google Scholar]
- Jasinsky S, Piazza P, Craft J, Hay A, Woolley L, Rieu I, Phillips A, Hedden P, Tsiantis M (2005) KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr Biol 15: 1560–1565 [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Kim BC, Soh MS, Hong SH, Furuya M, Nam HG (1998) Photomorphogenic development of the Arabidopsis shy2-1D mutation and its interaction with phytochromes in darkness. Plant J 15: 61–68 [DOI] [PubMed] [Google Scholar]
- King EK, Moritz T, Harberd NP (2001) Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159: 767–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser HMO, Pickett FB, Dharmasiri S, Estelle M (1996) Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J 10: 403–413 [DOI] [PubMed] [Google Scholar]
- Ljung K, Bhalerao RP, Sandberg G (2001) Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J 28: 465–474 [DOI] [PubMed] [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW (2000) AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol 123: 563–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J (2004) Interdependency of brassinosteroids and auxin signaling in Arabidopsis. PLoS Biol 2: 1460–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo P, Ozga JA, Reinecke DM (2002) Specificity of auxin regulation of gibberellin 20-oxidase gene expression in pea pericarp. Plant Mol Biol 49: 439–448 [DOI] [PubMed] [Google Scholar]
- Nordström A, Tarkowski P, Tarkowska D, Norbaek R, Åstot C, Dolezal K, Sandberg G (2004) Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: a factor of potential importance for auxin-cytokinin-regulated development. Proc Natl Acad Sci USA 101: 8039–8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill DP, Ross JJ (2002) Auxin regulation of the gibberellin pathway in Arabidopsis. Plant Physiol 130: 1974–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet F, Overvoorde PJ, Theologis A (2001) IAA17/AXR3: biochemical insight into and auxin mutant phenotype. Plant Cell 13: 829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozga JA, Reinecke DM (1999) Interaction of 4-chloroindole-3-acetic acid and gibberellin in early pea fruit development. Plant Growth Regul 27: 33–38 [Google Scholar]
- Ozga JA, Yu J, Reinecke DM (2003) Pollination-, development-, and auxin-specific regulation of gibberellin 3β-hydroxylase gene expression in pea fruit and seeds. Plant Physiol 131: 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinecke DM, Ozga JA, Magnus V (1995) Effect of halogen substitution of indole-3-acetic acid on biological activity in pea fruit. Phytochemistry 40: 1361–1366 [Google Scholar]
- Remington DL, Vision TJ, Guilfoyle TJ, Reed JW (2004) Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol 135: 1738–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo MJ, García Martínez JL, Santes CM, Gaskin P, Hedden P (1997) The role of gibberellins A1 and A3 in fruit growth of Pisum sativum L. and the identification of gibberellins A4 and A7 in young seeds. Planta 201: 446–455 [Google Scholar]
- Ross JJ, O'Neill DP (2001) New interactions between classical plant hormones. Trends Plant Sci 6: 2–4 [DOI] [PubMed] [Google Scholar]
- Ross JJ, O'Neill DP, Smith JJ, Kerckhoffs LHJ, Elliott RC (2000) Evidence that auxin promotes gibberellin A1 biosynthesis in pea. Plant J 21: 547–552 [DOI] [PubMed] [Google Scholar]
- Saibo NJ, Vriezen WH, Beemster GT, Van Der Straeten D (2003) Growth and stomata development of Arabidopsis hypocotyls are controlled by gibberellins and modulated by ethylene and auxins. Plant J 33: 989–1000 [DOI] [PubMed] [Google Scholar]
- Schomburg FM, Bizzell CM, Lee DJ, Zeevaart JAD, Amasino RM (2003) Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15: 151–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Haegman M, Kurepa J, Van Montagu M, Van Der Straeten D (1997) Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proc Natl Acad Sci USA 94: 2756–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Parry G, Graham N, Allen T, Bennett M (2002) Auxin cross-talk: integration of signaling pathways to control plant development. Plant Mol Biol 49: 411–426 [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT (2004) MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16: 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Reed JW (1999) Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126: 711–721 [DOI] [PubMed] [Google Scholar]
- Tian Q, Uhlir NJ, Reed JW (2002) Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14: 301–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart J (2005) The Botany Array Resource: e-Northerns, Expression Angling, and promoter analyses. Plant J 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ (1997) ARF1, a transcription factor that binds to auxin response elements. Science 276: 1865–1868 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ (1995) Composite structure of auxin response elements. Plant Cell 7: 1611–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Huizen R, Ozga JA, Reinecke DM (1997) Seed and hormonal regulation of gibberellin 20-oxidase expression in pea pericarp. Plant Physiol 115: 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Huizen R, Ozga JA, Reinecke DM, Twitchin B, Mander LN (1995) Seed and 4-chloroindole-3-acetic regulation of gibberellin metabolism in pea pericarp. Plant Physiol 109: 1213–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivian-Smith A, Koltunow AM (1999) Genetic analysis of growth-regulator-induced parthenocarpy in Arabidopsis. Plant Physiol 121: 437–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriezen WH, Achard P, Harberd NP, Van Der Straeten D (2004) Ethylene-mediated enhancement of apical hook formation in etiolated Arabidopsis thaliana seedlings is gibberellin dependent. Plant J 37: 505–516 [DOI] [PubMed] [Google Scholar]
- Wolbang CM, Chandler PM, Smith JJ, Ross JJ (2004) Auxin from the developing inflorescence is required for the biosynthesis of active gibberellins in barley stems. Plant Physiol 134: 769–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolbang CM, Ross JJ (2001) Auxin promotes gibberellin biosynthesis in decapitated tobacco plants. Planta 214: 153–157 [DOI] [PubMed] [Google Scholar]
- Yang X, Lee S, So J-h, Dharmasiri S, Dharmasiri N, Ge G, Jensen C, Hangarter R, Hobbie L, Estelle M (2004) The IAA1 protein is encoded by AXR5 and is a substrate of SCFTIR1. Plant J 40: 772–782 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Christensen S, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.