Abstract
Plant growth stages are identified as distinct morphological landmarks in a continuous developmental process. The terms describing these developmental stages record the morphological appearance of the plant at a specific point in its life cycle. The widely differing morphology of plant species consequently gave rise to heterogeneous vocabularies describing growth and development. Each species or family specific community developed distinct terminologies for describing whole-plant growth stages. This semantic heterogeneity made it impossible to use growth stage description contained within plant biology databases to make meaningful computational comparisons. The Plant Ontology Consortium (http://www.plantontology.org) was founded to develop standard ontologies describing plant anatomical as well as growth and developmental stages that can be used for annotation of gene expression patterns and phenotypes of all flowering plants. In this article, we describe the development of a generic whole-plant growth stage ontology that describes the spatiotemporal stages of plant growth as a set of landmark events that progress from germination to senescence. This ontology represents a synthesis and integration of terms and concepts from a variety of species-specific vocabularies previously used for describing phenotypes and genomic information. It provides a common platform for annotating gene function and gene expression in relation to the developmental trajectory of a plant described at the organismal level. As proof of concept the Plant Ontology Consortium used the plant ontology growth stage ontology to annotate genes and phenotypes in plants with initial emphasis on those represented in The Arabidopsis Information Resource, Gramene database, and MaizeGDB.
Plant systems are complex, both structurally and operationally, and the information regarding plant development requires extensive synthesis to provide a coherent view of their growth and development. The difficulty of developing such a synthesis is exacerbated by the deluge of new technologies such as high-throughput genotyping, microarrays, proteomics, transcriptomics, etc., that generate large amounts of data rapidly. The speed and magnitude of data deposition challenges our ability to represent and interpret this data within the context of any particular biological system (Gopalacharyulu et al., 2005). The ability to extract knowledge from historical sources and integrate it with new information derived from global datasets requires a sophisticated approach to data mining and integration.
Historically, the growth and development of cultivated plants have been monitored at the whole-plant level with the help of scales of easily recognizable growth stages. Consequently, there exist large volumes of literature detailing growth stages for individual plant species or closely related groups of species. For example, Zadok's scale (Zadok et al., 1974) was developed for the Triticeae crops and is widely used to stage the growth and development of cereal crops in the United States. The flexibility of this scale has allowed it to be extended to other cultivated plants, and a uniform code called the Biologische Bundesanstalt, Bundessortenamt, and Chemical Industry (BBCH) code was developed from it (Meier, 1997). The BBCH scale is quite generic and encompasses multiple crops, including monocot and eudicot species. It offers standardized descriptions of plant development in the order of phenological appearance, and has coded each stage for easy computer retrieval. It should be noted that Arabidopsis (Arabidopsis thaliana), as a representative of the Brassicaceae and by virtue of not being a cultivated species, did not have a specific growth stage vocabulary or scale until 2001 when Boyes et al. (2001) developed an experimental platform describing the Arabidopsis growth stages using the BBCH scale. This work created a crucial semantic link between Arabidopsis and cultivated plants. In addition to facilitating the description and synthesis of large amounts of data within a crop species, vocabularies like the BBCH and Zadok's scale also make possible transfer of information among researchers and provide a common language for comparative purposes (Counce et al., 2000).
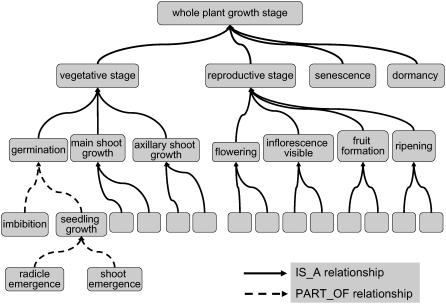
In the post genomic era, these scales have proved inadequate to handle the deluge of information that required large-scale computation for comparative analysis. This called for the conversion of existing scales into ontology that have an advantage over simple scales because their hierarchical organization facilitates computation across them. Terms in an ontology are organized in the form of a tree, the nodes of the tree represent entities at greater or lesser levels of detail (Smith, 2004). The branches connecting the nodes represent the relation between two entities such that the term radicle emergence stage is a child of the parent term germination stage (Fig. 1). Individual stages of a scale are then parts that can be related to the whole by their order of appearance during plant growth. Each term carries a unique identifier and strictly specified relationships between the terms allow systematic ordering of data within a database, this in turn improves input and retrieval of information (Bard and Rhee, 2004; Harris et al., 2004).
Figure 1.
The parent and child term organization in the whole-plant GSO. The solid curved lines joining the terms represent IS_A relationship and the dotted curved lines suggest a PART_OF relationship between the child and the parent terms. A term may or may not have a child term. In this example, germination IS_A vegetative stage and flowering IS_A reproductive stage. Similarly vegetative stage, reproductive stage, senescence, and dormancy are subtypes (IS_A) of whole-plant growth stage. Root emergence and shoot emergence are PART_OF the seedling growth stage. The seedling growth stage and imbibition are PART_OF germination. In this image not all the children terms are shown for every parent term in the GSO.
Consequently, several species-specific databases converted BBCH and other scales into formal ontologies (controlled vocabularies) to facilitate the annotation of genetic information. For example, the Gramene database (Jaiswal et al., 2006) designed its cereal growth stage ontology based on the stages described in the standard evaluation system for rice (Oryza sativa; INGER, 1996) and those described by Counce et al. (2000) for rice, by Zadok et al. (1974) for Triticeae (wheat [Triticum aestivum], oat [Avena sativa], and barley [Hordeum vulgare]), and by Doggett (1988) for sorghum. Except for the sorghum, which is a less studied crop, these species had fairly well-described growth staging vocabularies. MaizeGDB (Lawrence et al., 2005) developed a very extensive controlled vocabulary from a modified version of that described by Ritchie et al. (1993). The Arabidopsis Information Resource (TAIR; Rhee et al., 2003) developed the Arabidopsis growth stage ontology from the scale described by Boyes et al. (2001). However, ontologies created in these projects remained restricted to particular species or families, whereas comparative genomics requires that a common standard vocabulary be applied to a broad range of species. The uniform BBCH scale (Meier, 1997) appeared to be a suitable model to develop a unified ontology since this scale had already synthesized monocot and eudicot crop stages into a single vocabulary.
The Plant Ontology Consortium (POC) was inaugurated in 2003 for the purpose of developing common ontologies to describe the anatomy, morphology, and growth stages of flowering plants (Jaiswal et al., 2005). Its primary task was to integrate and normalize existing species-specific ontologies or vocabularies that had been developed by several major databases for the purpose of annotating gene expression and mutant phenotype. The plant ontology (PO) is divided into two aspects. The first is the plant structure ontology (PSO), a vocabulary of anatomical terms (K. Ilic, E.A. Kellogg, P. Jaiswal, F. Zapata, P. Stevens, L. Vincent, S. Avraham, L. Reiser, A. Pujar, M.M. Sachs, S. McCouch, M. Schaeffer, D. Ware, L. Stein, and S.Y. Rhee, unpublished data), which, since its release to the public domain in 2004, has become widely used by plant genome databases (Jaiswal et al., 2005). The second aspect is the plant growth and developmental stages ontology. This component of PO is further divided into the whole-plant growth stage ontology (GSO) developed by this project and the plant part developmental stages. This article focuses on the whole-plant GSO; we will discuss the history, design, and applications of the GSO and show how it simplifies the description of a continuous and complex series of events in plant development. The plant part developmental stages will be reviewed elsewhere.
RESULTS
The GSO was developed over a period of two years (2004–2006) by a team of plant biologists comprising systematists, molecular biologists, agronomists, plant breeders, and bioinformaticians. We worked to develop a set of terms to describe plant development from germination to senescence that would be valid across a range of morphologically distinct and evolutionarily distant species. Although the rate of addition of new terms to the GSO has slowed since its initial stages of development, it is still under active development as we refine the ontology in response to user input and feedback from database curators.
