Abstract
Successful application of posttranscriptional gene silencing (PTGS) for gene function study in both plants and animals depends on high target specificity and silencing efficiency. By computational analysis with genome and/or transcriptome sequences of 25 plant species, we predicted that about 50% to 70% of gene transcripts in plants have potential off-targets when used for PTGS that could obscure experimental results. We have developed a publicly available Web-based computational tool called siRNA Scan to identify potential off-targets during PTGS. Some of the potential off-targets obtained from this tool were tested by measuring the amount of off-target transcripts using quantitative reverse transcription-PCR. Up to 50% of the predicted off-target genes tested in plants were actually silenced when tested experimentally. Our results suggest that a high risk of off-target gene silencing exists during PTGS in plants. Our siRNA Scan tool is useful to design better constructs for PTGS by minimizing off-target gene silencing in both plants and animals.
Posttranscriptional gene silencing (PTGS), also known as RNA interference (RNAi) in animals, cosuppression in plants, and RNA quelling in fungi, is an epigenetic phenomenon that results in sequence-specific degradation of endogenous mRNAs (Cogoni and Macino, 2000). PTGS is mediated by 21- to 24-nucleotide (nt) double-strand RNA (dsRNA) molecules, termed short/small interference RNAs (siRNAs). The siRNAs are incorporated into a multisubunit protein complex, the RNAi-induced silencing complex, which directs the siRNA to and degrades the complementary mRNA (Baulcombe, 2004). Because PTGS allows silencing of a specific target gene, it has become a popular tool to study gene function in plants and animals (Kamath et al., 2003; Baulcombe, 2004; Hannon and Rossi, 2004; Kuttenkeuler and Boutros, 2004).
In plants, PTGS can be induced by antisense and sense transgenic technology and it is also achieved by expressing dsRNA through stable or transient transformation with RNAi constructs (binary vectors) to knock down the expression of target genes (Wesley et al., 2001; Miki and Shimamoto, 2004). In addition, a transient PTGS of plant genes by recombinant viruses carrying a near-identical sequence was adapted by a process called virus-induced gene silencing (VIGS; Baulcombe, 1999; Burch-Smith et al., 2004). Both PTGS approaches, RNAi and VIGS, are becoming powerful tools in functional genomic studies of plants. However, to infer gene function through PTGS, it is essential to determine the specificity of mRNAs that are targeted for silencing.
Theoretically, PTGS functions in a siRNA-specific rather than a target-specific manner. However, analyses of mammalian cells transfected with different siRNAs against a target gene by two different research groups led to contradictory conclusions about silencing unintended genes (Chi et al., 2003; Jackson et al., 2003; Jackson and Linsley, 2004). The reasons for these contradictory results are probably due to the differences in experimental designs or microarray analyses. In plants, several RNAi and VIGS studies successfully targeted specific members of gene families for silencing without affecting the transcript level of the most closely related family members or simultaneously silenced a few family members to overcome functional redundancy (Burch-Smith et al., 2004; Hwang and Gelvin, 2004). However, PTGS relies upon sequences of contiguous nucleotide identity and does not target genes simply based on family relationships. A dsRNA expressed in plants from a binary or virus vector is usually chosen to be identical to a partial or full-length sequence of the target gene. Many distinct siRNAs of 21 to 24 nt can be derived from the cleavage of the dsRNA by Dicer. This may improve the efficiency of RNA silencing, but could also increase the opportunity to suppress unintended genes, the off-targets, containing sequences identical to some of these siRNAs.
Investigating off-target gene silencing is crucial for accurate interpretation of gene function by PTGS and for use of PTGS application in agriculture. In this study, we estimate potential off-targets based on sequence identity for 25 plant species whose genomic or expressed sequence tag (EST) sequences are publicly available. Experimentally, we investigated the expression level of several potential off-target genes in an RNAi transgenic Arabidopsis (Arabidopsis thaliana) line and in gene-silenced (by VIGS) Nicotiana benthamiana plants. To assist the design of PTGS constructs to minimize off-target gene silencing or to identify potential off-targets from a particular PTGS construct, we have developed a Web-based computational tool. The tool provides an integrated sequence similarity search environment for plant and animal species, identifies potential off-targets, and predicts putatively effective siRNAs from the target query sequences.
RESULTS
Computational Analysis of Potential Off-Targets during PTGS
PTGS in plants is mediated by siRNAs derived from the cleavage of dsRNA produced through binary vector-based RNAi or VIGS constructs. The siRNAs recognize their targets by base pairing. Therefore, the potential off-targets are identified by a direct sequence identity and reverse complimentary sequence identity search. In this study, we define the cDNA producing dsRNA for silencing of the intended target gene (from which the cDNA comes) as the trigger and the unintended genes that share a contiguous ≥21-nt region of identity or reverse complementary identity to the trigger as off-targets. The continuous identical or reverse complementary sequence to a trigger region in an off-target is defined as an off-target region.
Using the full-length cDNA sequence of every transcript in the Arabidopsis genome mRNA dataset (ATH1) as a trigger, we estimated the patterns of potential off-targets by searching for identical or reverse complementary regions of ≥21 nt between a trigger sequence and all the other genes, except the target itself. Differently spliced transcripts from the same locus in the genome were not counted as additional off-targets. The potential off-target region was scanned along the full length of all released transcript sequences. Off-target regions can be in the 5′ and 3′ untranslated regions or coding regions because all these regions have been shown to be targeted by siRNAs (McManus et al., 2002; Yu et al., 2002; Yokota et al., 2003; Shirane et al., 2004). By using results obtained from Arabidopsis data analyses, we compared the distributions of off-target/trigger, off-target site/off-target, and length of off-target site/off-target site (Fig. 1) with their statistical averages (Table I). The histograms shown in Figure 1 have skewed distributions, with a majority of the trigger genes having similar potential off-targets as the average (Fig. 1A), a majority of the off-targets having approximately the same number of off-target sites as the average (Fig. 1B), and a majority of off-target sites having almost the same length as the average (Fig. 1C). The average values for off-targets, off-target sites, and lengths of off-target sites are also close to the peak values (medians).
Figure 1.
Histograms showing distributions of the numbers of silencing triggers and off-targets in Arabidopsis. A, Number of silencing triggers versus the number of off-targets. B, Number of off-targets versus the number of off-target sites. C, Number of off-target sites versus the length of off-target sites. y axis is in log10 scale.
Table I.
