Abstract
Steroidogenic factor 1 (SF-1), also known as adrenal 4-binding protein, is a member of the nuclear hormone receptor family that regulates transcription of genes encoding hormones and steroidogenic enzymes important to the function of the hypothalamic-pituitary-gonadal axis. The mammalian Ftz-F1 gene encodes SF-1 and is required for development of adrenal glands and gonads. To better understand the mechanisms regulating this gene in the gonads, we have examined its expression in the testis and characterized the promoter region for SF-1 in two testicular cell types. SF-1 promoter activity was examined in primary cultures of Sertoli cells and cell lines representative of Sertoli and Leydig cells. Deletion mutagenesis of the promoter identified several regions: both 5′ and 3′ to the transcriptional start sites that are important for transcriptional activity. Two elements, an E box and a CCAAT box, were found to be important for SF-1 transcription in the testis. An oligodeoxynucleotide containing both of these elements bound three specific protein complexes. The binding of one complex required only sequences within the E box and cross-reacted with antibodies against the basic helix-loop-helix ZIP proteins USF1 and USF2. A second specific complex required sequences within both the E box and CCAAT box for efficient binding, while a third complex predominantly interacted with sequences within the CCAAT motif. The presence of multiple protein complexes binding these sites suggests that regulation through these elements may involve interactions with different factors that depend on the state of the cell and its environment.
INTRODUCTION
Steroidogenic factor 1 (SF-1), also known as adrenal 4-binding protein (Ad4bp), is a transcription factor that has emerged as a key regulator of endocrine function and the development of adrenal glands and gonads [1–6]. SF-1 is a member of the nuclear hormone receptor family that was initially recognized for its role in the tissue-specific expression of the cytochrome P450 steroid hydroxylase genes [7, 8]. The promoters of these genes contain a common regulatory motif (AGGTCA) through which SF-1 binds and activates transcription. More recently, SF-1’s role in endocrine regulation has expanded to include genes acting at multiple levels of the hypothalamic-pituitary-gonadal and -adrenal axes [6, 9–13]. Although SF-1 has been implicated in the development of adrenal glands and gonads, little is known about the developmentally critical events regulated by this transcription factor or those leading to its induction.
The gene encoding SF-1 most closely resembles the Drosophila FTZ-F1 gene from which two developmentally regulated nuclear receptors are expressed [14, 15]. The mouse gene encoding SF-1 was designated Ftz-F1 and has been shown to express four distinct transcripts specified as ELP1, ELP2, ELP3, and SF-1/Ad4bp [16, 17]. The various transcripts arise from distinct promoters and contain both shared and isoform-specific regions. The expression profiles of the transcripts vary, suggesting that the proteins differ in their biological function [17].
Four different Ftz-F1 knockout models have been created, and the findings were consistent between each [3, 5, 6, 18]. The Ftz-F1 null mice presented with external genitalia that were male-to-female sex reversed, and the mice died shortly after birth due to corticosteroid deficiency [3]. In addition, mice lacking the Ftz-F1 gene fail to form adrenal glands and gonads because of an arrest of these developmental pathways in the embryo [3]. Although multiple transcripts are generated from the Ftz-F1 gene, several observations indicate that SF-1 is the gene product responsible for the defects observed in the knockout animals. Importantly, one of the animal models was generated by mutating only the initiating methionine of the SF-1 protein while leaving ELP1 and ELP2 intact. As indicated above, these animals had the same phenotype as the other knockout models. In addition, expression data on SF-1 and the other transcripts were consistent with SF-1’s role in the function of these tissues early in embryogenesis as well as in the adult animal [4].
In the embryo, SF-1 was first observed in what appeared to be a single population of cells in the urogenital ridge and later resolved into two discrete populations that gave rise to adrenocortical and gonadal cells [19]. As development proceeded, SF-1 exhibited a sexually dimorphic pattern of expression in the gonads, with predominant mRNA levels in the testis and little or no expression in the ovary [19, 20]. In the embryonic testis, SF-1 transcripts were found in the interstitial region and within the seminiferous cords, indicating that both Leydig (interstitial) and Sertoli (cords) cells express this protein [19, 21]. SF-1 expression continued after birth in the adrenal gland, testis, and ovary, where it has been detected in adrenocortical cells, testicular Leydig and Sertoli cells, and ovarian theca and granulosa cells [19, 22, 23]. In males, SF-1 expression decreased after the first wave of spermatogenesis, while in females, expression increased with the onset of folliculogenesis. Expression in adrenocortical cells persisted after birth at comparable levels in males and females. Additional studies have led to the identification of SF-1 transcripts in the ventromedial hypothalamic nucleus (e11.5, mouse), the developing pituitary gland (e13.5, mouse), the spleen, and the placenta [5, 6, 10, 13, 17, 18, 24–26].
The pivotal nature of SF-1 in endocrine function and development underscores the importance of understanding the mechanisms that control its expression. Thus identifying proteins that regulate SF-1 transcription will expand our knowledge of the events leading to adrenal and gonad development and those needed for steroid biosynthesis. Studies on transcriptional regulation of SF-1 are limited and have focused primarily on expression in the adrenocortical cell line Y1 and the pituitary gonadotroph cell line αT3 [27–29]. Collectively, these studies have identified an E box, a CCAAT box, and an Sp1 site that are important for promoter activity. The E box was shown to have functional significance in both Y1 and αT3 cells, while the importance of the CCAAT box and Sp1 site was determined only in Y1 cells [27–29]. Protein/DNA binding studies indicated that the transcription factors USF1 and USF2 interacted with the SF-1 E box in αT3 and Y1 cells and that the CCAAT binding factor (CBF/NF-Y) bound the CCAAT motif [27, 28]. Despite its critical role in the development and function of the gonads, there are no known reports identifying elements in the SF-1 gene needed for expression in cells of these tissues. Here, we report the characterization of SF-1 promoter activity in two cell types of the testis, Leydig cells and Sertoli cells. We have found that transcriptional regulation of SF-1 in Leydig and Sertoli cells shares several important regulatory features with expression in Y1 cells, including the use of the E box and CCAAT box elements. In addition, our studies suggest that interactions between proteins binding these elements are important in transcriptional regulation of SF-1 in these cells.
MATERIALS AND METHODS
SF-1 Promoter Clones
Genomic DNA isolated from Sprague-Dawley (Harlan Sprague Dawley, Indianapolis, IN) rats was used as a template in polymerase chain reaction (PCR) to generate a DNA fragment spanning from 734 base pairs (bp) 5′ of exon 1 in the rat Ftz-F1 gene to 60 bp in the 3′ direction (−734/+60). The forward primer used, which contains an added XhoI site, was 5′ GGGGCTCGAGATCCGTCTA-GGCCAGTTCAG 3′. The reverse primer used, which contains an added HindIII site, was 5′ GGGGAAGCTTCTATCGGGCTGTCAGGAACT 3′. The resulting DNA fragment was subcloned into the HindIII/XhoI sites of pGL3-Basic (Promega, Madison, WI) upstream of the luciferase reporter gene. SF-1 deletion mutants SF-1 (−232/+60), SF-1 (−87/+60), and SF-1 (−80/+60) were generated by digestion of SF-1 (−734/+60) with restriction endonucleases XhoI and either PvuII (−232), PstI (−87), or PmlI (−80). The digested promoter/reporters were treated with Klenow to generate blunt ends and ligated together. SF-1 block-replacement mutants were generated by PCR mutagenesis as described elsewhere [30, 31]. Amplified DNA fragments containing mutations were cloned into the HindIII/PstI sites of the SF-1 (−734/+60) plasmid to yield the desired block mutant. All clones were confirmed by sequencing using the ABI Prism dRhodamine Terminator Cycle Sequence Ready Reaction Kit (PE Applied Biosystems, Foster City, CA).