Currently, the GSO has a total of 112 active terms, each organized hierarchically (Figs. 1 and 2A) and associated with a human-readable definition. Although we started with existing systems such as the BBCH (Meier, 1997) as well as the controlled vocabularies developed for Arabidopsis by TAIR, for rice, Triticeae (wheat, oat, and barley), and sorghum by Gramene database, and for maize (Zea mays) by MaizeGDB, the current version of GSO is quite distinct from its predecessors. We will first discuss the major design issues we dealt with during the development of the GSO, and then describe the structure of the GSO and its applications to real-world problems. The ontology terms, database, and gene annotation statistics provided here are based on the April, 2006 release of the POC database.
Figure 2.
The GSO as seen on the ontology browser available at http://www.plantontology.org/amigo/go.cgi. A, For browsing, simply click on the [+] icon before the term name plant growth and developmental stages, and then on the [+] next to whole-plant growth stages (GSO). This will expand the tree by opening the children terms. The PO ID is the term's accession number, and the number followed by the term name is the total number of associations that have been curated to the genes for a given term. This number will change depending on the gene product filter a user may have chosen. Users can also get a pie chart showing the distribution of data associations to a term's children term. In this image, the general level (top level) terms in the GSO are “A_Vegetative growth,” “B_Reproductive growth,” “C_Senescence,” and “D_Dormancy.” The substages of “A_Vegetative growth” are “0_Germination,” “1_Main Shoot Growth,” and “2_Formation of Axillary Shoot,” while the substages of “B_Reproductive growth” are “3_Inflorescence Visible,” “4_Flowering,” “5_Fruit Formation,” and “6_Ripening.” Neither “C_Senescence” nor “D_Dormancy” currently has substages beneath them. The alphanumeric prefixes serve to make the substages appear in the order in which they occur during the plant's life cycle. If the temporal order is not defined consistently in all plants, the terms may not have these prefixes. The prefixes are usually abbreviations of the term name; for example, LP is for leaf production, SE is for stem elongation. The numerical portion uses double digits starting with 01, 02, and so on. Each of the substages may have more specific stages beneath it. When a term is retired or superseded, it is considered Obsolete. Such terms are moved to a location in the hierarchy underneath a term named “obsolete_growth_and_developmental_stage.” B, A detailed view of the substage PO:0007133, “Leaf production” and its children. Children terms up to 20 leaves visible were added to accommodate the growth stage requirements of the maize plant.
Architecture of the Ontology
We chose to make use of the data model originally developed for the gene ontology (GO) to describe the GSO. This data model uses a directed acyclic graph (DAG) to organize a hierarchy of terms such that the most general terms are located toward the top of the hierarchy while the most specific ones are located at the bottom of the hierarchy (Figs. 1 and 2, A and B). Each parent term has one or more children, and the relationship between a parent and one of its children is named either IS_A to indicate that the child term is a specific type of the parent term, or PART_OF to indicate that the child term is a component of the parent term (Smith, 2004). For example, the reproductive growth and flowering terms are related by IS_A, because flowering is a type of reproductive growth. On the other hand, seedling growth is related to its children terms radicle emergence and shoot emergence by PART_OF, because seedling growth is comprised of the two processes of radicle emergence and shoot emergence (Figs. 1 and 2A).
Each term is given a unique accession number named PO:XXXXXXX where the series of X's is a seven-digit number (Fig. 2A). Accession numbers are never reused, even when the term is retired or superseded. Obsolete terms are instead moved to a location in the hierarchy underneath a term named obsolete_growth_and_developmental_stage (Fig. 2A). This ensures that there is never any confusion about which term an accession number refers to. Each term also has a human-readable name like seedling growth, a paragraph-length definition that describes the criteria for identifying the stage, and citations that attribute the term to a source database, journal article, or an existing staging system. Many terms also have a synonym list; these are described in more detail below.
Our choice of the GO data model was driven by numerous practical considerations, foremost of which was the fact that the data model is supported by a rich set of database schemas, editing tools, annotation systems, and visualization tools.
Naming of Plant Growth Stages
The next issue we dealt with was how to name plant growth stages. Although development in any organism is a continuous process, it is important to have landmarks that identify discrete milestones of the process in a way that is easily reproducible. Extant systems either name growth stages according to a landmark (e.g. three-leaf stage) or by assigning a number or other arbitrary label to each stage. We chose to define growth stages using morphological landmarks that are visible to the naked eye (Counce et al., 2000), because such descriptive terms are more intuitive, self explanatory to the users, and easy to record in an experiment. To minimize differences among species, we were able to describe many growth stages using measurements/landmarks that are in proportion to the fully mature state. For example, the inflorescence stages are described in progression starting from the “inflorescence just visible,” “1/4 inflorescence length reached,” “1/2 inflorescence length reached,” to “full inflorescence length reached.” This provides an objective measurement of the degree of maturation of the inflorescence in a way that is not dependent on the absolute value of the inflorescence length.
Synonyms
Because the GSO crosses species and community boundaries, we needed to acknowledge the fact that each community has its own distinct vocabulary for describing plant structures and growth stages. To accommodate this, we made liberal use of the GO data model's synonym lists, which allow any GSO term to have one or more synonyms from species-specific vocabulary that are considered equivalent to the official term name. The GSO currently contains 997 synonyms taken from several plant species. On an average there are about nine synonyms per GSO term (Table I). Like terms, we attribute synonyms to the database, literature reference, or textbook from which they were derived.
Table I.
A summary of the number of synonyms integrated into the GSO from each species/family/source
The integration of synonyms for Soybean and Solanaceae is in progress.
| Species/Source | No. of Synonyms |
|---|---|
| Arabidopsis | 93 |
| Rice | 23 |
| Maize | 162 |
| Wheat | 65 |
| Oat | 65 |
| Barley | 65 |
| Sorghum | 13 |
| BBCH and Zadok scales | 381 |
| Soybean | 79 |
| Solanaceae (mainly tomato) | 51 |
| All the species | 997 |
| Average no. of synonyms per GSO term | About 9 |
As an example of a synonym, consider “dough stage in wheat” and “kernel ripening in maize,” both of which essentially refer to the fruit ripening stage. These terms are included as synonyms to the generic (species-independent) GSO term “ripening” (PO:0007010; http://www.plantontology.org/amigo/go.cgi?view=details&show_associations=terms&search_constraint=terms&depth=0&query=PO:0007010). From the end user's point of view, the synonyms can be used interchangeably with the generic terms when searching databases that use the PO. This means that data associated with the ripening stage of all plants is accessible even to a naïve user, irrespective of the variable terminology, diverse morphologies, and differing developmental time lines of plants such as wheat and maize.
By using synonyms, we were able to merge 98% of terms from the various species-specific source ontologies into unambiguous generic terms. In a few cases we encountered identical terms that are used by different communities to refer to biologically distinct stages. We resolved such cases by using the sensu qualifier to indicate that the term has a species-specific (not generic) meaning. One example of this, described in more detail later, is “inflorescence visible” (to the naked eye) versus “inflorescence visible (sensu Poaceae).” In most plants the inflorescence becomes visible to the naked eye soon after it forms, whereas in Poaceae (grasses), the inflorescence only becomes visible much later in its development, after emergence from the flag leaf sheath.
Spatiotemporal Representation
Less satisfactory is the design compromise that we reached to represent the spatial and temporal ordering of terms. The existing plant growth scales are organized by the temporal progression of developmental events. However, the GO data model presents unique challenges in designing an ontology that represents the temporal ordering of terms across multiple species that display small but key variations in that ordering.