Estimated off-target patterns during PTGS in plants
| Species | Source Sequence Set | Release Date | Total No. Unique Gene Transcripts | No. Triggers Predicted to Silence Off-Targets (% Total) | Average No. Predicted Off-Targets/Trigger | Average No. Predicted Off-Target Regions/Off-Target | Average Length (nt)/Off-Target Region | Average No. Efficient siRNAs/Trigger |
|---|---|---|---|---|---|---|---|---|
| Arabidopsis | ATH1 | 6/10/04 | 28,952 | 19,882 (68.7) | 3.9 | 2.9 | 41.2 | 35.2 |
| Arabidopsis | AGI 11 | 1/29/04 | 45,683 | 37,250 (81.5) | 4.3 | 3 | 99.2 | 147.5 |
| Capsicum annuum | CaGI 1 | 6/4/04 | 10,712 | 27,88 (26.0) | 1.9 | 2.2 | 40.7 | 12.5 |
| Gossypium spp. | CGI 5 | 9/16/03 | 24,350 | 14,270 (58.6) | 6.9 | 2.3 | 74 | 87.5 |
| Chlamydomonas reinhardtii | ChrGI 4 | 1/31/04 | 30,339 | 16,579 (54.6) | 6.1 | 1.9 | 40.2 | 12.5 |
| Glycine max | GmGI 11 | 9/16/03 | 67,826 | 49,658 (73.2) | 9.4 | 2.1 | 77.9 | 108.4 |
| Helianthus annuus | HaGI 3 | 9/16/03 | 20,520 | 11,961 (58.3) | 15.8 | 1.4 | 34.6 | 38.8 |
| Hordeum vulgare | HvGI 8 | 1/30/04 | 49,190 | 32,297 (65.6) | 16.3 | 2.6 | 99.4 | 205.9 |
| Tomato | LeGI 9 | 5/19/03 | 31,012 | 15,340 (49.5) | 2.4 | 2.3 | 60.8 | 30.9 |
| Lotus japonicus | LjGI 2 | 5/16/03 | 11,025 | 2,951 (26.8) | 2.6 | 1.7 | 35.6 | 10 |
| Lettuce | LsGI 2 | 2/3/04 | 22,185 | 12,345 (55.6) | 24.1 | 1.4 | 30.4 | 26.8 |
| Mesembryanthemum crystallinum | McGI 4 | 5/15/03 | 8,455 | 3,459 (42) | 13.3 | 2 | 33 | 29.4 |
| Medicago truncatula | MtGI 7 | 5/19/03 | 36,976 | 19,468 (52.7) | 5.6 | 2.2 | 49.4 | 50.6 |
| Nicotiana benthamiana | NbGI 1 | 1/28/04 | 6,118 | 31,99 (52.3) | 2.8 | 3.8 | 53.4 | 42.1 |
| Nicotiana tabacum | NtGI 1 | 6/4/04 | 10,232 | 2,184 (21.3) | 1.8 | 3.1 | 38.3 | 15.2 |
| Allium cepa | OnGI 1 | 9/17/03 | 11,726 | 3,735 (31.9) | 1.7 | 2.3 | 54.2 | 20.2 |
| Rice | OsGI 15 | 5/27/04 | 88,765 | 45,406 (51.2) | 22.5 | 4.4 | 41.7 | 174.3 |
| Pinus spp. | PGI 4 | 1/28/04 | 31,771 | 16,487 (51.9) | 11.7 | 5.5 | 38.4 | 27.5 |
| Rye | RyeGI 3 | 1/29/04 | 5,347 | 1,472 (27.5) | 2.7 | 6 | 32.4 | 10.7 |
| Sorghum bicolor | SbGI 8 | 5/26/04 | 39,148 | 22,039 (56.3) | 6.4 | 1.9 | 70.3 | 56.9 |
| Sugarcane | SoGI 1 | 1/30/04 | 95,884 | 55,269 (57.6) | 24.1 | 3.5 | 69.1 | 205.4 |
| Solanum tuberosum | StGI 9 | 5/26/04 | 32,553 | 20,469 (62.9) | 4.4 | 2.9 | 46.2 | 42.2 |
| Triticum aestivum | TaGI 8 | 1/29/04 | 12,3807 | 89,862 (72.6) | 19.5 | 2.7 | 52.4 | 102.2 |
| Theobroma cacao | TcaGI 1 | 6/4/04 | 2,539 | 531 (20.9) | 2.6 | 1.9 | 37.4 | 13 |
| Vitis vinifera | VvGI 3 | 11/14/03 | 23,109 | 10,548 (45.6) | 8.8 | 1.8 | 40.3 | 37.1 |
| Zea mays | ZmGI 14 | 1/2/04 | 56,364 | 40,028 (71) | 8.9 | 3 | 63.1 | 96.9 |
On average, 68.7% of Arabidopsis transcripts can potentially silence 3.9 off-target genes (Table I). Each candidate off-target has an average of three off-target regions with an average length of over 40 nt (Table I). However, this could be an overestimation because not all siRNAs derived from the cleavage of dsRNAs are efficient at silencing (Khvorova et al., 2003; Schwarz et al., 2003). Because little is known about the efficacy of specific siRNAs in plants, we adapted the rules developed by Ui-Tei et al. (2004) to predict whether and how many of the trigger sequences mentioned above produce efficient siRNAs to silence their potential off-targets. Specifically, the rules for a 21-nt siRNA duplex with a 2-nt overhang include: (1) antisense strand starts with A/U; (2) sense strand starts with G/C; (3) at least five of the first seven residues at the 5′ terminus of the antisense strand should be A/U; and (4) there is no G/C stretch of more than 9 nt in length (Ui-Tei et al., 2004). As shown in Table I, 68.7% of all trigger sequences in the ATH1 database are predicted to generate an average of 35 efficient 21-nt siRNAs to cause potential off-target silencing.
In addition to Arabidopsis, we also estimated the potential off-targets in 24 other plant species, using Gene Indices assembled from ESTs and other expressed transcripts provided by The Institute for Genomic Research (TIGR; Quackenbush et al., 2001). We first compared the results from Arabidopsis Gene Index version 11 (AGI 11) with the results from the genome mRNAs described above (ATH1). Slightly more trigger sequences were predicted to have potential off-targets when the AGI 11 dataset was analyzed (Table I). This is because numerous unique gene transcripts in AGI 11 are not represented in the current genome sequence. Estimations of off-targets in other plant species also indicated high likelihood of off-target silencing (Table I). The potential off-target effects for plants like rye (Secale cereale), pepper (Capsicum annuum), and Lotus japonicus may be underestimated in this analysis because of limited sequence availability. For example, the percentage of target sequences predicted to silence off-targets is approximately 26% for these species. When those species with <20,000 EST sequences in their gene indices are excluded, 50% to 70% of all gene transcripts in each of 17 plant species used as silencing triggers can cause potential off-target silencing. The average numbers of off-targets for each trigger sequence range from 2.4 in tomato (Lycopersicon esculentum) to 24 in lettuce (Lactuca sativa) and sugarcane (Saccharum officinarum; Table I).