DNA Preparation
All plasmid DNAs were prepared from overnight bacterial cultures using Qiagen DNA plasmid columns according to the supplier’s protocol (Qiagen, Chatsworth, CA). Oligodeoxynucleotides were purchased from either Life Technologies (Gaithersburg, MD) or Genosys (The Woodlands, TX).
Cell Culture Conditions and Transfection Analysis
Preparation of primary rat Sertoli cells and culture conditions for the MSC-1 mouse Sertoli cell line have been previously described [30]. Culture conditions for MA-10 cells, a mouse Leydig tumor cell line, have been described [32]. For transient transfection studies, Sertoli cells were plated by testis weight (1.7 mg/well) onto 12-well dishes 3 days prior to transfection. On the morning of transfection, medium was changed to Dulbecco’s modified Eagle’s medium (Life Technologies, Gaithersburg, MD) without additives. Four to six hours later, cells were transfected using 0.2 μg of pGL3-reporter vector and 45 ng of pRL-TK in combination with 1 μl lipofectamine and 6 μl Plus reagent according to the manufacturer’s recommendations (Life Technologies); pRL-TK expresses Renilla luciferase from the herpes simplex virus thymidine kinase promoter and was included to control for transfection efficiency. Sixty hours after transfection, the cells were lysed and assayed for both firefly and Renilla luciferase using the Dual-Luciferase Reporter Assay System (Promega). MSC-1 cells were plated in 24-well plates at 52 000 cells per well and then transfected using 0.3 μg pGL3-reporter vector, 10 ng pRL-TK, and 2 μl lipofectamine. Specifics of the transfection procedure are described elsewhere [30, 31]. MA-10 cells were plated at a density of 26 000 cells per well and transfected using 0.1 μg pGL3-reporter, 10 ng pRL-TK, and 1.5 μl lipofectamine. Lipid/DNA complexes were removed after 12–14 h, and the cells were re-fed with complete medium (Waymouth’s supplemented with 20 mM Hepes, 15% horse serum, and 50 μg/ml gentamicin). MSC-1 and MA-10 cells were assayed as described for primary Sertoli cells. For each promoter construct, at least two independent clones were examined. The transfection data were represented as the firefly luciferase/Renilla luciferase activity of each construct, which was normalized to the firefly luciferase/Renilla luciferase activity of either pGL3-Basic or the wild-type SF-1 (−734/+60 bp) promoter construct. The values were then averaged over a minimum of three independent experiments.
Determination of Transcriptional Start Sites and PCR Analysis of Ftz-F1
Transcriptional start sites were mapped by rapid amplification of cDNA ends (RACE) as described elsewhere [33,34]. Primers used for the 5′ RACE were generated against exon 3C from both mouse and rat Ftz-F1 [17,29]. RNA was isolated from primary Sertoli cells, MSC-1 cells, and MA-10 cells using Trizol reagent according to manufacturer recommendations (Life Technologies). Complementary DNA was generated from total RNA using the most 3′ primer SF1.7 (5′-CTTGCAGCTCTCGCACGT-3′). Amplified products from two rounds of PCR using the Ftz-F1-specific primers SF1.8 (5′-GAGCAGCCCGTAGTGGTAG-3′) and SF1.9 (5′-GCCGAAGCTTACACACTGGACACAGCTCGTC-3′), respectively, were subcloned, and individual clones were sequenced. Additional conformation of Ftz-F1 gene products was determined by PCR using primer sets that are specific for amplifying specific Ftz-F1 gene products—for ELP1 (ELP1F: 5′-AGAATCGGGGTTTTGTTCTC-3′, ELP1R: 5′-CAGCCTCGAGAGGCCTG-3′), ELP2 (ELP2F: 5′-CTGTCACTCCTAGCCTCTGAT-3′ and SF1.7), ELP3 (ELP3F: 5′-GGGTTCGCGAAGCGCG-3′ and SF1.7), and SF-1 (SF3: 5′-GGGGTCTAGAAGTTTGCAGTCCGCCGCT-3′ and SF1.7). Amplified PCR products were gel purified and sequenced to confirm the identity of the products.
Electrophoretic Mobility Shift Analysis
To prepare nuclear extracts, cells were washed with ice-cold buffer (25 mM Hepes, pH 7.4, 1 mM dithiothreitol [DTT], 1.5 mM EDTA, 10% glycerol) and scraped from plates into the buffer specified above with 0.5 mM PMSF added. Cells were lysed using 30 strokes of a Dounce homogenizer (B pestle), and the nuclei were pelleted by centrifugation (16 000 × g) for 1 min. The supernatant was removed and nuclei were resuspended in extraction buffer (25 mM Hepes, pH 7.9, 1 mM DTT, 1.5 mM EDTA, 10% glycerol, and 0.5 M KCl). Nuclei were extracted on ice for 10 min and then frozen on dry ice. Nuclei were then thawed and centrifuged (85 000 × g) for 6 min. Supernatants were immediately aliquoted and placed at −80°C. Protein concentration was determined by the BCA method (Pierce, Rockford, IL) using BSA as a standard.
For electrophoretic mobility shift assays, nuclear extracts (6–10 μg protein) were incubated with 25 fmol of radiolabeled double-stranded oligonucleotide in the presence of 10 mM Hepes, pH 7.9, 3 mM MgCl2, 30 mM KCl, 0.5 mM DTT, 12% glycerol, 0.6 mM EDTA, 0.2 mM PMSF, 50 ng salmon sperm DNA, 1 μg dIdC, 10 μM ZnCl, and 1 μg/ml BSA in a 20-μl reaction volume as described elsewhere [30]. Addition of competitors or antibodies to the reaction immediately preceded the addition of extract. Antibodies for USF1 (C-20)X, USF2 (C-20)X, and c-Myc (N-262)X were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). These antibodies are supplied as rabbit polyclonal IgG and were used directly as supplied by the manufacturer at 1 μg IgG per binding reaction.
Reactions were incubated on ice for 10 min prior to addition of probe and then for an additional 30 min on ice before being loaded onto the gel unless otherwise noted. Protein/DNA complexes were resolved on a 4% polyacrylamide gel (acrylamide:bisacrylamide, 40:1) run in 25 mM Tris (pH 8.5), 190 mM glycine at 250 volts for 1.5 h at 4°C. Gels were dried and analyzed by autoradiography.
RESULTS
Expression of Ftz-F1 Transcripts in Sertoli and Leydig Cells
The Ftz-F1 gene has eight exons and encodes four distinct transcripts designated ELP1, ELP2, ELP3, and AD4BP/SF-1. Each transcript is generated from a distinct transcriptional start site and differs with respect to its expression profile [17, 29]. The structure of the Ftz-F1 gene and its four transcripts is depicted in Figure 1A, which shows that both promoter usage and alternative splicing are responsible for differences in the derived mRNAs (adapted from Ninomiya et al. [17]). RACE was used to determine the transcriptional start sites utilized by the Ftz-F1 gene in Sertoli and Leydig cells, the two testicular cell types known to express this gene. 5′ RACE was performed using primers specific for mouse and rat Ftz-F1 sequences within exon 3B, an exon common to each transcript; and RNA was isolated from primary rat Sertoli cells, a mouse Sertoli cell line (MSC-1), and a mouse Leydig cell line (MA-10). Sequence analysis of the RACE products indicated that the large majority of transcripts were generated from the SF-1 promoter (Fig. 1B). In addition, for each cell type, the transcriptional start sites for SF-1 clustered around the +1 and +6 positions relative to that previously reported in adult rat testis, where transcripts were most likely from expression in Leydig cells (Fig. 2) [29]. The 5′ RACE strategy also identified other Ftz-F1 transcripts that appeared to be less abundant as evidenced by the number of sequenced RACE products (data summarized in Fig. 1B).