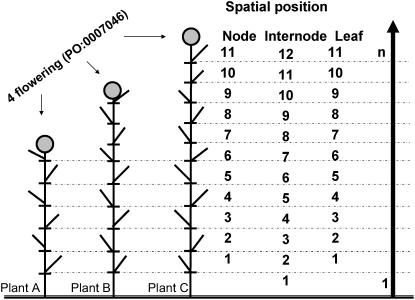
In particular, the GO data model does not have a standard mechanism for representing organisms' developmental time lines. This has forced each organism database that has sought to represent developmental events using the GO model to grapple with the issue of representing a dynamic process in a static representation. Some animal model organism databases, such as WormBase for Caenorhabditis elegans, Flybase for Drosophila, Zfin for Zebra fish, have developed developmental stage ontologies (OBO, 2005) in which temporal ordering is represented using either the DERIVED_FROM, DEVELOPS_FROM, or OCCURS_AT_OR_AFTER relationship to indicate that one structure is derived from another or that one stage follows another. However, we found these solutions to be unworkable for the GSO because of the requirement that the ontology must represent growth stages across multiple species. For example, consider the process of main shoot growth. In the wheat plant, main shoot growth may be completed at the nine-leaf stage, while in rice and maize, shoot growth may be completed at the 11- and 20-leaf stages, respectively, and this varies with different cultivars/germplasms (Fig. 3). Transition to the subsequent stage of reproductive growth is thus staggered for each species, and cannot be accurately described by an ontology in which each stage rigidly follows another.
Figure 3.
Corresponding growth stages in different plants and advantages of using broad and granular terms for annotations. In this example one can say flowering occurs in plant A at the six-leaf visible stage, in plant B at the nine-leaf visible stage, and in plant C at the11-leaf visible stage. Plants A to C represent either different germplasm accessions/cultivars of the same species or accessions/cultivars from different species. This nomenclature allows the researcher to record when a gene is expressed or a phenotype is observed by following the gradual progression of the plant's life cycle. For example, if a gene is expressed at the six-leaf or the fifth-internode stage, the meaning is now clear, while in the past, the information had to be recorded as the fifth leaf from the top of the plant. Such annotation required that one wait until the plant completed its life cycle to count the number of leaves from the top, or that one make an assumption how many leaves there would be in the plant/population used in the study. Note: the number of nodes and the number of leaves is always less than the number of internodes by one. The arrow pointing upwards suggests that the numbers are counted in that direction in ascending order starting with 1 and going up to n, where n can be any number depending on the plant.
Our compromise is to visually order the display of terms in a temporal and spatial fashion, but not to build this ordering into the structure of the ontology itself. In practice, what we do is add alphabetic and numeric prefixes to each term. When terms are displayed the user interface tools sort them alphabetically so that later stages follow earlier ones (Fig. 2A). This compromise is similar to the one taken by the Drosophila developmental stages ontology (Flybase, http://flybase.org/; OBO, 2005).
As an example of how this works, we describe the stages of leaf production using terms named “LP.01 one leaf visible,” “LP.02 two leaves visible,” “LP.03 three leaves visible,” and so forth. When displayed using the ontology web browser, the terms appear in their natural order (Fig. 2B). However, there is nothing hard wired into the ontology that indicates that LP.01 one leaf visible precedes LP.02 two leaves visible.
A related issue is the observation that during plant maturation, multiple developmental programs can proceed in parallel. For instance, the processes of leaf production and stem elongation, although coupled, are temporally overlapping and can proceed at different relative rates among species and among cultivars within a species. We represent such processes as independent children of a more generic term. In the case of the previous example, both leaf production and stem elongation are represented as types of main shoot growth using the IS_A relationship (Figs. 1 and 2A).
Description of the Ontology
The four main divisions of the GSO are “A_Vegetative growth,” “B_Reproductive growth,” “C_Senescence,” and “D_Dormancy” (Fig. 2A). As described earlier, the alphabetic prefix is there to force these four divisions to be displayed in the order in which they occur during the plant's life cycle in general. The substages of “vegetative growth” are “0_Germination,” “1_Main Shoot Growth,” and “2_Formation of Axillary Shoot,” while the substages of “reproductive growth” are “3_Inflorescence Visible,” “4_Flowering,” “5_Fruit Formation,” and “6_Ripening.” Neither “C_Senescence” nor “D_Dormancy” currently has substages beneath them. Again, the numeric prefixes are there only to make the substages appear in a logical order. Each of the substages has multiple, more specific stages beneath it.
Although the BBCH scale (Meier, 1997) was the starting point for the GSO, we have diverged from it in many important aspects. A major difference is the number of top-level terms (Figs. 1 and 2A). The BBCH scale has 10 principle stages as its top-level terms, but the GSO only has four. We collapsed four BBCH top-level stages (germination, leaf development, stem elongation, and tillering) into our top-level vegetative growth term, and collapsed another six BBCH top-level terms (booting, inflorescence emergence, flowering, fruit development, and ripening) into reproductive growth. We felt justified in introducing the binning terms vegetative growth and reproductive growth for several reasons; (1) to help annotate genes that act throughout these phases; (2) persistent use in current scientific literature, especially when the specific stage of gene action or expression remains unclear; and (3) they were requested by our scientific reviewers to enhance the immediate utility of the ontology.
We now look in more detail at some of the more important parts of the ontology.
Germination (PO:0007057)
This node in the GSO has eight children that are broadly applicable to seed germination. The stages under “seedling growth” and “shoot emergence” are not given numerical prefixes, as it is not clear which event precedes the other among the various species. Only events of seed germination were considered in this ontology, whereas the BBCH scale equates seed germination with germination of vegetatively propagated annual plants and perennials such as bud sprouting. The two processes are in fact quite distinct in terms of organs developing at this stage, the physiology, and various metabolic processes, and thus we felt that combining them was inappropriate.
Main Shoot Growth (PO:0007112)
This refers to the stage of the plant when the shoot is undergoing rapid growth. It can be assessed in different ways depending on the species and the interests of the biologist. Plants may be equally well described in terms of leaves visible on the main shoot or in terms of the number of nodes detectable (Zadok et al., 1974), and biologists studying Arabidopsis commonly assess the size of the rosette. To accommodate existing data associated to these terms we created three instances of main shoot growth, namely the “leaf production,” “rosette growth,” and “stem elongation,” with a strong recommendation to use “leaf production” wherever possible.
Leaf Production (PO:0007133)
Leaves are produced successively so that the progression through this stage can be measured by counting the number of visible leaves on the plant (Figs. 2B and 3). In any species, leaves are always counted in the same way (Meier, 1997; described in detail later). In plants other than monocotyledons, leaves are counted when they are visibly separated from the terminal bud. The recognition of the associated internode (below) follows the same rule (Fig. 3). Leaves are counted singly unless they are in pairs or whorls visibly separated by an internode, in which case they are counted as pairs or whorls. In taxa with a hypogeal type of germination, the first leaf on the epicotyl is considered to be leaf one and in grasses the coleoptile is leaf one.
In the GSO the stages of leaf production continue up to 20 leaves/pairs/whorls of visible leaves (Fig. 2A), but this can be emended to accommodate higher numbers, as new species are included. This is unlike the BBCH scale (Meier, 1997), where only nine leaves can be counted and all the rest would be annotated to nine leaves or more. This was done to accommodate the leaf development stages of maize, where depending on cultivars the number of leaves can be few as five or have 20 or more leaves. The maize community and the MaizeGDB database (Lawrence et al., 2005) use a modified version of Ritchie's scale (Ritchie et al., 1993) in which the stages of the maize plant are measured solely by counting the leaves from the seedling through the vegetative stages, and the nodes are not counted.