Off-target silencing, however, may be desirable in PTGS applications to generate a loss-of-function phenotype if the target and off-targets are functionally redundant. For example, the same family members or genes encoding proteins that share conserved functional protein domains may be simultaneously silenced. Arabidopsis gene family information at The Arabidopsis Information Resource (TAIR) Web site (http://www.arabidopsis.org) was used to evaluate the chance that potential off-targets belong to the same gene family as the target gene. The data from the TAIR ftp site, after removing ambiguous entries and single-member families, contain 5,842 genes from 674 families (see Supplemental Table S1), with family size ranging from two to 307 members. Our analysis indicates that 4,677 gene sequences (79.9% of 5,842 genes) of these families may trigger off-target silencing of other members of the same family when a full-length sequence is used as a trigger. Among the families with different numbers of gene members, the number of off-targets was different but generally fewer than six (Fig. 2; Supplemental Table S1). Moreover, only 418 genes from 192 families containing fewer than 10 members share an identical region of ≥21 nt with all other members in the same gene family, so only these transcripts used as trigger sequences can silence all family members. No single trigger sequence from any family with more than 10 members was predicted to silence all members of the family. In addition, the percentage of gene members to be silenced decreases from approximately 79% to <9% as family size increases from two to >35. The family CYP705A in the P450 superfamily is an exception. This family has 24 gene members, and >75% of them were predicted to be potential off-targets when any one of its members is used as an RNAi trigger (Fig. 2).
Figure 2.
Average number of off-target gene family members predicted to be silenced by a trigger from the same family in Arabidopsis. Data for gene families with the same size are combined and averaged.
An Integrated Search Environment for PTGS
An online tool (available at http://bioinfo2.noble.org/RNAiScan.htm) has been developed to evaluate and minimize the risk of off-target effects during PTGS. The tool is a search environment with several integrated components, including a sequence similarity search to identify potential off-targets, efficiency estimation of siRNAs, and functional analysis of off-target genes. In the sequence similarity search, the query sequence is compared with a gene transcript dataset using the BLASTn program (Altschul et al., 1997) with user-specified mismatches that may be tolerable in siRNA target recognition (Saxena et al., 2003). The datasets include Arabidopsis and rice (Oryza sativa) genome mRNAs and all TIGR gene indices for plants and animals, and they are updated regularly. All possible siRNAs derived from the query are then evaluated for efficiency using Ui-Tei's rule (Ui-Tei et al., 2004). The predicted off-target genes can be subject to further functional analysis by identifying Pfam domains (Sonnhammer et al., 1998) and associated gene ontology assignments. The tool can also be used to estimate off-targets during siRNA-induced gene silencing in animals. In addition to displaying the potential siRNAs and off-target candidates, the results page also displays all the potential siRNAs with no sequence similarity to any other genes in the searched database. Therefore, the RNAi scan tool can be used to design siRNAs with minimal off-targets.
Experimental Verification of Off-Target Silencing in Plants
Expression of at Least Three Off-Target Genes Was Knocked Down in BTI1 RNAi Transgenic Lines
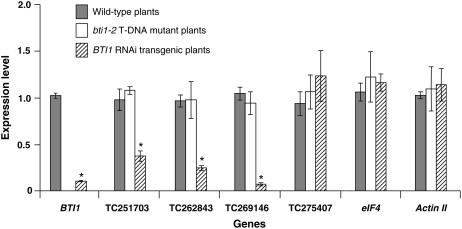
BTI1 is an Arabidopsis protein that interacts with the Agrobacterium tumefaciens VirB2 protein (Hwang and Gelvin, 2004). A functional study of BTI1 through RNAi transgenic Arabidopsis lines has been published, where dsRNA covering the coding region of BTI1 was produced from the RNAi vector pFGC5941 (Hwang and Gelvin, 2004). We used the coding region of BTI1 as a trigger sequence to search for potential off-targets against the TIGR AGI 12.1 database with the Web-based tool siRNA Scan described above. We further confirmed the annotations of these off-targets by BLAST search in the National Center for Biotechnology (NCBI). In addition to one family member, BTI2, 13 other genes were found that share at least 21-nt continuous direct identity or reverse complementary identity to the BTI1 coding sequence (Table II). These 14 genes have the potential to be targeted by the siRNAs generated from BTI1-derived dsRNA. Twelve of them were selected for the analysis of their expression levels in BTI1 RNAi transgenic lines (Table II). They are TC251703 (BTI2), TC251496 (putative 3-isopropylmalate dehydratase large subunit), TC256637 (encodes a probable Ser/Thr kinase), TC258543 (RTNLB6), TC255665 (encodes a zinc-finger motif protein), TC262843 (encodes a hypothetical protein), TC263798 (encodes a protein of unknown function), TC269146 (encodes a hypothetical protein), TC275407 (encodes a putative Ser/Thr kinase), TC265975 (encodes a probable membrane protein), TC275528 (encodes a maturase-related protein), and TC275625 (encodes protein of unknown function). In addition, expression of the other 22 genes that share almost 21- or 22-nt continuous identity to the BTI1 coding region, but with one mismatch, was also analyzed. The expression levels of these genes, as well as the target gene BTI1, were measured by real-time quantitative reverse transcription-PCR (qRT-PCR) and compared with their expression in nontransgenic plants. The Arabidopsis elongation factor-1α (EF-1α) gene was used as the endogenous control to normalize the relative transcripts in the reactions. The results showed about 90% down-regulation of target gene BTI1 in one RNAi transgenic line (Fig. 3). Down-regulation of some of the selected potential off-targets, TC262843 and TC269146, and a family member, TC251703 (BTI2), was detected in the same RNAi transgenic line. They share at least 23 contiguous nucleotide identity with the BTI1 coding region (Table II). About 75% and 50% down-regulation occurred to TC262843 and TC251703, respectively (Fig. 3). Surprisingly, expression of TC269146 is about 95% down-regulated, a little more than that of the target gene. TC262843 and TC269146 share the same 23-nt identity with the BTI1 gene. An oligo DNA with this 23-nt sequence was synthesized and labeled for detection of the potential siRNA involved, but it was not detectable. Nevertheless, a low amount of siRNA could be detected when a 200-bp DNA probe of BTI1 containing this 23-nt sequence was used for hybridization (data not shown). No significant difference was seen in the expression of other analyzed genes with at least 21-nt identity or reverse complementarity to BTI1, whereas very low (unquantifiable) amounts of transcripts were detected for TC255665 in all the tested leaf tissues (data not shown). Also, no significant differences were seen in the expression of the selected 22 genes that share almost 21- or 22-nt continuous identity to the BTI1 coding region, but with one mismatch (Supplemental Fig. S1). Similar results were seen for all the above-mentioned experiments from two other independent BTI1 RNAi transgenic lines tested (data not shown).
Table II.