FIG. 1.
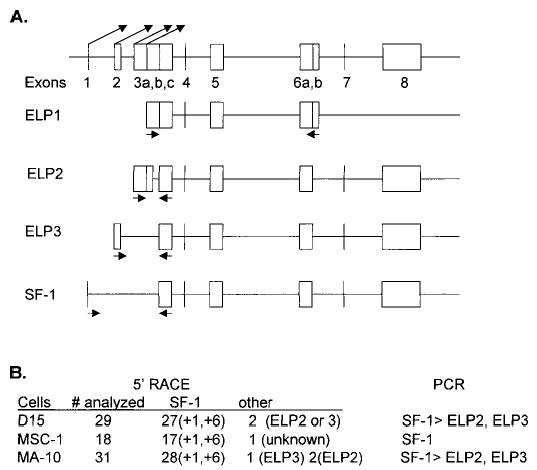
Analysis of Ftz-F1 gene isoforms and transcriptional start sites in Sertoli and Leydig cells. A) Schematic diagram of the Ftz-F1 gene adapted from Ninomiya et al. [17]. The angled arrows above the diagram indicate the transcriptional start sites for SF-1, ELP3, ELP2, and ELP1, left to right, respectively. Primers (not shown) used in 5′ RACE were specific for exon 3C. Arrows under each depicted Ftz-F1 transcript represent the location of primers used in RT-PCR. B) Results from 5′ RACE and RT-PCR. The number (#) analyzed indicates the total number of clones sequenced for which transcripts could be distinguished. Of that total, the number of either SF-1-derived transcripts or other transcripts is indicated. The predominant transcriptional start site for SF-1 is indicated in parentheses and is relative to that reported for the rat [29]. Results from the RT-PCR corroborated the 5′ RACE data. RT-PCR products were isolated and verified by sequencing.
FIG. 2.
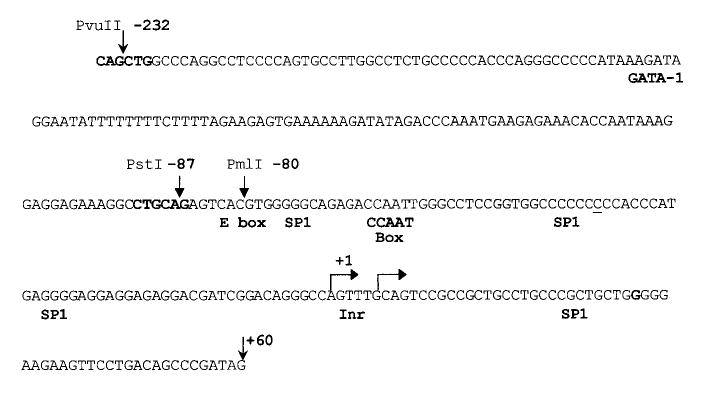
Nucleic acid sequence of the rat SF-1 promoter region. Sequence of the rat promoter region from −235 to +60. PCR was used to amplify the rat SF-1 promoter region using genomic DNA isolated from Sprague-Dawley rats. The sequence corresponded to that previously reported for the rat with the exception of a single-base insertion at position −41 (underlined). This change was confirmed in four independent PCR reactions. The major transcriptional start sites are indicated as bent arrows. Restriction enzyme sites used in cloning deletion mutants and potential transcription factor regulatory elements identified through the Transfac database are indicated above and below the sequence, respectively [58].
In addition, RT-PCR was used to investigate Ftz-F1 expression by employing primers that specifically amplify each transcript. The arrows under each diagramed transcript indicate the location of the primers (Fig. 1A). ELP2, ELP3, and SF-1 transcripts were identified in primary Sertoli cells and MA-10 cells, while in MSC-1 cells, only SF-1 transcripts were found. Comparison of the amplified products after various rounds of amplification (15, 20, 30, 40 cycles) supported the 5′ RACE observation that SF-1 is the predominant transcript in these cells.
Importance of Several Regions of the SF-1 Promoter for Transcriptional Activity in Sertoli and Leydig Cells
To examine transcriptional regulation of SF-1 in the testis, we cloned a fragment of the rat SF-1 promoter spanning from 734 bp in the 5′ direction to 60 bp in the 3′ direction relative to the major transcriptional start site (+1 in Fig. 2). The sequence of the SF-1 promoter from −232 to +60 is depicted in Figure 2. Also shown are sites corresponding to the start of transcription, several restriction enzyme recognition sequences used for promoter analysis, and consensus binding sites for relevant transcriptional regulators. The SF-1 5′ flanking sequence (−734/+60) was placed upstream of the firefly luciferase reporter gene (SF1 [−734/+60] Luc) and transiently transfected into primary Sertoli cells, MSC-1 cells, or MA-10 cells to evaluate promoter function. SF-1 promoter activity was 60-fold greater than that of a promoterless control (pGL3-Basic) in primary cultures of Sertoli cells, while in MSC-1 and MA-10 cells, activity was 18- and 10-fold higher, respectively (Fig. 3A). Relative activity of the SF-1 promoter was therefore greater in primary Sertoli cells than in either of the two cell lines tested. SF1 promoter activity was also examined in the adrenocortical cell line Y1, where it was found to be approximately 45 times greater than that of pGL3-Basic (data not shown).
FIG. 3.
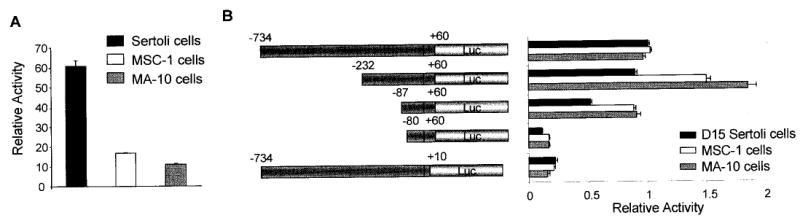
Regions both 5′ and 3′ to the start of transcription were required for SF-1 promoter activity. A region of the Ftz-F1 gene from −734 to +60 bp, relative to the SF-1 transcriptional start site (+1), was cloned upstream of the firefly luciferase reporter gene. Promoter activity was determined by transient transfection analysis in either primary rat Sertoli cells, MSC-1 cells, or MA-10 cells as described in Material and Methods. Promoter constructs were cotransfected with pRL-TK, which expresses Renilla luciferase from the thymidine kinase promoter, and was used to control for transfection efficiency. In both A and B, firefly luciferase activity from the different promoter constructs was normalized to Renilla luciferase values. In A, the firefly/Renilla luciferase activity of the SF-1 (−734/+60) construct was assayed in each of the cell lines and reported as the activity relative to the firefly/Renilla luciferase activity of the promoterless control vector pGL3-Basic. In B, various promoter deletion mutants are indicated; the activity of each, determined as described above, was analyzed in each of the cell types. Here, the data represent the firefly/Renilla luciferase activity of each SF-1 promoter made relative to the firefly/Renilla luciferase activity of the SF-1 (−734/+60) Luc construct. Transfections were done a minimum of three times. Error bars represent the SEM.