Stem elongation can be assessed by the number of visible nodes; this metric is commonly applied to the Triticeae, for which the Zadok's (Zadok et al., 1974) or BBCH scales (Meier, 1997) were originally developed. Stem elongation begins when the first node becomes detectable. This is usually equivalent to node number seven (the number varies in different cultivars), since earlier nodes are not detectable before elongation commences in the grasses. Boyes et al. (2001) considers Arabidopsis rosette growth analogous to stem elongation in the grasses, and uses leaf expansion as the common factor linking the rosette growth and stem elongation stages. In our model, “rosette growth” (PO:0007113) and “stem elongation” (PO:0007089) are treated as separate instances of sibling stages (Figs. 1 and 2A), mainly to provide language continuity for users, rather than for biological reasons.
Reproductive Growth (PO:0007130)
Reproductive growth and its child terms are organized a little differently from vegetative growth. Reproductive growth has four instances: “inflorescence visible” (PO:0007047), “flowering” (PO:0007026), “fruit formation” (PO:0007042), and “ripening” (PO:0007010; Fig. 2A). The “inflorescence visible (sensu Poaceae)” (PO:0007012) specific to grass family is an instance of the generic term “inflorescence visible” (PO:0007047). This in turn has two instances: “booting” (PO:0007014) and “inflorescence emergence from flag leaf sheath” (PO:0007041). As described earlier, the generic “inflorescence visible” stage is considered separate from “inflorescence visible (sensu Poaceae),” as the former includes all plants where inflorescence formation and visibility coincide, while in members of the Poaceae, many developmental events in the reproductive phase start during the vegetative phase but manifest themselves as visible morphological markers much later.
Other stages are similar in their organization to the existing scales, but as we continue including various species from families Solanceae and Fabaceae, we anticipate that changes in the organization may be required to accommodate them into the GSO.
User Interface
The GSO terms are in a simple hierarchy that is intuitive to use. The GSO is a relatively small ontology and has a total of 112 terms, excluding the obsolete node. It has four top nodes, 15 interior nodes (terms associated with children terms), and 88 leaf nodes (terms without any children terms; Fig. 2A). New terms are added based on user requests after thorough discussions. A researcher can browse the GSO using the ontology browser available at http://www.plantontology.org/amigo/go.cgi. This is a Web-based tool for searching and browsing ontologies and their associations to data. It has been developed by the GO consortium (http://www.geneontology.org/GO.tools.shtml#in_house) and modified to suit our needs. To browse, clicking on the [+] sign in front of the term expands the tree to show children terms (Fig. 2A). This view provides information on the PO identification (ID) of the GSO term, term name, followed by a number of associated data such as genes. For every green-colored parent term a summary of the data associated to its children terms is presented as a pie chart. The user has an option to filter the number of associated data displayed based on species, data sources, and evidence codes. The icons for [i] and [p] suggest the relationship types between the parent and child term as described in the legend. While browsing, a user can click on the term name to get the details at any time (Fig. 4B). The users will see the icon [d] for develops_from relationship type. This relationship type is used strictly in the PSO and not GSO. It suggests that a plant structure develops from another structure (Jaiswal et al., 2005).
Figure 4.
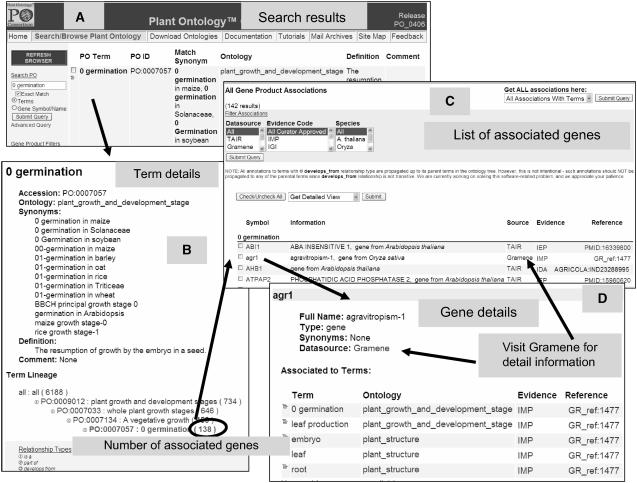
An example of a GSO search using the ontology browser and search Web interface. A, Ontology search results for 0 germination by using the exact match and terms filter. To start searching, visit the www.plantontology.org Web site and click on the “Search and Browse Plant Ontology” link on the page menu. An ontology browser page opens that has a search option on the left-hand side. Type the term name of interest, such as “germination” for a generic search or “0 germination” for an exact match. Select the term filter and submit query. Click on the term name to visit the term detail page or browse the lineage of this term in the ontology by clicking the tree icon next to the check box. B, The term detail page provides information on the term name, accession/ID, synonyms, definition, comments, and associations to genes. C, The list of genes associated to the term are listed in the bottom half of the term detail page. A default list gives all the genes with every type of evidence code and source. The evidence type, species, and source filters can be used to generate the list as desired. The list provides the gene symbol, name, source, evidence, and a citation. The gene symbol links to the gene detail page and the source links to the original record in the contributor's database (e.g. TAIR/Gramene), the evidence code links to its details and the reference links to the original citation referred to by the contributor for inferring the ontology association to the gene. D, The gene detail page provides information on the symbol, name, synonym, source, a list of all the terms in the GSO and PSO, evidence, and the citations. This view suggests where and when a gene is expressed and/or an associated phenotype is observed.
In addition to the browse utility, users may search by entering the name of a term or a gene. For example, querying with “germination” results in three terms, of which two are from the GSO section of the plant growth and development stage ontology and one from the PSO. To avoid getting a large list, users may choose the exact match option before submitting the query. A search for “0 germination” choosing exact match gives one result (Fig. 4A). A user may browse the parents and children of this term by clicking on the blue-colored tree icon and following the [+] sign next to the term name, which suggests that there are additional terms under this term, or simply clicking on the term name “0 germination” for more details. The term detail page (Fig. 4B) provides information on the ID, aspect ontology (plant structure or growth and development), species-specific synonyms, if any, definition, external references and links, if any, and the associated data. The association section allows a user to select the source database, species name, and the evidence code (Table II) used to make the annotation to limit the data displayed. For example, there are 138 gene associations to the term “0 germination” (PO:0007057; Fig. 4B). The list of associated data (Fig. 4C) gives information about the name, symbol, type (e.g. gene), the source, and the species, in addition to the evidence used for inferring the association to the term. The gene symbol provides a hyperlink to the gene detail page (Fig. 4D), and the data source links to the same entry on the provider's Web site. This allows a user to search for extended details that may not be provided in the POC database, such as information on genome location, biochemical characterization, associations to the GOs, etc. For help at any time, users can click on the help menu at the bottom of the browser page or visit the link http://www.plantontology.org/amigo/docs/user_guide/index.html.
Table II.