Result from the siRNA Scan search with Arabidopsis BTI1 coding sequence as a trigger
TC numbers in bold are the ones used for quantitative RT-PCR.
| Off-Target ID | Identical (I) or Reverse Complementary (RC) Sequence | BLAST Search |
|---|---|---|
| NP167859 | I, AGCCTGTTCATAAGGTTCTCGG | Arabidopsis reticulon family protein (RTNLB6) (At3g61560). |
| NP302761 | I, AGAAGAAGAAGACTAAGAAGCI, AGCCTGTTCATAAGGTTCTCGG | Contains similarity to DnaJ gene YM8520.10 gb|825566 from Saccharomyces cerevisiae cosmid gb|Z49705. ESTs gb|Z47720 and gb|Z29879 come from this gene. |
| TC251496 | RC, TGATTCTTCTTCGTCTTCTTC | GB|AAM51226 Unknown protein {Arabidopsis}; similar to UP|Q6Z702 (Q6Z702) putative 3-isopropylmalate dehydratase large subunit. |
| TC251703 | I, CCTGTTCATAAGGTTCTCGGCG; I, TGGTCTAATGCCACTATGTTCATT;I, CCAAAGATTCCTGAAGTTCATATCCCTGAAGAACCT;I, TTCTTGACATTGGCATACATAGCTCT | GB|AAP47461.1|32331867|AY164887 RTNLB2 {Arabidopsis}, complete. Recently identified as BTI2. |
| TC255665 | I, TCTTCGTCTTCTTCATCTTCT | GB|AAD31078.1|4850408|F3F19 Contains PF|00097 zinc finger (C3HC4) ring finger motif. {Arabidopsis}, complete. |
| TC256637 | RC, TTCTTCTTCGTCTTCTTCATC | PIR|B96716 Probable Ser/Thr kinase F23O10.20 [imported]—Arabidopsis. |
| TC258543 | I, AGCCTGTTCATAAGGTTCTCGG | GB|AAP47457.1|32331859|AY164883 RTNLB6 {Arabidopsis}, partial (90%). |
| TC262843 | I, CTTCTTCGTCTTCTTCATCTTCT | UP|O49467 (O49467) Hypothetical protein F24J7.50 (hypothetical protein AT4g19490). |
| TC263798 | RC, TTCTTCTTCGTCTTCTTCATC | GB|AAO50470 Unknown protein {Arabidopsis}. |
| TC265975 | I, AGAAGAAGAAGACTAAGAAGC | UP|080799 (080799) T8F5.5 protein; weakly similar to PIR|S64314 probable membrane protein YGR023w—yeast (S. cerevisiae). |
| TC269146 | I, CTTCTTCGTCTTCTTCATCTTCT | UP|O49467 (O49467) Hypothetical protein F24J7.50 (hypothetical protein AT4g19490). |
| TC275407 | RC, TTCTTCTTCGTCTTCTTCATC | UP|Q8VYC1 (Q8VYC1) Putative Ser/Thr kinase. |
| TC275528 | I, GAGAAGAAGAAGACTAAGAAG | UP|Q9FJR9 (Q9FJR9) Similarity to maturase-related protein, complete. |
| TC275625 | RC, TTCTTCTTCGTCTTCTTCATCTTCT | GB|AAM51439 Unknown protein {Arabidopsis}; weakly similar to UP|081812(081812) auxilin-like protein, partial (13%). |
Figure 3.
Comparison of the expression levels of BTI1 and some of its potential off-targets in wild-type, bti1-2 mutant, and BTI1 RNAi transgenic Arabidopsis plants. Initiation factor-4 (eIF4) and actin II genes were used as controls to show equal loading. Error bars are the sds of three biological replicates of quantitative RT-PCR. *, Expression level of the gene in the BTI1 RNAi transgenic line is significantly lower than that in the wild-type or bti1-2 mutant plants with 98% confidence by t test.
To rule out the possibility that the lowered expression of some of these selected genes might be due to the loss of function of the BTI1 protein instead of off-target silencing, we analyzed the expression levels of all the above investigated genes in an Arabidopsis bti1-2 null mutant line (T-DNA knockout line; Salk-032220). As expected, the expression of BTI1 was below the detectable levels in the mutant plants, but the expression levels of all the other genes tested were similar to those in nontransgenic Arabidopsis plants (Fig. 3). Expression of two additional housekeeping genes, eIF4 and actin II, was also investigated for the confirmation of equal RNA amounts used for real-time qRT-PCR. Expression levels were similar among the samples from the three groups, further confirming the equal loading of total RNA in the reactions (Fig. 3). Thus, it is off-target silencing caused by BTI1-derived dsRNA that down-regulated the expression of three of the five selected potential off-targets in BTI1 RNAi transgenic plants. In addition, there is a striking phenotypic difference between BTI1 RNAi transgenic lines and the bti1-2 mutant line. Plants from four independent BTI1 RNAi transgenic lines flowered earlier than the wild-type and bti1-2 mutant plants (Fig. 4) and this is probably due to off-target gene silencing in the RNAi plants.
Figure 4.
Comparison of phenotypes exhibited by BTI1 RNAi transgenic lines, bti1-2 mutant plants, and wild-type nontransgenic plants. All plants were grown in short-day conditions for 5 weeks and moved to a growth chamber in long-day conditions for 1 week. A, Plants from four independent BTI1 RNAi transgenic lines. B, bti1-2 mutant plants. C, Wild-type plants.
Expression of Some Potential Off-Targets Was Down-Regulated in N. benthamiana Plants Infiltrated with VIGS Constructs
Computational analysis showed that more than one-half of the EST contigs (TCs) in the N. benthamiana database have multiple potential off-targets during PTGS (Table I). We randomly chose two genes for experimental analyses. They were TC381 and TC1146 encoding the U2 small nuclear ribonucleoprotein A (snRNAP A) and pyruvate decarboxylase, respectively. Partial fragments from both genes were separately cloned into the tobacco rattle virus (TRV) RNA2 vector (Liu et al., 2002) for VIGS analysis (their sequences can be found in Supplemental Fig. S2). The search with siRNA Scan using both fragments as trigger sequences revealed several potential off-targets (Table III). Clone TC381 shares contiguous identity of at least 21-nt identity to the following unique sequence entries in the current N. benthamiana database: CK286172, CK288691, CK289650, TC10748, and TC7796. These genes encode proteins that are unrelated to snRNAP A. Three of these potential off-targets, CK286172, CK288691, and TC10748, were selected for expression analysis by real-time qRT-PCR in TC381 VIGS-silenced plants. Expression levels of these genes, as well as that of the target gene TC381, were compared with their expression in the plants infiltrated with TRV1 (RNA1) + TRV2∷00 (RNA2 empty vector). The EF-1α gene was used as the endogenous reaction control to normalize the relative quantity and expression of the β-tubulin gene was investigated to confirm the equal loading of total RNA. The results showed a greater silencing of TC10748 than the target gene TC381 and very little silencing (not statistically significant) for CK286172 and CK288691 genes (Fig. 5A). The analysis was repeated twice with two different groups of plants and similar results were observed.
Table III.