To help identify elements in the SF-1 proximal promoter region that are required for activity in Sertoli and Leydig cells, a series of deletion mutants were constructed and transiently transfected into primary Sertoli cells, MSC-1 cells, and MA-10 cells (Fig. 3B). The results indicated that regions both 5′ and 3′ to the transcriptional start sites are important for full promoter activity in all cells studied (Fig. 3B). Deletion from −734 bp to −232 bp had only a modest effect on promoter activity in primary Sertoli cells, while in the cell lines the transcriptional activity increased, suggesting that this region contained elements that repressed expression in these cells (Fig. 3B). The next deletion removed a region from −232 bp to −87 bp and resulted in approximately a 50% reduction in promoter activity relative to the −232/+60 construct. This effect was observed in each of the cell types. Further deletion to −80 bp, which removes half of a consensus E box element, caused the most dramatic reduction in promoter activity. This element has been shown to be important for SF-1 expression in Y1 and alpha T3 cells [27, 28] and appears to be a key element in SF-1 expression in the testis. Evaluation of the SF-1 promoter by deletion mutagenesis also included evaluation of the region 3′ to the transcriptional start site. Deletion of the promoter from +60 bp to +10 bp resulted in a dramatic reduction of promoter activity in all cell types analyzed.
Importance of an E Box and a CCAAT Box for SF-1 Promoter Activity in the Testis
We extended the analysis of the SF-1 promoter by constructing a series of block-replacement mutations to help identify elements important for SF-1 expression in the testis (Fig. 4). Six different block-replacement mutants were generated and placed in the context of the −734/+60-bp promoter. Transient transfection analysis in primary Sertoli cells, MSC-1, and MA-10 cells identified two regions important for promoter activity. One mutation, μ1, disrupts bases within the consensus E box element (CACGTG to agttct). This mutation had the greatest impact on promoter activity in both Sertoli and Leydig (MA-10) cells. Mutations μ3 and μ4 identified a second element important for promoter activity. Within the region mutated by μ4, there was a consensus sequence for a CCAAT box, a regulatory motif commonly found in the promoters of mammalian genes. Our studies indicate that the CCAAT motif and sequences flanking its 5′ side are important for promoter function. Mutations μ2, μ5, and μ6 had no significant impact on promoter function. Thus examination of the region between −83 and −51 revealed the use of two promoter elements, an E box and a CCAAT box, in the regulation of SF-1 transcription in Sertoli and Leydig cells.
FIG. 4.
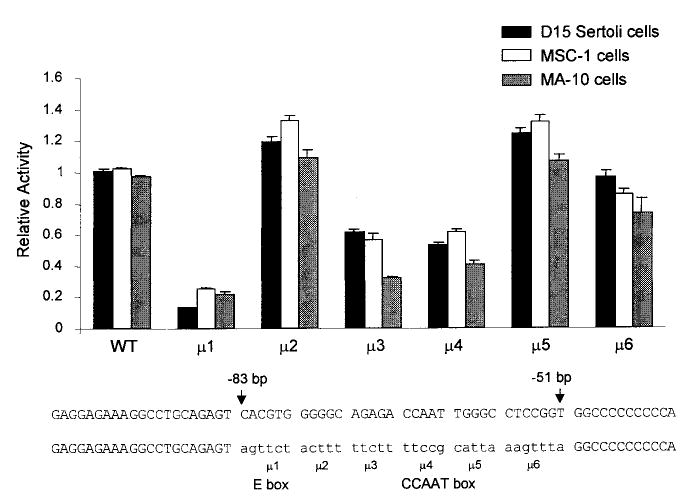
An E box and a CCAAT box were important for transcription of SF-1 in Sertoli and Leydig cells. Six block-replacement mutants spanning the region between −83 and −51 bp of the SF-1 promoter were generated and placed into the context of the SF-1 (−734/+60) Luc vector. The sequences of the regions mutated and the respective replacement sequences (lowercase) are shown at the bottom of the figure. Mutations 1 and 4 contain sequences for E box and CCAAT box elements, respectively. Promoter activity was determined in primary rat Sertoli cells, MSC-1 cells, and MA-10 cells by transient transfection analysis as described in the legend to Figure 3. The data represent the firefly/Renilla luciferase activity of each construct normalized to the firefly/Renilla luciferase activity of the wild-type −734/+60 promoter construct. Transfections were done a minimum of three times. Error bars represent the SEM.
Occurrence of Cooperative Interactions Between CCAAT Box and E Box Binding Proteins
To examine complexes binding the E box/CCAAT box region of the promoter, an electrophoretic mobility shift assay (EMSA) was performed using an oligodeoxynucleotide probe that encompassed both functional sites of the promoter (E/C; for sequence see Fig. 6B). With use of nuclear extracts from either Sertoli cells or MA-10 cells, three major complexes were observed (Fig. 5, bands 1–3). These complexes bound specifically to the E/C probe as revealed by the ability of specific (E/C) but not nonspecific (NS) sequences to compete for complex binding. Migration of these complexes was similar in Sertoli and Leydig cells, suggesting that related complexes bind these important response elements in the two testicular cell types. MSC-1 nuclear extracts also had three specific complexes, but only two migrated similarly (bands 2 and 3) to complexes observed in Sertoli and MA-10 nuclear extracts. Complexes that formed band 3 appeared more diffuse and tended to run more slowly in the gel when Sertoli cell (or MSC-1) nuclear extracts were used as compared to MA-10 nuclear extracts. A fourth complex that ran below band 3 was also observed with extracts from MSC-1 cells that was not present in Sertoli or MA-10 cells.
FIG. 6.
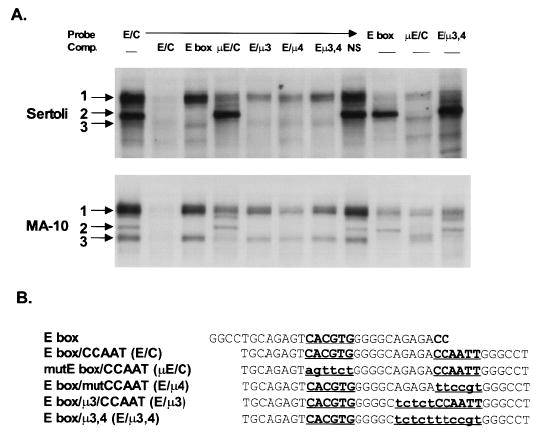
Three complexes having distinct binding requirements interacted with the E box/CCAAT box elements in the SF-1 proximal promoter region. A) EMSAs were performed as described in the legend to Figure 5 using various probes and competitors as indicated at the top of the figure. Assays were performed with nuclear extracts from either primary rat Sertoli cells (top) or MA-10 cells (bottom). B) Sequences of the probes and competitors used in the EMSAs. Bold letters indicate the element of interest; lowercase lettering indicates a change in the sequence from wild type. These mutations are consistent with the mutations tested functionally by transient transfection.
FIG. 5.
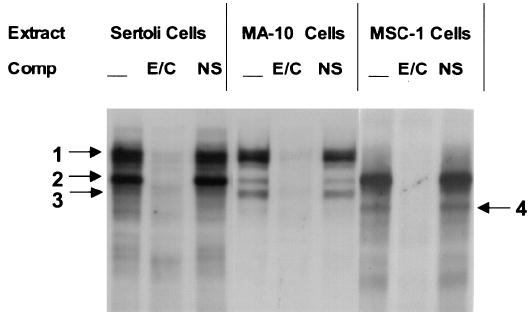
Several complexes bound the SF-1 promoter region containing the E box and CCAAT box. A double-stranded radiolabeled oligodeoxynucleotide probe containing both the E box and CCAAT box sequences (E/C, 5′-TGCAGAGTCACGTGGGGGCAGAGACCAATTGGGCCT-3′) was used in an electrophoretic gel shift assay together with nuclear extracts from either primary Sertoli cells, MA-10 cells, or MSC-1 cells. Radiolabeled probe (25 fmol; E/C) was incubated with 6–10 μg of nuclear extract and resolved on a 4% polyacrylamide gel as described in Material and Methods. Where indicated, unlabeled homologous (E/C) or nonspecific (NS; 5′ CTAGAGTCGACCTGCAGGCATGCAAGCTTGGCATTC-3′) oligodeoxynucleotide competitors were added to the reaction at a concentration equal to 80-fold that of the labeled probe. The arrows with corresponding numbers indicate the major complexes.