List of evidence codes for use in annotations to GSO
These are used in building the annotation inferences that indicate the type of experiment cited by the researcher whose data was used to determine the protein and/or transcript expression and phenotype of mutant(s) or quantitative trait loci.
| Evidence Code | Name |
|---|---|
| IC | Inferred by curator |
| IDA | Inferred from direct assay |
| IEA | Inferred from electronic annotation |
| IEP | Inferred from expression pattern |
| IMP | Inferred from mutant phenotype |
| IGI | Inferred from genetic interaction |
| IPI | Inferred from physical interaction |
| ISS | Inferred from sequence or structural similarity |
| NAS | Nontraceable author statement |
| TAS | Traceable author statement |
Annotations to GSO
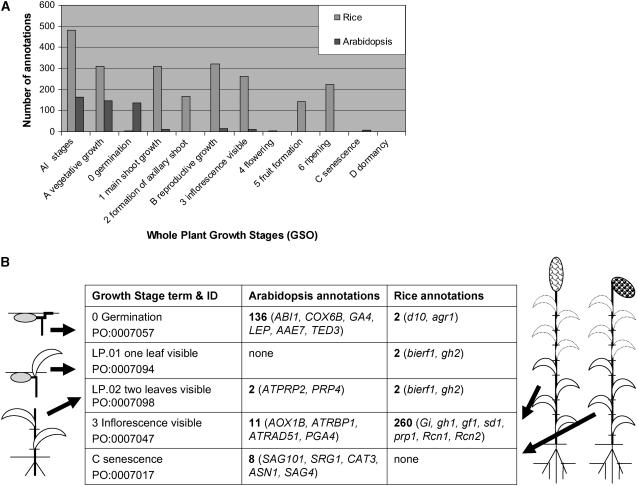
Annotation is the process of tagging snippets of information to the genomic element by skilled biologists to extract its biological significance and deepen our understanding of the biological processes (Stein, 2001). The curator attributes the added information to its source by the use of evidence codes (http://www.plantontology.org/docs/otherdocs/evidence_codes.html) indicating the kind of experiment that was carried out to infer the association to a GSO term, such as inferred from expression pattern (IEP) involving northern, western, and/or microarray experiment, or inferred from direct assay (IDA) such as isolated enzyme and/or in situ assays, etc. (Table II). The user interface has query filter options to search for genes annotated with a given type of evidence code. Explicit spatiotemporal information related to the whole plant is extracted from literature by a curator and described using terms from the GSO. The current build of the GSO has over 600 genes associated to it from the TAIR and Gramene databases (Fig. 5A). Analysis of the data at this point may not be entirely reflective of current research in Arabidopsis and rice, as manual curation is a dynamic and evolving process and will necessarily lag behind the actual state of research. In TAIR, about 130 gene associations to whole-plant growth stages carry the evidence code IEP, while in the Gramene database, a majority of GSO annotations (about 480) carry the evidence code inferred from mutant phenotype (IMP) and a smaller number of IEP and inferred by genetic interaction (IGI) associations. A closer look at the number of genes associated to various terms and their immediate parents (Fig. 5, A and B) reveals that many of these genes with GSO annotations in TAIR are associated to germination stages, which is a vegetative stage. Similarly the vegetative stages, particularly five- to six-leaf stages (children of leaf production) and reproductive stages, namely the inflorescence visible (sensu Poaceae), a child of inflorescence visible, fruit formation, and ripening stages in the rice plant are of particular importance (Fig. 5B). The Solanaceae Genome Network (SGN) has adapted the GSO and has created a mapping file for Solanaceae (tomato [Lycopersicon esculentum]) synonyms that is used to associate their data. Tomato mutants are initially being curated to these terms and, predictably, a large number of mutants will be associated to the ripening stages (data not shown). As we continue to solicit data from collaborating databases and annotate using the GSO, we obtain a global view of how data is associated with different stages of plant growth (Fig. 5B).
Figure 5.
Summary of the Arabidopsis and rice gene annotations to the GSO. A, Growth stage-specific gene annotations from Arabidopsis and rice. The stages prefixed with A to D are the top most categories of the growth stages, namely vegetative, reproductive, senescence, and dormancy. The stages prefixed with 0 to 2 are vegetative substages, and those with 3 to 6 are reproductive substages. All stages means all the GSO terms. B, A list of selected Arabidopsis and rice genes annotated to five specific growth stage terms, suggesting the current state of annotations and not the actual growth stage-specific profile. A similar list can be generated to get growth stage-specific gene expression profiles for a given species. In columns 2 and 3, the numbers (written in bold) appearing before the parentheses are the total number of gene annotations; species-specific genes are written in italics.
Application
This section provides examples of genetic analyses that typically use whole-plant ontogeny as a feature of the experimental design and data analysis. It indicates some of the difficulties of extracting spatiotemporal information from the literature and shows the advantages of curating genomic information using the plant ontologies (GSO and PSO), which allow the users to query when and where a gene is assayed, expressed, or its effects become visible during the life cycle of a plant. In addition the PO database supports queries such as what are the genes that are expressed during the germination stage in Arabidopsis and rice or show me all of the phenotypes in the reproductive stages of a rice plant when mutated.
Annotation Examples of Mutant Phenotypes
The primary description of phenotypic data is usually at the whole-plant level and it is rarely a straight-forward exercise of term-to-term association for the curator. For example, characterization of dwarf mutants is done in different ways, most often by the leaf or node number that is affected, counted either top down or bottom up; in this system the leaf and the internode below it can be used to define the same stage. This is distinct from node visible stages that are less reliable, as the first node that is visible is a variable number in grasses (Fig. 3).
An example is provided by recording of internode elongation, the main morphological feature that is affected in dwarf plants, is attributed among others to the effect of gibberellin and brassinosteroids (Chory, 1993; Ashikari et al., 1999). Yamamuro et al. (2000) show that brassinosteroid plays important roles in internode elongation in rice and have characterized dwarf mutants based on the specific internode that is affected. In the dn-type mutant all the nodes are uniformly affected (the total number of nodes in a given mature rice plant). However, in the nl-type mutant, only the fourth internode is affected, while in the case of the sh-type mutant, only the first internode is affected. However, in this case, the authors of the study number the internodes from top down (the uppermost internode below the panicle is the first internode). To be consistent with the GSO, these numbers have to be converted to the appropriate leaf/node counting from the base of the shoot (Fig. 3). This has to be achieved by the curator's personal knowledge of the plant, from legacy information available for the species and germplasm accession, or by contacting the authors. Unlike the above example, generally leaves are counted from below and the curator extracts information from statements such as when the plant is at the three-leaf stage. This permits an immediate visualization of the morphological appearance of the plant to the researcher and curator as well as the user (Fig. 3). Currently by using the IMP filter, more than 500 genes annotated to different growth stages are available in the PO database.
Cross-Database Comparison of Gene Annotations
Almost all organismal databases are mutually exclusive and provide little or no overlap in their schemas with other databases. Thus they cater to exclusive user communities. To illustrate how the use of ontologies can overcome database interoperability problems, we compare the related processes of flowering time in Arabidopsis and heading date in rice (Fig. 6). The gene network underlying the photoperiodic flowering response involves photoreceptors, circadian clock systems, and floral regulator genes (Yanovsky and Kay, 2002; Izawa et al., 2003; Putterill et al., 2004; Searle and Coupland, 2004). Interestingly, the molecular components that underlie the transition from vegetative to reproductive growth are conserved in Arabidopsis and rice (Hayama and Coupland, 2004; Putterill et al., 2004).
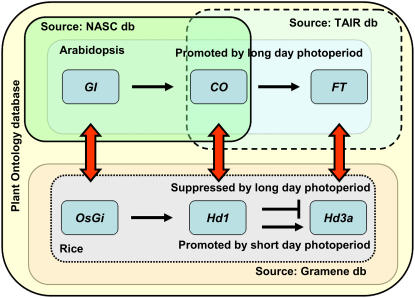
Figure 6.
Genes participating in the flowering time pathway. This image illustrates the flowering time pathway genes from Arabidopsis, GI, CO and FT, and rice, OsGI, Se1 (Hd1), and Hd3a. In the PO database, the annotation for these genes is provided by three databases, the National Arabidopsis Stock Centre, TAIR (for Arabidopsis), and Gramene (for rice). The curators have used terms (Table III) from the whole GSO and PSO to suggest when and where in a plant these genes were expressed or their phenotype was observed. Based on the experiment types (evidence codes) and citation evidences, the databases recorded information about the mutant/gene/gene product to the GSO and the PSO terms. Compared to the short-day length promotion of flowering in rice, flowering is promoted by long-day exposure in Arabidopsis. When rice is exposed to long days, it leads to a down regulation of the Hd3a gene by Se1 (Hd1), leading to a delayed transition of the vegetative shoot apical meristem to the reproductive inflorescence meristem. In other words, the growth stage inflorescence visible (sensu Poaceae), which is synonymous with heading stage, is delayed. The double-headed arrows suggest that the Arabidopsis and rice genes are orthologous. The colored boxes around the genes represent the databases that provided the gene annotations. In the PO database, the putative orthology of these genes cannot currently be determined or displayed, but it can be inferred by visiting either the Gramene or the TAIR database.