Result from the siRNA Scan search for potential off-targets with the cloned sequences of N. benthamiana TC381 and TC1146 (Supplemental Fig. S1) as trigger sequences
TC numbers in bold are the ones used for quantitative RT-PCR.
| Target ID | Annotation of the Target | Potential Off-Target | Identical (I) or Reverse Complementary (RC) Sequence | Annotation of the Potential Off-Target |
|---|---|---|---|---|
| TC381 | Similar to TIGR_Osa1|9630.m01295 U2 snRNAP protein, Arabidopsis, partial 79% | CK286172 | RC, GGGTGTATTCTGGCCCGGGCC TGRC, TATCTTGTATAGTAGTTATTAGTATAGT | Similar to UP|O93419 (O93419) collagen XVIII precursor, partial (1%). |
| CK288691 | RC, TATCTTGTATAGTAGTTATTAGTATAGT | Annotation not available. | ||
| CK289650 | I, TTCGCGGTCCCGGGCTTCGTG | Similar to TIGR_Osa1|9631.m00421 CYFIP2, partial (9%). | ||
| TC10748 | I, TATCTTGTATAGTAGTTATTA GTATA | Similar to TIGR_Ath1|At5g13210.1 68418.m01516 expressed protein, partial (17%). | ||
| TC7796 | I, TATCTTGTATAGTAGTTATTA TGTATAG | Weakly similar to UP|Q75NB3 (Q75NB3) Cys proteinase, partial (34%). | ||
| TC1146 | Similar to UP|Q8H9C6 (Q8H9C6) pyruvate decarboxylase (fragment), partial (33%) | CK282591 | RC, ATCGCTTTGCGAACCCGACTAG | Similar to UP|Q6Z3U3 (Q6Z3U3) VP1/ABI3 family regulatory protein-like, partial (5%). |
| CK287535 | RC, ATCGCTTTGCGAACCCGACTAG | Similar to TIGR_Ath1|At5g65980.1 68418.m08307 auxin efflux carrier family protein contains auxin efflux carrier domain, Pfam:PF03547, partial (14%). | ||
| CK292351 | RC, TTTGTTCTCGGGCCTTTACCAG | Similar to UP|CP22_HORVU (P55748) Ser carboxypeptidase II-2 precursor (CP-MII.2) (fragment), partial (30%). | ||
| CK296810 | RC, ATCGCTTTGCGAACCCGACTAG | Similar to TIGR_Ath1|At5g65980.1 68418.m08307 auxin efflux carrier family protein contains auxin efflux carrier domain, Pfam:PF03547, partial (11%). | ||
| TC8666 | RC, ATCGCTTTGCGAACCCGACTAG | Similar to TIGR_Ath1|At2g17500.1 68415.m02022 auxin efflux carrier family protein contains auxin efflux carrier domain, Pfam:PF03547, partial (41%). |
Figure 5.
Off-target gene silencing in N. benthamiana by VIGS. A, Comparison of the expression levels of TC381, CK286172, TC10748, CK288691, and β-tubulin in TC381 silenced and nonsilenced N. benthamiana. B, Comparison of the expression levels of TC1146, CK282591, CK287535, CK292351, CK296810, TC8666, and β-tubulin in the N. benthamiana plants silenced with the TRV2∷00 or TRV2∷TC1146 construct. Error bars are the sds among three technical replicates of real-time qRT-PCR. *, Expression level of the gene in the TRV2∷TC381- or TRV2∷TC1146-infiltrated plant is significantly lower than that in the TRV2∷00-infiltrated plant with 98% confidence by t test. Both of the experiments in A and B were repeated twice with two individual silenced plants and the results were similar.
A similar scenario was found in TRV2∷TC1146-infiltrated plants. Five unique EST entries share at least 21 nt of contiguous reverse complementary sequences to the cloned TC1146 fragment (Table III). They are CK282591 encoding a protein similar to the VPI/ABI3 family regulatory protein, CK292351 encoding a Ser carboxypeptidase II-2 precursor, and CK287535, CK296810, and TC8666 encoding three different members of the auxin efflux protein family. The expression levels of these genes and TC1146 in silenced plants were compared between the silenced plants and TRV-RNA1 + TRV2:00-infiltrated plants. The expression of target gene TC1146 was decreased to about 50% in TC1146-silenced plants, whereas among the five analyzed potential off-targets, CK287535, CK296810, and TC8666 were down-regulated to a greater extent than TC1146 (Fig. 5B). These three genes all belong to the auxin efflux protein family and have the same 22-nt reverse complementary sequences to TC1146. The expression level of CK282591 was slightly reduced, whereas there was no significant change in the expression of CK292351 in TC1146-silenced plants. These experiments were repeated twice with two individual silenced plants and the results were similar. In conclusion, some of the potential off-target genes were silenced to different degrees when VIGS was used to silence target genes of interest in N. benthamiana. Because we do not have null mutations of these target genes in N. benthamiana, we cannot rule out the possibility that the reduced transcripts of the off-target genes result from the reduction in the amount of target protein.
DISCUSSION
The use of RNA silencing/interference for suppressing gene expression has become a powerful and promising approach in gene function exploration and disease treatment in both plants and animals. Its successful application relies on specific and efficient silencing of particular genes or gene families. Exquisite specificity of RNAi through siRNA in animal cells has been supported by several studies (Tuschl et al., 1999; Elbashir et al., 2001; Chi et al., 2003; Semizarov et al., 2003). However, some contradictory reports indicated that siRNA used for RNA silencing can cause off-target suppression at both transcriptional and translational levels in animals. For example, siRNA causes silencing of unintended genes that lack complete sequence identity and sometimes induces nonspecific interferon responses in animals (Holen et al., 2002; Amarzguioui et al., 2003; Bridge et al., 2003; Saxena et al., 2003; Sledz et al., 2003; Jackson and Linsley, 2004; Scacheri et al., 2004). Recently, a computational study using the genome and transcriptome sequence data of Homo sapiens, Caenorhabditis elegans, and Schizosaccharomyces pombe suggested that the risk of transcriptional off-target silencing by siRNA is considerable in all of these organisms (Qiu et al., 2005). However, to date there is no experimental evidence in plants to show that off-target silencing of unrelated genes can occur as a result of nucleotide sequence similarity with siRNA.
Direct introduction of siRNA into plant protoplasts for PTGS was reported, but it is difficult to apply in intact plant tissues (Vanitharani et al., 2003). The functions of various plant genes revealed by PTGS have been studied mostly through VIGS or by generating stable transgenic lines that express antisense RNA or dsRNA that is identical or complementary to the partial or full-length sequences of target genes. Cleavage of the expressed dsRNAs by Dicer produces many siRNAs. Because siRNAs of 21 to 26 nt have been reported in plants (Hamilton et al., 2002; Llave et al., 2002; Tang et al., 2003; Qi et al., 2005), theoretically, 21-nt identity or reverse complementarity between a trigger sequence and a target could be the minimal requirement for successful RNAi. Our computational analyses indicate a high risk of off-target gene silencing among different plant species (Table I). Results from our experimental analyses with both an RNAi transgenic line and VIGS further confirmed the silencing of some potential off-targets that share at least 22 nt of direct identity or reverse complementary identity to the trigger sequences (Figs. 3 and 5). We were not able to detect the particular siRNAs that have caused off-target silencing through normal northern-blot analysis. This may be due to low abundance or rapid degradation of one particular siRNA molecule. Nevertheless, the obvious down-regulation of some predicted off-targets that only occurred in BTI RNAi transgenic lines, but not in bti1 mutant plants, is compelling. In other studies, a stretch ≥23 nt of perfect sequence identity was found necessary to silence a green fluorescent protein transgene (Thomas et al., 2001) and heterologous silencing occurred when at least 23- or 24-nt identity existed between the RNAi trigger sequence and the intended gene (Ekengren et al., 2003; Liu et al., 2004). In our analysis, 22-nt sequence identity was sufficient to cause off-target silencing. None of the investigated genes with 21- or 22-nt identity, but containing one mismatch to the trigger sequence, was silenced. Thus, at least 22-nt identity may be required for off-target silencing to occur. However, we only analyzed 19 of 70 predicted off-target candidates with 22-nt identity, but containing one mismatch identified by siRNA Scan. Hopefully, a systematic experimental analysis to determine the minimal sequence identity for PTGS between trigger and target sequences will help to set a baseline for potential off-target searching with a particular trigger sequence in the future.