Various DNA sequences containing mutations corresponding to those functionally tested by transient transfection were used as competitors in an EMSA to help delineate the sequence requirements of the complexes binding the SF-1 promoter. In both Sertoli and Leydig cells, band 2 appeared to be the only complex whose binding depended solely on bases in the E box (Fig. 6). All competitor DNA sequences containing an intact E box (E/C, E box, E/μ3, E/μ4, and E/μ3,4) were able to compete and thus bind band 2, while those lacking the E box (μE/C, NS) were not. In addition, corresponding complexes migrating similarly to band 2 were observed with all DNA probes containing an intact E box (E/C, E box, E/μ3,4), but not with one that lacked this sequence (μE/C). On the other hand, in both Sertoli and Leydig cells, the binding of complex 3 depended mostly on the presence of sequences within regions 3 and 4 of the promoter. Thus DNA lacking these sequences (E box, E/μ3, E/μ4, E/μ3,4) failed to fully compete for band 3 binding, and probes that lacked these sequences (E box, E/μ3,4) did not have complexes of similar migration to that of band 3. Interestingly, band 1 binding appeared to depend on the presence of both the E box and region 3/4. In this instance, sequences lacking either an E box or regions 3 or 4 failed to fully compete for band 1 binding. Note, however, that the presence of one of these regions was sufficient for partial competition of this band (μE/C, E/μ3, E/μ4, E/μ3,4), but competition was significantly compromised when compared to that of the wild-type competitor (E/C). In addition, probes containing only one of the binding sites (E box, μE/C E/μ3,4) bound significantly less of the complexes in band 1 than the probe containing both sites. These studies indicate that complexes binding the SF-1 promoter E box/CCAAT box region are similar in Sertoli and Leydig cells. In addition, the binding studies indicated that complexes in band 2 depended only on sequences in the E box, while the complexes in band 3 required sequences in the element defined by regions 3 and 4. Furthermore, a third complex (band 1) required sequences within both the E box and region 3/4 for full binding activity, suggesting a cooperative interaction between proteins binding these two elements.
USF1 and USF2 As Components of the Complexes Binding the E Box in Sertoli and Leydig Cells
The E box is a response element with the consensus sequence CANNTG. This element is typically recognized by proteins in the basic helix-loop-helix (bHLH) family that are most often associated with control of cell growth and differentiation. The sequence of the SF-1 E box, CACGTG, often binds regulators in a subclass within this family called bHLH-ZIP proteins that include USF, TFE3, TFEB, c-Myc, Max, and Mad. To determine whether these proteins are components of the complexes formed on the E/C probe, we included antibodies against USF1, USF2, and c-Myc in an EMSA and examined their ability to cross-react with proteins in the complexes (Fig. 7). In Sertoli and Leydig cell nuclear extracts, both the USF1 and USF2 antibodies cross-reacted with proteins in band 2, as indicated by the loss or reduction of bound complex. No cross reactivity was observed with proteins in either band 1 or 3. Antibodies against c-Myc failed to cross-react with complexes in any of the major bands. Thus, USF1 and USF2 formed a complex that binds the SF-1 E box independently of the downstream element but interestingly does not appear to be part of the E box binding complex requiring the downstream region for full binding activity.
FIG. 7.
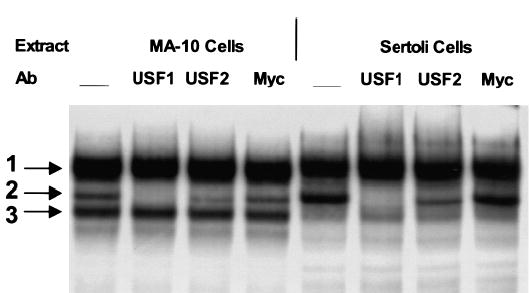
USF1 and USF2 were shown to be components of a complex binding the SF-1 promoter in primary Sertoli cells and MA-10 cells. A radiolabeled probe containing both the E box and CCAAT box elements (E/C, 5′-TGCAGAGTCACGTGGGGGCAGAGACCAATTGGGCCT-3′) was utilized in an electrophoretic gel shift assay with nuclear extracts from either primary rat Sertoli cells or MA-10 cells. Assays were done as described in the legend to Figure 5. Where indicated, 1 μg of rabbit polyclonal IgGs against USF1, USF2, or c-Myc was added prior to addition of the E/C radiolabeled probe.
DISCUSSION
SF-1 is a member of the nuclear hormone receptor family that is an important transcriptional regulator of several steroid biosynthetic enzymes and is necessary for the differentiation of adrenal glands and gonads [7, 8]. SF-1’s pivotal role in endocrine regulation and development emphasizes the importance of understanding the events that regulate the Ftz-F1 gene. To date, our knowledge of the mechanisms needed for SF-1 transcription is limited, and has stemmed predominantly from studies in adrenocortical and gonadotroph cell lines [27–29]. Thus, despite its critical role in testis development and function, little is known about the transcriptional mechanisms responsible for SF-1 expression in testicular tissue. Here, we report our findings on the Ftz-F1 gene expression pattern and activity of the SF-1 promoter in testicular Sertoli and Leydig cells.
Since the gene encoding SF-1 (Ftz-F1) encodes four transcripts that arise from distinct transcriptional start sites, 5′ RACE was used to determine the Ftz-F1 start sites employed in Sertoli and Leydig cells. The results indicated that SF-1 is the major transcript made from the Ftz-F1 gene in both testicular cell types. In addition, the same two major transcriptional start sites were identified in Sertoli and Leydig cells. These two sites were equally represented in the RACE results, and the most 5′ site corresponded to that previously identified in adult rat testis and adrenal [29]. However, since SF-1 expression is significantly decreased in Sertoli cells of the adult testis, these earlier results likely represented only Leydig cell transcripts [21]. Interestingly, the transcriptional start site for SF-1 in mouse EC cells and Y1 cells was mapped to a position just downstream of the +6 site at position +9 [17, 22].
The use of multiple transcriptional start sites is a common observation for genes, like SF-1, that lack a TATA box. In many promoters that lack a TATA motif, an initiator element (Inr) is critical in positioning RNA polymerase II [35, 36]. Several Inr elements have been described and classified according to sequence homology and are generally found to span the transcriptional start sites [35–37]. An Inr consensus sequence (PyPyA+1NT/ApyPy) is found in the SF-1 promoter spanning the first transcriptional start site (Fig. 2; [36]). Further analysis of the gene is required to determine whether this putative Inr is functional. Frequently Inrs work in conjunction with upstream elements to help position the polymerase. These elements are often rich in cytosine and guanine (GC rich) and bind the transcription factor Sp1 [35]. Interestingly, the SF-1 promoter has several GC-rich regions. One of these regions was mutated in our study (μ2) but had no effect on promoter function. However, an Sp1 binding site at position −30 was shown to be important for transcription in Y1 cells [28]. A role for Sp1 binding sites at position −30 in SF-1 regulation in the testis is currently under investigation.
The 5′ RACE studies also helped to assess the relative amounts of Ftz-F1 transcripts in the testis. In these studies, primers were used that bound within an exon present in each of the Ftz-F1 gene products. In both MA-10 and Sertoli cells, less than 10% of the RACE products were from Ftz-F1 transcripts other than SF-1, suggesting that SF-1 is the predominant gene product expressed in these cells. This observation was also supported by RT-PCR studies in which amplification of SF-1 transcripts was detectable after only 20 cycles, while 30 and 40 cycles were required to observe ELP2 and ELP3.