The three key regulatory genes in Arabidopsis are GIGANTEA (GI), CONSTANS (CO), and FLOWERING TIME (FT), and in rice they are Oryza sativa Gigantea (OsGI), Photosensitivity (Se1; synonymous with Heading date 1 [Hd1]), and Hd3a (Hayama et al., 2003; Fig. 6). GI is an activator of CO (Izawa et al., 2000) and literature provides evidence that the Se1 (Hd1) gene from rice is an ortholog of a CO family member in Arabidopsis (Putterill et al., 1995; Yano et al., 2000). Furthermore, an allele at the Hd3a locus in rice promotes the transition to floral development (Kojima et al., 2002) and it appears to be an ortholog of FT (Kardailsky et al., 1999; Kobayashi et al., 1999). Thus, the relationship of OsGI to Se1 (Hd1) and that of Se1 (Hd1) to Hd3a in rice is similar to GI, CO, and FT in Arabidopsis, despite the fact that Arabidopsis is a long-day plant while rice is a short-day plant (Kojima et al., 2002; Hayama et al., 2003; Fig. 6).
At present all the above genes are available in the PO database, annotated either or to both GSO and PSO terms (Table III). The Arabidopsis databases, National Arabidopsis Stock Centre and TAIR, have used IMP, IEP, IDA, and traceable author statement (TAS) evidence codes to annotate GI, CO, and FT genes to the exact plant structure where they are expressed. Gramene database has used the IMP and IGI evidence codes to annotate OsGi, Se1 (Hd1), and Hd3a. For rice the IGI code was used to describe the epistatic interaction between Se1 (Hd1) and Hd3a. Table III also includes the annotation of the same genes to the GO. Although this information is not provided by the POC database, it can be retrieved by visiting the respective source databases TAIR and Gramene from the gene detail pages. The information on GO annotations further suggests the biochemical roles of these genes and their functional similarity or dissimilarity.
Table III.
Annotation of the three orthologous sets of flowering time pathway genes from Arabidopsis and rice
The GSO and PSO annotations for Arabidopsis GI, CO, and FT, and rice OsGI, Se1 (Hd1), and Hd3a genes were imported from the PO database. The annotations to GO (columns 4–6) were imported from TAIR and Gramene databases to give an overview on the functional characteristics of the orthologous genes. The curators assigned the ontology terms from the whole growth stage (GSO) and plant structure (PSO) aspect to suggest when and where in a plant these genes were expressed or phenotyped. Depending on the experiment type (evidence code) and citation evidence (references), the databases recorded information about the mutant/gene/gene product to GSO and PSO terms. The same procedure was used for GO annotations. Columns 2 to 6 provide information on the term names, ID, and the evidence code.
| Genes | Plant Growth Stage (GSO) | Plant Structure (PSO) | GO: Cellular Component | GO: Molecular Function | GO: Biological Process |
|---|---|---|---|---|---|
| Arabidopsis | |||||
| GI | Whole plant (PO:0000003; TAS) | Flower (PO:0009046; IMP) | Nucleoplasm (GO:0005654; IDA) and nucleus (GO:0005634; IDA) | Unknown | Response to cold (GO:0009409; IMP), flower development (GO:0009908; TAS), regulation of circadian rhythm (GO:0042752; IMP), positive regulation of long-day photoperiodism, and flowering (GO:0048578; IMP) |
| CO | Not available | Flower (PO:0009046; IMP) | Nucleus (GO:0005634; NAS) | Transcription factor activity (GO:0003700; ISS) | Regulation of flower development (GO:0009909; IMP) |
| FT | Not available | Leaf (PO:0009025; TAS) and shoot apex (PO:0000037; IDA) | Unknown | Phosphatidylethanolamine binding (GO:0008429; ISS) and protein binding (GO:0005515; IPI) | Positive regulation of flower development (GO:0009911; IMP) |
| Rice | |||||
| OsGi | Inflorescence emergence from flag leaf sheath (PO:0000003; IMP) | Inflorescence (PO:0009049; IMP) | Nucleus (GO:0005634; IEP) | Unknown | Inflorescence development (GO:0010229; IMP) |
| Se1 (Hd1, Fl1) | Inflorescence emergence from flag leaf sheath (PO:0007041; IGI, IMP), stem elongation (PO:0007089; IMP), and FR.04 fruit ripening complete (PO:0007038; IMP) | Floret (sensu Poaceae; PO:0006318; IMP), inflorescence (PO:0009049; IGI, IMP), inflorescence meristem (PO:0000230; IMP), and seed (PO:0009010; IMP) | Nucleus (GO:0005634; IEP) | DNA binding (GO:0003677; ISS), transcription factor activity (GO:0003700; ISS), and zinc ion binding (GO:0008270; ISS) | Inflorescence development (GO:0010229; IGI), long-day photoperiodism (GO:0048571; IEP, IGI), and short-day photoperiodism (GO:0048572; IEP, IGI) |
| Hd3a (Fl32a) | Inflorescence emergence from flag leaf sheath (PO:0007041; IMP), stem elongation (PO:0007089; IMP), and FR.04 fruit ripening complete (PO:0007038; IMP) | Floret (sensu Poaceae; PO:0006318; IMP), inflorescence (PO:0009049; IMP), inflorescence meristem (PO:0000230; IMP), seed (PO:0009010; IMP), and sporophyte (PO:0009003; IEP) | Unknown | Phosphatidylethanolamine binding (GO:0008429; ISS) | Inflorescence development (GO:0010229; IMP), short-day photoperiodism (GO:0048572; IMP), and regulation of timing of transition from vegetative to reproductive phase (GO:0048510; IMP) |
Cross-database querying is often difficult because of the way the stage of plant growth is described or the way a trait or phenotype is assayed and curated in species-specific databases. In Arabidopsis the time of flowering is indicated by the number of rosettes on a plant (Samach et al., 2000), while it is indicated by the number of days between planting (or transplanting) and heading of the primary panicle in rice (Yano et al., 2000). The phenology or growth stage studied in both plants is the same (appearance of reproductive structure), but the annotation typically used to identify that growth stage is very different. Once generic terminology describing plant phenology/growth stages is agreed upon and consistently utilized in database curation, these kinds of results will become more readily accessible with fewer queries.
Standard Growth Stage Vocabulary in Experimental Description and Design
Associated with the problem of database curation is the problem of data collection in laboratories and research groups, where data related to plant growth stages are typically collected based on chronological age alone such as 5 d after germination, 10 d after flowering, 1-month-old plant, leaf tissue was harvested in the spring of 2005, etc. The widely differing developmental timelines do not allow meaningful comparisons, even among members of the same species, particularly when environmental conditions vary. However, if critical studies can be performed on a few model genotypes from the same species across various environments they can serve as a reference. This kind of data has been described for 24 rice cultivars, including Nipponbare, Azucena, IR36, IR64, Koshihikari, etc. (Yin and Kropff, 1996), for 19 genotypes of maize, including B73, Mo17, hybrid B73xMo17, and 16 additional hybrids (Padilla and Otegui, 2005), and a comparative study including wheat, barley, and maize (McMaster et al., 2005). The overall outcome of all these studies suggested that although genotypes may differ in their growth profiles in terms of growth rate or flowering time as a result of environmental variables (i.e. light, temperature, or water-deficit conditions), the targeted vegetative growth stages recorded by counting the number of leaves almost always followed a predictable pattern for a given genotype. The responses to variables such as increase or decrease of growth rate or stem elongation, versus the leaf numbers, were not interdependent. This further proved that such experiments can be used by researchers to estimate the growth stage profile based on counting the number of leaves and that this estimate of growth stage was independent of the environment as long as the genotype is known. Thus, data collected with reference to a commonly defined series of whole-plant growth stages such as the ones described in the GSO will provide greater coherence and facilitate comparisons between and within species (Boyes et al., 2001).