Statistically, longer siRNAs should be less likely to silence unintended genes by chance. Therefore, predicted off-target risk might be overestimated for siRNAs longer than 21 nt. However, because on average each predicted off-target for all plant species analyzed here has multiple off-target regions and each average off-target region is longer than the longest siRNAs reported in plants (Table I), the overall trend of off-target risk predicted from this study should be valid for longer siRNAs such as 22 to 26 nt. More importantly, because siRNAs of various lengths simultaneously coexist within the plant cell (Hamilton et al., 2002), off-target risk should be estimated with the shortest siRNAs. In addition, mismatches in siRNA target recognition are tolerable and these siRNAs can cause RNA degradation and translational repression in animals (Saxena et al., 2003). It is not known whether this applies to plants. In the case of BTI1 RNAi transgenic lines, transcript accumulation of all the investigated potential off-targets with contiguous ≥22-nt identity or with 21- or 22-nt identity, but containing one mismatch to the expressed dsRNA, was not affected (Supplemental Fig. S1). However, this is just one particular case. If mismatches of siRNAs can be tolerated in plant PTGS, our computational analysis results may represent the lower limit of off-target effects as only 21-nt identical regions were counted as potential off-target sites in this study. Nevertheless, our siRNA Scan tool includes the option of searching potential off-targets with complete sequence identity or reverse complementary identity of 18 to 29 nt to the trigger sequence, as well as allowing a few mismatches to the potential siRNAs.
Although our computational analysis showed a high risk of off-target gene silencing during PTGS in plants, the efficiency of off-target gene silencing should be the main factor that affects the functional analysis of a particular target gene. Results from our experimental analyses showed a varied reduction of expression levels of the potential off-targets in both Arabidopsis and N. benthamiana. The reduction ranges from none to a greater reduction than that of the target gene expression. The underlying mechanisms for this variation in expression levels of off-targets are not yet clear. Gene silencing efficiency is correlated with siRNA sequence-specific features (Khvorova et al., 2003; Schwarz et al., 2003; Amarzquioui and Prydz, 2004; Reynolds et al., 2004; Ui-Tei et al., 2004), location of the complementary sequence of siRNA in the target (Birmingham et al., 2006), and target accessibility (Luo and Chang, 2004; Pancoska et al., 2004; Brown et al., 2005). At present, target accessibility cannot be reliably predicted because sequences may be partial for most datasets, which are assembled from ESTs and other transcripts. Besides, the in vivo mRNA secondary structure is unknown for the datasets in our analysis. Therefore, we only used siRNA sequence properties to assess gene silencing efficiency. Specifically, siRNA efficacy was predicted according to the rules by Ui-Tei et al. (2004). A recent comparison study indicates that, among currently available siRNA efficacy predictors, Ui-Tei's rules are stable and high performance (Saetrom and Snove, 2004). Although Ui-Tei's rules have not been experimentally tested in plants, they agree with the requirement for thermodynamic features of an efficient siRNA duplex, which favors siRNA unwinding by helicases. However, the siRNAs predicted to cause off-target gene silencing had different efficiencies in silencing the target and off-target genes in our experimental analysis. This could be due to the different accessibility of the mRNAs to the siRNA. Earlier, it was shown that siRNAs against different regions of genes display marked variation in their potency in mediating mRNA degradation (Thomas et al., 2001). In addition, strong RNAi effects, as assessed by phenotypic analysis, were found to correlate with high expression levels of the targeted genes or higher expression of RNAi trigger sequences in C. elegans and plants (Chuang and Meyerowitz, 2000; Cutter et al., 2003; Hu et al., 2004; Kerschen et al., 2004). The structure of siRNA may also have an effect on its efficiency to induce gene silencing. These complex factors together may result in different silencing efficiencies among target and off-target genes.
Short reverse complementary sequences (over 21 nt) were found in some completely unrelated genes in our computational analyses. In animals, siRNAs can simultaneously induce sequence-specific degradation of two endogenous mammalian transcripts oriented in opposite directions (Hu et al., 2004). Our experimental results indicate that either strand of the siRNA duplex may silence the genes that contain identical sequences. It is not known whether these endogenous short reverse complementary sequences in different genes have any biological function.
CONCLUSION
The understanding of off-target silencing is crucial for accurate interpretation of gene function by PTGS. Our computational analyses with the genome and transcriptome sequences from 25 plant species showed a high risk of off-target gene silencing when a full-length sequence of each transcript entry in the datasets was used as an RNAi trigger. This off-target gene silencing risk was confirmed by our experimental analysis with both RNAi transgenic Arabidopsis lines and N. benthamiana plants infected by VIGS constructs. So far, the contribution of silenced off-targets to the silencing phenotype is not known and needs to be analyzed in exploring target gene function. For example, although the BTI1 RNAi lines had off-target gene silencing, the transformation recalcitrant phenotype of these lines is not in question because it was confirmed by null mutant lines of the target gene (Hwang and Gelvin, 2004). Nevertheless, no approach used to inactivate gene function is free from potential problems, and the reality of off-target silencing does not override the enormous potential of RNAi as a tool for individual or high-throughput studies of gene function. On the other hand, off-target silencing in PTGS provides an advantage in overcoming gene functional redundancy and in its potential to be applied in heterologous gene silencing across species. Hopefully, a further understanding of the molecular mechanisms of RNAi will add more restraining rules for off-target prediction and reveal possible approaches to overcome off-target effects for target specificity. At this point, our siRNA Scan should provide an extremely useful tool in searching for the potential off-targets of an RNAi trigger of interest and will also help to design more specific RNAi triggers and appropriate controls in experiments.
MATERIALS AND METHODS
Datasets
Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa subsp. japonica) genome mRNAs were downloaded from TIGR (http://www.tigr.org). Gene Indices for 25 plant organisms were downloaded from TIGR. The Gene Ontology (GO) database was provided by the GO Consortium (http://geneontology.org). Arabidopsis gene family information and GO annotation were downloaded from TAIR (http://www.arabidopsis.org). Protein domain data (http://pfam.wustl.edu) was also downloaded for off-target function analysis.