Transcriptional regulation of SF-1 was examined in primary cultures and cell lines of the testis. Promoter activity was notably more robust, relative to that of a promoterless control, in primary Sertoli cells than it was in MSC-1 cells or MA-10 cells. However, examination of the SF-1 promoter in the adrenocortical cell line, Y1, showed that relative promoter activity approached that observed in primary Sertoli cell cultures (data not shown). Our current understanding of these differences is limited; and except in the case of MA-10 cells, promoter activity in the various cells appears to follow the corresponding cellular expression of SF-1 protein and mRNA (unpublished results). Thus, additional genomic sequence may be required for full promoter activity in Leydig cells.
Deletion mutagenesis of the −734/+60 promoter identified several regions that contain important regulatory elements. Deletion of the region between −734 and −232 caused a notable increase in promoter activity in MA-10 and MSC-1 cells, while no significant effect was observed in primary Sertoli cells. The data suggest that a repressor interacts with an element in this region and may partially explain why relative activity of the −734 promoter appeared lower in these cell lines. A second region between −232 and −87 was identified as having a positive regulatory element(s), as deletion of this region resulted in an approximate 2 times decrease in promoter activity in each cell type examined. Published studies using αT3 and Y1 cells, however, did not show a similar effect [27, 28] whereas studies using a granulosa cell line, DC3, did [28]. This suggests that the elements in this region may function specifically in the gonads; but more direct studies are required to confirm this. Analysis of the sequence within this region revealed potential binding sites for several transcription factors, including GATA-1 and HNF3. More refined analysis of this area will help identify the elements and transcription factors important for SF-1 transcription.
Deletion analysis also supported a role for the 3′ promoter region in SF-1 regulation. Removal of sequences between +10 and +60 severely compromised activity and suggested that important elements reside 3′ to the start of transcription. A functional role for cis-acting elements located downstream of the transcriptional start site has been reported for several genes [38–43]. Interestingly, specific downstream elements known as DPEs have been implicated as interacting with the Inr [42, 43]. Inspection of the SF-1 promoter does not reveal any sequences having the required position and sequence constraints of a DPE. However, many TATA-less genes have also been reported to have a common 3′ element referred to as MED-1 [41]. A sequence similar to the MED-1 consensus (GCTCCC/G) is located in the 3′ end (+16 to +21) of the SF-1 promoter. Interestingly, the human Wilms’ tumor gene, a gene that is expressed similarly to SF-1 in the testis, also has a MED-1 sequence in the 3′ region of its promoter [41, 44]. The molecular mechanism by which these 3′ sequences enhance transcription is largely unknown, but they may act similarly to 5′ promoter elements to help recruit components of the general transcription machinery or may function as a core promoter element by facilitating the binding and selection of the TFIID complex.
Site-directed mutagenesis of the region between −83 and −51 identified two response elements, the E box and the CCAAT box, as important for the regulation of SF-1 in Sertoli and Leydig cells. Both of these elements have been shown to be important for SF-1 promoter activity in Y1 cells, revealing an important commonality in the mechanisms regulating this gene in adrenocortical, Leydig, and Sertoli cells [28, 29]. The E box was also shown to be important for SF-1 expression in the gonadotroph cell line αT3 [27]. The sequence of the SF-1 E box (CACGTG) has been shown to bind a number of transcriptional regulators in mammalian cells, all of which tend to be in the bHLH-Zip class of transcription factors [45–51]. These include USF1, USF2, TFEB, TFE3, c-Myc, Max, and Mad; and resolving the specificity of these transcriptional regulators has been complicated by the fact that both protein binding and sequence requirements are sensitive to the in vitro binding conditions [52]. Studies in Y1, αT3, and granulosa cells identified USF as the major SF-1 E box binding complex [27, 53].
The CCAAT box is one of the most common elements in eukaryotic promoters and is found at slightly higher frequency in TATA-less promoters than in TATA-containing ones [54]. This sequence often binds the ubiquitous transcription factor NF-Y (also called CBF, αCP1, CP1), which is a heteromeric protein composed of three subunits, NF-YA, NF-YB, and NF-YC [55]. This protein requires not only the pentanucleotide CCAAT core for efficient binding but also sequences flanking the 5′ side and 3′ side of the core. The sequence of the SF-1 CCAAT element matches well with the consensus NF-Y binding site [54]. Our studies support a role for both the CCAAT motif and bases flanking the 5′ side of this sequence. Furthermore, our preliminary studies suggest that mutation of either the flanking sequence or the CCAAT motif only partially disrupts binding of an important regulator, as disruption of both sites had significantly greater impact on promoter activity than either single mutation. However, mutation of bases to the 3′ side had no impact, suggesting either that they are not important for factor binding or that the mutated bases were within the sequence constraints for factor binding. Studies in Y1 cells showed that a protein binding this element cross-reacted with an antibody against CBF (NF-Y), implicating this as a possible candidate for binding to the CCAAT box in testicular cells [28].
We used EMSA to characterize proteins binding to the E box and CCAAT box regions of the SF-1 promoter and observed three major specific complexes that migrated similarly in Sertoli and MA-10 cells. One of the binding complexes required only sequences within the E box for efficient binding and cross-reacted with antibodies generated against USF1 and, to a lesser degree, USF2 (Fig. 7). However, in addition to USF1/USF2 binding we identified a second, more substantial, complex that interacts with the E box in Sertoli and Leydig cells (Figs. 6A and 7, complex 1). Interestingly, for efficient binding, this complex not only required sequences within the E box but also those within the CCAAT motif (Fig. 6A). Although we currently do not know the identity of the proteins in this complex, it does not likely contain USF1, USF2, or c-Myc, as antibodies against these proteins did not affect formation of the complex or result in the production of a supershifted complex. In an attempt to identify components of this complex, we used many (> 20) different commercially available antibodies, including one directed against NF-YA (Rockland, Inc., Gilbertsville, PA), and found no significant effect on complexes binding the E/C probe (data not shown). Studies using a radiolabeled probe containing a consensus binding site for NF-Y (5′-CTGTGGCATGCTCTAACCAATTAGAGAA-3′) confirmed the presence of NF-Y and the specificity of the NF-YA antibodies under our EMSA binding conditions. Therefore NF-Y does not appear to be a component of the complexes binding the E/C probe.
SF-1 is expressed in a limited number of tissues and cells, and the mechanisms that direct this tissue-specific expression are not well understood. Identifying the proteins that are responsible for SF-1 transcriptional activity and its restriction to particular cell types will be critical to our understanding of SF-1 regulation and will help unravel the genetic pathways leading to adrenal and gonad formation. Previous studies have shown that the −92-bp SF-1 promoter is expressed in a cell-specific manner, indicating that elements within this region of the promoter help restrict SF-1 expression [27–29]. Within this region of the promoter, only three elements have been identified in the regulation of SF-1 transcription: the E box, the CCAAT box, and one of the potential Sp1 sites. With respect to the E box, this element has been shown to be important for SF-1 transcription in adrenocortical cells, gonadotroph cells, and now Sertoli and Leydig cells. Furthermore, the CCAAT box has been shown to be important for SF-1 transcription in adrenocortical, Sertoli, and Leydig cells, while a role for the Sp1 site closest to the transcription start site has been established only in adrenocortical cells.