DISCUSSION
The GSO is meant to link genetic and molecular information along the ontogenetic trajectory of plant growth, from germination to senescence in developmental time and space. Development is the execution of the genetic program for the construction of a given organism. The morphological structure is the product of many hundreds or thousands of genes that must be expressed in an orchestrated fashion to create any given tissue, body part, or multicellular structure (Davidson, 2001). Development is thus the outcome of a vast network of genes whose expression is regulated both spatially and temporally. Suites of genes are expressed only during specific times during the life cycle of a plant, while other genes are turned on and off intermittently throughout the life cycle. Effective annotation of growth stage-specific gene markers in plant genome databases requires the development and use of ontologies, such as the GSO described here. Many genetic and developmental studies are initially conducted using a specific model system that is rich in genomic resources, but validation of hypotheses often depends on investigation of multiple plant systems (Cullis, 2004). Incorporation of information from multiple sources requires integration and synthesis of data across species and database boundaries. The use of common terminology to describe homologous features in diverse species is the first step. Inclusion of synonyms for growth stages of every plant species offers an effective solution for the immediate term, but may become unwieldy in the future. It is analogous to the approach taken by the WORDNET project that defines words using sets of synonyms and currently covers 150,000 English words (Fellbaum, 1998). We are working with our software developers to provide tools that will categorize synonyms, eventually helping the user community to find the GSO terms that qualify as the growth stage terms for the plant species of their choice and automate the process of identifying derivative synonyms that can be queried in multiple ways. For example, a user may want to query on the terms sixth leaf/six leaves/6 leaves, all of which are derivatives of each other. Improvements in developer's tools will help prevent the ontology from becoming unwieldy and will greatly improve the efficiency of searches.
The GSO will also be valuable in describing high-throughput experimental designs, where plant development is typically analyzed using global patterns of gene expression at defined developmental stages (Schnable et al., 2004). We further anticipate that the design of an experiment is likely to influence the potential to conduct comparative analyses. For example, a problem may arise when a normalized set of tissue samples, e.g. from leaf tissue harvested at the three-, six-, and 10-leaf stages, is used to isolate a protein sample for a proteomics experiment or mRNA for either the microarray experiment or for constructing an expressed sequence tag/cDNA library. Unless each sequence from the library is associated with a particular source tissue and growth stage, it is very difficult to ascertain the actual growth stage at which the mRNA was expressed. Further in the PSO and GSO annotations it is not necessary that one gene is associated with only one plant structure and growth stage description. There can be multiple annotations to accommodate the necessary information about an expression profile, e.g. an expressed sequence tag accession can be expressed in leaf tissue at both the three- and 10-leaf stage but it may not be detected in the six-leaf stage. Hence, the use of well-defined GSO would be extremely useful to provide a framework for comparing gene expression patterns analyzed at different stages within and across species.
The generic design of the GSO aims to facilitate the process of integrating genomic information from diverse plant systems to deepen our understanding of plant form and function. Adoption of the ontology will contribute to its continued improvement and development and will promote an increasingly global view of plant biology. Members of the POC have used the emerging GSO to annotate genes and phenotypes in plants. As proof of concept, data associations from TAIR and Gramene are already available and users can now search over 600 annotated genes, updated on a monthly basis. The Gramene database (Jaiswal et al., 2006) will display the cereal GSO together with the GSO and eventually retire the cereal GSO, giving transition time for its users to familiarize themselves with the new terms. A similar approach will be taken by TAIR (Rhee et al., 2003), and MaizeGDB (Lawrence et al., 2005) is currently testing their annotations. Initially, emphasis was focused on the core databases but expanding use of the ontology by Soybase collaborators Rex Nelson and Randy Shoemaker and SGN collaborators Naama Menda and Lukas Mueller highlights its utility for comparative genomics. Soybase has adapted the GSO for description of soybean (Glycine max) data. SGN adapted the GSO for taxonomic family-wide description of Solanaceous plants and is currently testing it for tomato mutant description. In subsequent releases associations to maize and tomato will become available in the PO database, followed by soybean.
As our understanding of the gene networks and underlying molecular details regarding the origin and diversification of complex pathways such as flowering time grows, a challenge is presented to test the ability to place this knowledge into a framework that can accommodate the information as it emerges and place it into an appropriate comparative context. Similarly our current understanding of genetics and evolution in plants raises many questions about orthology, paralogy, and coorthology in diverse species (Malcomber et al., 2006). The functional relationships among these genes and gene families will be reflected in databases that annotate such information using precise morphological terms from the GSO and the PSO. The effective use of controlled vocabularies also helps identify problems and gaps in knowledge related to the curation of genes in different species where the evolutionary relationships are not entirely clear. Drawing from the experience of its core databases, the POC in the future will address the above issues by preparing and sharing annotation standards that can be used by other member databases to the benefit of the larger plant science community.
The current GSO design is based on annual plants, therefore discussions are underway with collaborators representing the poplar (Populus spp.) and citrus research communities to expand it to include perennials. We also hope that future software developments will allow us to hard wire temporal relationships into the ontology. We encourage databases and individual researchers to contact us if they are suggesting new terms, modification of existing definition(s), term-to-term relationships, or even interested in joining the POC by contributing the associations to their genes and mutant phenotypes by writing an e-mail to po-dev@plantontology.org. More information about joining POC can be found online (http://www.plantontology.org/docs/otherdocs/charter.html).
MATERIALS AND METHODS
Ontology Development
Biologists from University of Missouri at St. Louis and Missouri Botanical Gardens, and curators from the TAIR, MaizeGDB, and Gramene databases worked together to evaluate growth and development in Arabidopsis (Arabidopsis thaliana), maize (Zea mays), and rice (Oryza sativa), examining the vocabularies and models used to describe the whole-plant growth stages in each species. Growth stages of Arabidopsis were described by Boyes et al. (2001) based on the BBCH scale (Meier, 1997) that includes both monocot and nonmonocot species. The BBCH scale in turn is based on the Zadok scale, developed for Triticeae (Zadok et al., 1974), which forms one of the literature bases for the cereal GSO developed by Gramene database (Jaiswal et al., 2006). Rice terminology was derived from INGER (1996), for Triticeae from Zadok et al. (1974) and Haun (1973), and for sorghum from Doggett (1988). MaizeGDB (Lawrence et al., 2005) derives its growth stage vocabulary from a modified version of Ritchie's scale (Ritchie et al., 1993). The vocabulary developed by MaizeGDB was integrated into cereal GSO in the Gramene database as well. With these preexisting interconnections in the core databases, we were able to begin synthesizing them into a generic ontology. Similar growth stage concepts for the above species were identified and mapped to the generic growth stages and stored in mapping files. The mapping files are available at http://brebiou.cshl.edu/viewcvs/Poc/mapping2po/. More details about the project and ontology development is available on the documentation section of the PO Web site (http://www.plantontology.org/docs/docs.html).
Review of Ontology
All aspects of the ontologies developed by the POC, including the GSO, are a collaborative effort and involve evaluation and assessment by numerous external experts. Before each ontology is released to the public, the POC's internal board of senior editors provides critical assessments and offers suggestions for substantive changes that are thoroughly discussed and incorporated into a revised version of the ontologies. The revised ontologies are then released to database curators and developers, who check for inconsistencies and provide critical feedback about problems and/or advantages associated with use of the new ontologies. In the final phase, the ontologies are subjected to review (http://www.plantontology.org/docs/growth/growth.html) by an external panel of experts. Over 15 outside scientists with expertise in the growth and development of diverse plant species have provided valuable input to the development of this ontology (http://www.plantontology.org/docs/otherdocs/acknowledgment_list.html).