Computational Off-Target Estimation
The gene transcript sequence dataset for each organism was searched against itself for contiguous ≥21-nt identical or reverse complementary regions using the BLAST algorithm (Altschul et al., 1997). The BLAST output was loaded into a local data warehouse into which GO and gene family databases were also integrated. Patterns of potential off-targets were obtained by querying the database. Possible distinct siRNAs were enumerated from each identical region between a query and an off-target, and efficient siRNAs were also predicted according to the rules by (Ui-Tei et al. 2004). The rules include a 5′ antisense strand, starting with an A or U base, a 5′ sense strand, starting with G or C, and the first seven bases from a 5′ antisense strand end containing at least three to five A/U bases. The procedures of the above analyses were modified and implemented in an online tool that is publicly accessible (http://bioinfo2.noble.org/RNAiScan.htm). The input parameters of the tool for analysis of siRNAs and off-targets can be adjusted by users. Additionally, gene function prediction is provided for off-targets by aligning them to Pfam-A seed sequences using the BLASTx program. The E-value cutoff is set to 10−5. When a domain is identified, a local GO database is searched for its available GO annotations in molecular function, biological process, and cellular component.
Plant Materials and Growth Conditions
Arabidopsis (ecotype Columbia) and BTI1 RNAi transgenic lines (Hwang and Gelvin, 2004) and BTI1 T-DNA insertion mutants (Salk-032220, bti1-2, ecotype Columbia) were grown in a growth chamber at 22°C with 1 h daylight. Nicotiana benthamiana and the Agrobacterium tumefaciens strain GV2260 were used for VIGS analysis. The planting conditions were the same as described previously (Ryu et al., 2004).
Plasmid Construction
The sequences of TC381 and TC1146 were obtained from the TIGR database of N. benthamiana. cDNA fragments, including the sequences with >20-nt continuous identity or complementarity to their potential off-target genes, were amplified by RT-PCR. Primers for the amplification of specific TC381 and TC1146 fragments are shown in Supplemental Table S2. About 5 μg total RNA from N. benthamiana leaves were used for RT at 42°C with NNpoly(dT)20 as the primer for 2 h. The RT product was used for PCR amplification for TC381 and TC1146 fragments, respectively, in a PTC-100 Peltier thermal cycler (M.J. Research). The PCR products were cloned into the pGEM-T-easy vector (Promega). Clones with inserts that are identical to the area in the target genes were amplified by PCR with primers that contain attb recombination sequences adapted to the previous primer pairs. PCR products were purified, sequenced, and cloned into the pTRV2 VIGS vector using the GATEWAY cloning system with the protocol from the manufacturer (Invitrogen). The recombinant plasmids were named TRV2∷TC381 and TRV2∷TC1146.
VIGS
Agrobacterium strain GV2260 containing pTRV1, TRV2∷00, TRV2∷TC381, and TRV2∷TC1146 were grown in an incubator at 28°C on Luria-Bertani broth with 10 μg L−1 rifampicin and 50 μg L−1 kanamycin for 2 d. Inoculum was prepared with the protocol published previously (Ryu et al., 2004). Leaves of 2-week-old N. benthamiana plants (two- to three-leaf stage) were infiltrated with a 1:1 Agrobacterium mixture of either TRV1+TRV2∷00 or TRV1+TRV2∷TC381 or TRV1+TRV2∷TC1146 as described earlier (Liu et al., 2002). The infiltrated plants were grown for 14 to 18 d for silencing to occur. Leaf samples were then collected for RT-PCR analysis.
RNA Extraction and Real-Time qRT-PCR Analysis
For Arabidopsis, total RNA was isolated from leaf tissues using TRIzol reagent (Invitrogen), followed by RNase-free DNase treatment (Promega). First-strand cDNA was synthesized with the Omniscript RT kit (Qiagen) using oligo(dT)15 according to the manufacturer's instructions. For qRT-PCR, real-time experiments were conducted in an ABI PRISM 7000 sequence detection system (Applied Biosystems), using the intercalation dye SYBR Green I as a fluorescent reporter. Quantification of PCR products was performed via a calibration curve procedure using EF-1α as an endogenous control. The ratio of gene-specific expression to the expression level of the designated calibrator was defined as relative expression using a standard curve method described in User Bulletin Number 2 (Applied Biosystems).
For N. benthamiana, total RNA from each plant at 15 d postinfiltration was extracted using TRIzol reagent with the protocol provided by the manufacturer (Molecular Research Center). First-strand cDNA was synthesized with 2 μg of total RNA using primer NNpoly(dT)20 as described above. Semiquantitative RT-PCR was performed with the modified program from the one described in “Plasmid Construction” with annealing temperature at 60°C and 20 cycles for TC381, 33 cycles for TC1146. The primer sequences are listed in Supplemental Table S2. Further real-time qRT-PCR analysis for each comparison was done with the total RNA of one TRV2∷00-infiltrated and one recombinant clone-infiltrated plant, as described above for Arabidopsis samples. The analysis was repeated twice with two different groups of plants for each combination. The primer sequences used for real-time qRT-PCR are listed in Supplemental Table S3.
Data Analysis
Data were subjected to ANOVA using Student's t test software of Excel 2003 with 98% confidence.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparison of the expression levels of three genes with 21- or 22-nt identity but containing one mismatch to BTI1 gene sequence in wild-type, bti1-2 mutant, and BTI1 RNAi transgenic Arabidopsis plants.
Supplemental Figure S2. List of the sequences that were cloned for VIGS analysis.
Supplemental Table S1. Gene family members as triggers and their off-targets in the same family in Arabidopsis.
Supplemental Table S2. Primers for cDNA cloning and semiquantitative RT-PCR.
Supplemental Table S3. Primers for real-time qRT-PCR assay.
Supplementary Material
Acknowledgments
We thank Dr. Stan Gelvin and Dr. Hau-Hsuan Hwang for sending us the Arabidopsis seeds of BTI1 RNAi lines and the bti1-2 T-DNA mutant; Dr. S.P. Dinesh-Kumar for providing GATEWAY-ready TRV-VIGS vectors; Dr. Choong-Min Ryu for help with infiltrations and discussions; and Dr. Elison Blancaflor, Dr. Stan Gelvin, and Dr. Jianzhong Liu for reviewing this manuscript.