Although USF1, USF2, NF-Y, and Sp1 have been shown to interact with these identified response elements, more direct functional data are required to determine whether these proteins regulate SF-1 transcription. Because of their ubiquitous nature, these factors are clearly not sufficient by themselves to direct tissue-specific expression of this gene. Thus, if these proteins are involved, additional mechanisms must be invoked. The possibility of different mechanisms and/or transcription factors regulating SF1 expression in adrenocortical cells—versus those regulating SF-1 expression in testicular cell types—is exemplified by the sexually dimorphic expression of SF-1 in male and female gonads. This sexual dimorphic expression is not found in the expression of SF-1 in the adrenal glands of either males or females. Possible mechanisms include cell-specific modifications or the use of specific coactivator proteins. However, analysis of the SF-1 promoter is incomplete, and therefore additional elements that work in conjunction with these may be responsible for the cell specificity of SF-1. Alternatively, proteins other than USF1, USF2, CBF, or Sp1 may interact with these elements and help direct cell-specific expression. One possibility is the relative ratio of ubiquitous transcription factors within an individual cell contributing to the cell-specific expression of a particular gene (reviewed in [56]). For example, the ratio of Sp1 to Sp3 within a cell corresponds to the susceptibility of differentiated epithelial cells to viral infection by human papillomavirus type 16 [57]. A mechanism such as this is especially intriguing in view of the number of Sp1 binding sites within the SF-1 promoter. Although beyond the scope of this study, preliminary results not presented here suggest that the Sp1 sites do play a role in SF-1 expression in testicular cell types as previously shown in Y1 cells. Unique to our study is the identification of a complex different from USF1 or USF2 that requires sequences in both the E box and CCAAT box for efficient binding. This is the major in vitro binding complex on the E box/CCAAT region of the SF-1 promoter and may represent a unique transcription complex that helps restrict SF-1 expression to particular cell types. Currently, it is unclear whether the complex represents a single protein that makes contacts with each site or distinct proteins that interact with and facilitate binding of the other. The presence of multiple protein complexes binding these sites suggests that regulation through these elements may involve different factors that function under distinct cellular conditions. Further studies are needed to help determine the identity of proteins in these other complexes and to determine the role of these, as well as of USF1, USF2, and Sp1, in the regulation of SF-1 in the testis.
Acknowledgments
We thank Jiang kai Chen for the preparation of primary Sertoli cell cultures and all cell lines used in this work.
Footnotes
This work was supported in part by NIH Grant R29HD-3521701A1 (to L.L.H.) and by an NICHD-supported Center of Reproductive Sciences Grant (HD-33994). M.A.F.D was supported by NIH/NICHD Grant (1 F32 HD08500-01).
References
- 1.Parker KL, Schimmer BP. Steroidogenic factor 1: a key determinant of endocrine development and function. Endocr Rev. 1997;18:361–377. doi: 10.1210/edrv.18.3.0301. [DOI] [PubMed] [Google Scholar]
- 2.Morohashi KI, Omura T. Ad4BP/SF-1, a transcription factor essential for the transcription of steroidogenic cytochrome P450 genes and for the establishment of the reproductive function. FASEB J. 1996;10:1569–1577. doi: 10.1096/fasebj.10.14.9002548. [DOI] [PubMed] [Google Scholar]
- 3.Luo X, Ikeda Y, Parker KL. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell. 1994;77:481–490. doi: 10.1016/0092-8674(94)90211-9. [DOI] [PubMed] [Google Scholar]
- 4.Luo X, Ikeda Y, Schlosser DA, Parker KL. Steroidogenic factor 1 is the essential transcript of the mouse FTZ-F1 gene. Mol Endocrinol. 1995;9:1233–1239. doi: 10.1210/mend.9.9.7491115. [DOI] [PubMed] [Google Scholar]
- 5.Sadovsky Y, Crawford PA, Woodson KG, Polish JA, Clements MA, Tourtellotte LM, Simburger K, Milbrandt J. Mice deficient in the orphan receptor steroidogenic factor 1 lack adrenal glands and gonads but express P450 side-chain-cleavage enzyme in the placenta and have normal embryonic serum levels of corticosteroids. Proc Natl Acad Sci USA. 1995;92:10939–10943. doi: 10.1073/pnas.92.24.10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shinoda K, Lei H, Yoshii H, Nomura M, Nagano M, Shiba H, Sasaki H, Osawa Y, Ninomiya Y, Niwa O, Morohashi KI, Li E. Developmental defects of the ventromedial hypothalamic nucleus and pituitary gonadotroph in the Ftz-F1 disrupted mice. Dev Dyn. 1995;204:22–29. doi: 10.1002/aja.1002040104. [DOI] [PubMed] [Google Scholar]
- 7.Rice DA, Mouw AR, Bogerd AM, Parker KL. A shared promoter element regulates the expression of three steroidogenic enzymes. Mol Endocrinol. 1991;5:1552–1561. doi: 10.1210/mend-5-10-1552. [DOI] [PubMed] [Google Scholar]
- 8.Morohashi K, Honda S, Inomata Y, Handa H, Omura T. A common trans-acting binding factor, Ad4-binding protein, to the promoters of steroidogenic P-450s. J Biol Chem. 1992;267:17913–17919. [PubMed] [Google Scholar]
- 9.Caron KM, Clark BJ, Ikeda Y, Parker KL. Steroidogenic factor 1 acts at all levels of the reproductive axis. Steroids. 1997;62:53–56. doi: 10.1016/s0039-128x(96)00159-6. [DOI] [PubMed] [Google Scholar]
- 10.Asa SL, Bamberger AM, Cao B, Wong M, Parker KL, Ezzat S. The transcription activator steroidogenic factor-1 is preferentially expressed in the human pituitary gonadotroph. J Clin Endocrinol Metab. 1996;81:2165–2170. doi: 10.1210/jcem.81.6.8964846. [DOI] [PubMed] [Google Scholar]
- 11.Duval DL, Nelson SE, Clay CM. A binding site for steroidogenic factor-1 is part of a complex enhancer that mediates expression of the murine gonadotropin-releasing hormone receptor gene. Biol Reprod. 1997;56:160–168. doi: 10.1095/biolreprod56.1.160. [DOI] [PubMed] [Google Scholar]
- 12.Halvorson LM, Ito M, Jameson JL, Chin WW. Steroidogenic factor-1 and early growth response protein 1 act through two composite DNA binding sites to regulate luteinizing hormone beta-subunit gene expression. J Biol Chem. 1998;273:14712–14720. doi: 10.1074/jbc.273.24.14712. [DOI] [PubMed] [Google Scholar]
- 13.Roselli CE, Jorgensen EZ, Doyle MW, Ronnekleiv OK. Expression of the orphan receptor steroidogenic factor-1 mRNA in the rat medial basal hypothalamus. Brain Res Mol Brain Res. 1997;44:66–72. doi: 10.1016/s0169-328x(96)00187-8. [DOI] [PubMed] [Google Scholar]
- 14.Lavorgna G, Ueda H, Clos J, Wu C. FTZ-F1, a steroid hormone receptor-like protein implicated in the activation of fushi tarazu. Science. 1991;252:848–851. doi: 10.1126/science.1709303. [DOI] [PubMed] [Google Scholar]
- 15.Ueda H, Sonoda S, Brown JL, Scott MP, Wu C. A sequence-specific DNA-binding protein that activates fushi tarazu segmentation gene expression. Genes Dev. 1990;4:624–635. doi: 10.1101/gad.4.4.624. [DOI] [PubMed] [Google Scholar]