Ontology Editing Tools and Web Interface
The plant ontologies are built and maintained using the DAG editor (DAG-edit) developed by the GO software group. It is open source software implemented in Java and installed locally; flat files are used to store the ontologies. DAG-edit permits creating and deleting new terms, and adding synonyms in categories such as exact, broad, narrow, or related synonyms. This software also supports a user-defined plug in for reading, saving, importing, and exporting (Harris et al., 2004; http://sourceforge.net/project/showfiles.php?group_id=36855). The ontologies are shown using a tree structure. As the GSO is a relatively small ontology, the DAG-edit shows a good overview of the expanded tree in one window. The tool DAG-edit was superseded by the Open Biomedical Ontology Editor (OBO-edit) in its recent release by the GO software group. The same will be used in the future development and maintenance of the GSO.
The PO uses the Amigo ontology browser as the Web interface for searching and displaying the ontologies (Fig. 4). Querying can be done using term names, numerical identifier, synonyms, or definitions. The associated annotations to terms from all the represented databases can be viewed on the term detail page (Jaiswal et al., 2005).
Acknowledgments
We thank Drs. Rex Nelson and Randy Shoemaker from Soybase and Naama Menda and Lukas Mueller of SGN for their participation in development and mapping of Soybean and Solanaceae terms to the GSO. We thank numerous researchers for reviewing the GSO and they are listed online (http://www.plantontology.org/docs/otherdocs/acknowledgment_list.html). We apologize to the countless scientists and farmers whose observations on growth stages and plant development were not incorporated into this manuscript for lack of space. We acknowledge the Gene Ontology Consortium for software support. We thank Drs. Dean Ravenscroft and Junjian Ni of Gramene database for reviewing the manuscript.
This work was supported by the National Science Foundation (grant no. 0321666).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Susan McCouch (srm4@cornell.edu).
Open Access articles can be viewed online without a subscription.
References
- Ashikari M, Wu J, Yano M, Sasaki T, Yoshimura A (1999) Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the alpha-subunit of GTP-binding protein. Proc Natl Acad Sci USA 96: 10284–10289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard JB, Rhee SY (2004) Ontologies in biology: design, applications and future challenges. Nat Rev Genet 5: 213–222 [DOI] [PubMed] [Google Scholar]
- Boyes BC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J (1993) Out of darkness: mutants reveal pathways controlling light-regulated development in plants. Trends Genet 9: 167–172 [DOI] [PubMed] [Google Scholar]
- Counce PA, Keisling TC, Mitchell AJ (2000) A uniform, objective, and adaptive system for expressing rice development. Crop Sci 40: 436–443 [Google Scholar]
- Cullis CA (2004) Plant Genomics and Proteomics. John Wiley & Sons, Hoboken, NJ
- Davidson HE (2001) Genomic Regulatory Systems, Ed 1. Academic Press, San Diego
- Doggett H (1988) Sorghum, Ed 2. John Wiley & Sons, New York
- Fellbaum C (1998) WordNet: An Electronic Lexical Database. MIT Press, Cambridge, MA
- Gopalacharyulu PV, Lindfors E, Bounsaythip C, Kivioja T, Yetukuri L, Hollmen J, Oresic M (2005) Data integration and visualization system for enabling conceptual biology. Bioinformatics (Suppl 1) 21: i177–i185 [DOI] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al (2004) The gene ontology (GO) database and informatics resource. Nucleic Acids Res 32: D258–D261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haun JR (1973) Visual quantification of wheat development. Agron J 65: 116–119 [Google Scholar]
- Hayama R, Coupland G (2004) The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol 135: 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422: 719–722 [DOI] [PubMed] [Google Scholar]
- INGER (1996) Standard Evaluation System for RICE. International Rice Research Institute, Manila, Philippines, pp 1–52
- Izawa T, Oikawa T, Tokutomi S, Okuno K, Shimamoto K (2000) Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant). Plant J 22: 391–399 [DOI] [PubMed] [Google Scholar]
- Izawa T, Takahashi Y, Yano M (2003) Comparative biology comes into bloom: genomic and genetic comparison of flowering pathways in rice and Arabidopsis. Curr Opin Plant Biol 6: 113–120 [DOI] [PubMed] [Google Scholar]
- Jaiswal P, Avraham S, Ilic K, Kellogg EA, Pujar A, Reiser L, Seung RY, Sachs MM, Schaeffer M, Stein L, et al (2005) Plant ontology (PO): a controlled vocabulary of plant structures and growth stages. Comp Funct Genomics 6: 388–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal P, Ni J, Yap I, Ware D, Spooner W, Youens-Clark K, Ren L, Liang C, Zhao W, Ratnapu K, et al (2006) Gramene: a bird's eye view of cereal genomes. Nucleic Acids Res 34: D717–D723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43: 1096–1105 [DOI] [PubMed] [Google Scholar]
- Lawrence CJ, Seigfried TE, Brendel V (2005) The maize genetics and genomics database: the community resource for access to diverse maize data. Plant Physiol 138: 55–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcomber ST, Preston JC, Reinheimer R, Kossuth J, Kellogg EA (2006) Developmental gene evolution and the origin of grass inflorescence diversity. Adv Bot Res 44: 423–479
- McMaster GS, Wilhelm WW, Frank AB (2005) Developmental sequences for simulating crop phenology for water-limiting conditions. Aust J Agric Res 56: 1277–1288 [Google Scholar]
- Meier U (1997) Growth Stages of Mono- and Dicotyledonous Plants (Biologische Bundesanstalt, Bundessortenamt und Chemische Industrie [BBCH]-Monograph). Blackwell Wissenschafts-Verlag, Berlin
- OBO (2005) Open Biological Ontologies. http://obo.sourceforge.net/ (June 1, 2006)
- Padilla JM, Otegui ME (2005) Co-ordination between leaf initiation and leaf appearance in field-grown maize (Zea mays): genotypic differences in response of rates to temperature. Ann Bot (Lond) 96: 997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J, Laurie R, Macknight R (2004) It's time to flower: the genetic control of flowering time. Bioessays 26: 363–373 [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857 [DOI] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al (2003) The Arabidopsis information resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie SW, Hanway JJ, Benson GO (1993) How a Corn Plant Develops. CES Special Report No. 48. Iowa State University, Ames, IA, p 21
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Schnable PS, Hochholdinger F, Nakazono M (2004) Global expression profiling applied to plant development. Curr Opin Plant Biol 7: 50–56 [DOI] [PubMed] [Google Scholar]
- Searle I, Coupland G (2004) Induction of flowering by seasonal changes in photoperiod. EMBO J 23: 1217–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B (2004) Beyond Concepts: Ontology as Reality Representation. In AVaL Vieu, ed, International Conference on Formal Ontology and Information Systems. Proceedings of FOIS 2004, Turin, Italy
- Stein L (2001) Genome annotation: from sequence to biology. Nat Rev Genet 2: 493–503 [DOI] [PubMed] [Google Scholar]
- Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M (2000) Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12: 1591–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, et al (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA (2002) Molecular basis of seasonal time measurement in Arabidopsis. Nature 419: 308–312 [DOI] [PubMed] [Google Scholar]
- Yin X, Kropff MJ (1996) The effect of temperature on leaf appearance in rice. Ann Bot (Lond) 77: 215–221 [Google Scholar]
- Zadok JC, Chang TT, Konzak FC (1974) A decimal code for growth stages of cereals. Weed Res 14: 415–421 [Google Scholar]