This work was supported by The Samuel Roberts Noble Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Kirankumar S. Mysore (ksmysore@noble.org).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Altschul SF, Madden TL, Schäffer A, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarzguioui M, Holen T, Babaie E, Prydz H (2003) Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res 31: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarzquioui M, Prydz H (2004) An algorithm for selection of functional siRNA sequences. Biochem Biophys Res Commun 316: 1050–1058 [DOI] [PubMed] [Google Scholar]
- Baulcombe D (1999) Fast forward genetics based on virus-induced gene silencing. Curr Opin Plant Biol 2: 109–130 [DOI] [PubMed] [Google Scholar]
- Baulcombe D (2004) RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, et al (2006) 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods 3: 199–204 [DOI] [PubMed] [Google Scholar]
- Bridge AJ, Pebernard S, Ducraux A, Nicoulaz A-L, Iggo R (2003) Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet 34: 263–264 [DOI] [PubMed] [Google Scholar]
- Brown KM, Chu CY, Rana TM (2005) Target accessibility dictates the potency of human RISC. Nat Struct Mol Biol 12: 469–470 [DOI] [PubMed] [Google Scholar]
- Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP (2004) Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J 39: 734–746 [DOI] [PubMed] [Google Scholar]
- Chi J-T, Chang HY, Wang NN, Chang DS, Dunphy N, Brown PO (2003) Genomewide view of gene silencing by small interfering RNAs. Proc Natl Acad Sci USA 100: 6343–6346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C-F, Meyerowitz EM (2000) Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA 97: 4985–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C, Macino G (2000) Post-transcriptional gene silencing across kingdoms. Curr Opin Genet Dev 10: 638–643 [DOI] [PubMed] [Google Scholar]
- Cutter AD, Payseur BA, Salcedo T, Estes AM, Good JM, Wood E, Hartl T, Maughan H, Strempel J, Wang B, et al (2003) Molecular correlates of genes exhibiting RNAi phenotypes in Caenorhabditis elegans. Genome Res 13: 2651–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekengren SK, Liu Y, Schiff M, Dinesh-Kumar SP, Martin GB (2003) Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J 36: 905–917 [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Garborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498 [DOI] [PubMed] [Google Scholar]
- Hamilton A, Voinnet O, Chappell L, Baulcombe D (2002) Two classes of short interfering RNA in RNA silencing. EMBO J 21: 4671–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ, Rossi JJ (2004) Unlocking the potential of the human genome with RNA interference. Nature 431: 371–378 [DOI] [PubMed] [Google Scholar]
- Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H (2002) Positional effects of short interfering RNAs targeting the human coagulation trigger tissue factor. Nucleic Acids Res 30: 1757–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Hipolito S, Lynn R, Abraham V, Ramos S, Wong-Staal F (2004) Relative gene-silencing efficiencies of small interfering RNAs targeting sense and antisense transcripts from the same genetic locus. Nucleic Acids Res 32: 4609–4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H-H, Gelvin SB (2004) Plant proteins that interact with VirB2, the Agrobacterium tumefaciens pilin protein, mediate plant transformation. Plant Cell 16: 3148–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS (2003) Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol 21: 635–637 [DOI] [PubMed] [Google Scholar]
- Jackson AL, Linsley PS (2004) Noise amidst the silence: off-target effects of siRNAs? Trends Genet 20: 521–524 [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, et al (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237 [DOI] [PubMed] [Google Scholar]
- Kerschen A, Napoli CA, Jorgensen RA, Muller AE (2004) Effectiveness of RNA interference in transgenic plants. FEBS Lett 466: 223–228 [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216 [DOI] [PubMed] [Google Scholar]
- Kuttenkeuler D, Boutros M (2004) Genome-wide RNAi as a route to gene function in Drosophila. Brief Func Genomics Proteomics 3: 168–176 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2002) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2004) Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB transcription factors, COI1 and CTR1 in N-mediated resistance to tobacco mosaic virus. Plant J 38: 800–809 [DOI] [PubMed] [Google Scholar]
- Llave C, Kasschau KD, Rector MA, Carrington JC (2002) Endogenous and silencing-associated small RNAs in plants. Plant Cell 14: 1605–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo KQ, Chang DC (2004) The gene-silencing efficiency of siRNA is strongly dependent on the local structure of mRNA at the targeted region. Biochem Biophys Res Commun 318: 303–310 [DOI] [PubMed] [Google Scholar]
- McManus MT, Petersen CP, Haines BB, Chen J, Sharp PA (2002) Gene silencing using micro-RNA designed hairpins. RNA 8: 842–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D, Shimamoto K (2004) Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol 45: 490–495 [DOI] [PubMed] [Google Scholar]
- Pancoska P, Moravek Z, Moll UM (2004) Efficient RNA interference depends on global context of the target sequence: quantitative analysis of silencing efficiency using Eulerian graph representation of siRNA. Nucleic Acids Res 32: 1469–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Denli AM, Hannon GJ (2005) Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell 19: 421–428 [DOI] [PubMed] [Google Scholar]
- Qiu S, Adema CM, Lane T (2005) A computational study of off-target effects of RNA interference. Nucleic Acids Res 33: 1834–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J (2001) The TIGR gene indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res 29: 159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A (2004) Rational siRNA design for RNA interference. Nat Biotechnol 22: 326–330 [DOI] [PubMed] [Google Scholar]
- Ryu C-M, Anand A, Kang L, Mysore KS (2004) Agrodrench: a novel and effective agroinoculation method for virus-induced gene silencing in roots and diverse Solanaceous species. Plant J 40: 322–331 [DOI] [PubMed] [Google Scholar]
- Saetrom P, Snove O Jr (2004) A comparison of siRNA efficacy predictors. Biochem Biophys Res Commun 321: 247–253 [DOI] [PubMed] [Google Scholar]
- Saxena S, Jónsson SO, Dutta Z (2003) Small RNAs with imperfect match to endogenous mRNA repress translation. J Biol Chem 278: 44312–44319 [DOI] [PubMed] [Google Scholar]
- Scacheri PC, Rozenblatt-Rosen O, Caplen NJ, Wolfsberg TG, Umayam L, Lee JC, Hughes CM, Shanmugam KS, Bhattacharjee A, Meyerson M, et al (2004) Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc Natl Acad Sci USA 101: 1892–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD (2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208 [DOI] [PubMed] [Google Scholar]
- Semizarov D, Frost L, Sarthy A, Kroeger P, Halbert DN, Fesik SW (2003) Specificity of short interfering RNA determined through gene expression signatures. Proc Natl Acad Sci USA 100: 6347–6352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirane D, Sugao K, Namiki S, Tanabe M, Iino M, Hirose K (2004) Enzymatic production of RNAi libraries from cDNAs. Nat Genet 36: 190–196 [DOI] [PubMed] [Google Scholar]
- Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BRG (2003) Activation of the interferon system by short-interfering RNAs. Nat Cell Biol 5: 834–839 [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Eddy SR, Birney E, Bateman A, Durbin R (1998) Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res 26: 320–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Reinhart BJ, Bartel DP, Zamore PD (2003) A biochemical framework for RNA silencing in plants. Genes Dev 17: 49–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CL, Jones L, Baulcombe DC, Maule AJ (2001) Size constraints for targeting post-transcriptional gene silencing and for RNA-directed methylation in Nicotiana benthamiana using a potato virus X vector. Plant J 25: 417–425 [DOI] [PubMed] [Google Scholar]
- Tuschl T, Zamore PD, Lehmann R, Bartel DP, Sharp PA (1999) Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev 13: 3191–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui-Tei K, Naito Y, Takahashi F, Haraguchi T, Ohki-Hamazaki H, Juni A, Ueda R, Saigo K (2004) Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res 32: 936–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanitharani R, Chellappan P, Fauquet CM (2003) Short interfering RNA-mediated interference of gene expression and viral DNA accumulation in cultured plant cells. Proc Natl Acad Sci USA 100: 9632–9636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang M, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Yokota T, Sakamoto N, Enomoto N, Tanabe Y, Miyagishi M, Maekawa S, Yi L, Kurosaki M, Taira K, Watanabe M, et al (2003) Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO Rep 4: 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JY, DeRuiter SL, Turner DL (2002) RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc Natl Acad Sci USA 99: 6047–6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.