- 16.Oba K, Yanase T, Nomura M, Morohashi K, Takayanagi R, Nawata H. Structural characterization of human Ad4bp (SF-1) gene. Biochem Biophys Res Commun. 1996;226:261–267. doi: 10.1006/bbrc.1996.1343. [DOI] [PubMed] [Google Scholar]
- 17.Ninomiya Y, Okada M, Kotomura N, Suzuki K, Tsukiyama T, Niwa O. Genomic organization and isoforms of the mouse ELP gene. J Biochem (Tokyo) 1995;118:380–389. doi: 10.1093/oxfordjournals.jbchem.a124918. [DOI] [PubMed] [Google Scholar]
- 18.Ikeda Y, Luo X, Abbud R, Nilson JH, Parker KL. The nuclear receptor steroidogenic factor 1 is essential for the formation of the ventromedial hypothalamic nucleus. Mol Endocrinol. 1995;9:478–486. doi: 10.1210/mend.9.4.7659091. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda Y, Shen W-H, Ingraham HA, Parker KL. Developmental expression of mouse steroidogenic factor-1, an essential regulator of the steroid hydroxylases. Mol Endocrinol. 1994;8:654–662. doi: 10.1210/mend.8.5.8058073. [DOI] [PubMed] [Google Scholar]
- 20.Shen W-H, Moore CD, Ikeda Y, Parker KL, Ingraham HA. Nuclear receptor steroidogenic factor 1 regulates the Müllerian inhibiting substance gene: a link to the sex determination cascade. Cell. 1994;77:651–661. doi: 10.1016/0092-8674(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 21.Hatano O, Takayama K, Imai T, Waterman MR, Takakusu A, Omura T, Morohashi K. Sex-dependent expression of a transcription factor, Ad4BP, regulating steroidogenic P-450 genes in the gonads during prenatal and postnatal rat development. Development. 1994;120:2787–2797. doi: 10.1242/dev.120.10.2787. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda Y, Lala DS, Luo X, Kim E, Moisan M-P, Parker KL. Characterization of the mouse FTZ-F1 gene, which encodes a key regulator of steroid hydroxylase gene expression. Mol Endocrinol. 1993;7:852–860. doi: 10.1210/mend.7.7.8413309. [DOI] [PubMed] [Google Scholar]
- 23.Morohashi K, Hatano O, Nomura M, Takayama K, Hara M, Yoshii H, Takakusu A, Omura T. Function and distribution of a steroidogenic cell-specific transcription factor, Ad4BP. J Steroid Biochem Mol Biol. 1995;53:81–88. doi: 10.1016/0960-0760(95)00041-w. [DOI] [PubMed] [Google Scholar]
- 24.Ingraham HA, Lala S, Ikeda Y, Luo X, Shen W-H, Nachtigal MW, Abbud R, Nilson JH, Parker KL. The nuclear receptor steroidogenic factor 1 acts at multiple levels of the reproductive axis. Genes Dev. 1994;8:2303–2312. doi: 10.1101/gad.8.19.2302. [DOI] [PubMed] [Google Scholar]
- 25.Ikuyama S, Ohe K, Sakai Y, Nakagaki H, Fukushima T, Kato Y, Morohashi K, Takayanagi R, Nawata H. Follicle stimulating hormone-beta subunit gene is expressed in parallel with a transcription factor Ad4BP/SF-1 in human pituitary adenomas. Clin Endocrinol (Oxf) 1996;45:187–193. doi: 10.1046/j.1365-2265.1996.d01-1555.x. [DOI] [PubMed] [Google Scholar]
- 26.Ramayya MS, Zhou J, Kino T, Segars JH, Bondy CA, Chrousos GP. Steroidogenic factor 1 messenger ribonucleic acid expression in steroidogenic and nonsteroidogenic human tissues: Northern blot and in situ hybridization studies. J Clin Endocrinol Metab. 1997;82:1799–1806. doi: 10.1210/jcem.82.6.3967. [DOI] [PubMed] [Google Scholar]
- 27.Harris AN, Mellon PL. The basic helix-loop-helix, leucine zipper transcription factor, USF (upstream stimulatory factor), is a key regulator of SF-1 (steroidogenic factor-1) gene expression in pituitary gonadotrope and steroidogenic cells. Mol Endocrinol. 1998;12:714–726. doi: 10.1210/mend.12.5.0100. [DOI] [PubMed] [Google Scholar]
- 28.Woodson KG, Crawford PA, Sadovsky Y, Milbrandt J. Characterization of the promoter of SF-1, an orphan nuclear receptor required for adrenal and gonadal development. Mol Endocrinol. 1997;11:117–126. doi: 10.1210/mend.11.2.9881. [DOI] [PubMed] [Google Scholar]
- 29.Nomura M, Bartsch S, Nawata H, Omura T, Morohashi K. An E box element is required for the expression of the ad4bp gene, a mammalian homologue of ftz-f1 gene, which is essential for adrenal and gonadal development. J Biol Chem. 1995;270:7453–7461. doi: 10.1074/jbc.270.13.7453. [DOI] [PubMed] [Google Scholar]
- 30.Heckert LL, Daggett MA, Chen J. Multiple promoter elements contribute to activity of the follicle-stimulating hormone receptor (FSHR) gene in testicular Sertoli cells. Mol Endocrinol. 1998;12:1499–1512. doi: 10.1210/mend.12.10.0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heckert LL, Schultz K, Nilson JH. Different composite regulatory elements direct expression of the human α subunit gene to pituitary and placenta. J Biol Chem. 1995;270:26497–26504. doi: 10.1074/jbc.270.44.26497. [DOI] [PubMed] [Google Scholar]
- 32.Ascoli M. Characterization of several clonal lines of cultured Leydig tumor cells: gonadotropin receptors and steroidogenic responses. Endocrinology. 1981;108:88–95. doi: 10.1210/endo-108-1-88. [DOI] [PubMed] [Google Scholar]
- 33.Frohman MA. RACE: rapid amplification of cDNA ends. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: A Guide to Methods and Applications. San Diego: Academic Press, Inc.; 1990. pp. 28–38. [Google Scholar]
- 34.Frohman MA. Rapid amplification of complementary DNA ends for generation of full-length complementary DNAs: thermal RACE. Methods Enzymol. 1993;218:340–356. doi: 10.1016/0076-6879(93)18026-9. [DOI] [PubMed] [Google Scholar]
- 35.Weis L, Reinberg D. Transcription by RNA polymerase II: initiator-directed formation of transcription-competent complexes. FASEB J. 1992;6:3300–3309. doi: 10.1096/fasebj.6.14.1426767. [DOI] [PubMed] [Google Scholar]
- 36.Smale ST. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim Biophys Acta. 1997;1351:73–88. doi: 10.1016/s0167-4781(96)00206-0. [DOI] [PubMed] [Google Scholar]
- 37.Smale ST, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 38.Fornasari D, Battaglioli E, Flora A, Terzano S, Clementi F. Structural and functional characterization of the human alpha3 Nicotinic subunit gene promoter. Mol Pharmacol. 1997;51:250–261. doi: 10.1124/mol.51.2.250. [DOI] [PubMed] [Google Scholar]
- 39.Mariman E, Wieringa B. Expression of the gene encoding human brain creatinine kinase depends on sequences immediately following the transcription start point. Gene. 1991;102:205–212. doi: 10.1016/0378-1119(91)90079-q. [DOI] [PubMed] [Google Scholar]
- 40.Kharroubi AE, Martin MA. cis-Acting sequences located downstream of the human immunodeficiency virus type 1 promoter affect its chromatin structure and transcriptional activity. Mol Cell Biol. 1996;16:2958–2966. doi: 10.1128/mcb.16.6.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ince TA, Scotto KW. A conserved downstream element defines a new class of RNA polymerase II promoters. J Biol Chem. 1995;270:30249–30252. doi: 10.1074/jbc.270.51.30249. [DOI] [PubMed] [Google Scholar]
- 42.Burke TW, Kadonaga JT. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burke TW, Kadonaga JT. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- 44.Pelletier J, Schalling M, Buckler AJ, Rogers A, Haber DA, Housman D. Expression of the Wilm’s tumor gene WT1 in the murine urogenital system. Genes Dev. 1991;5:1345–1356. doi: 10.1101/gad.5.8.1345. [DOI] [PubMed] [Google Scholar]
- 45.Carr CS, Sharp PA. A helix-loop-helix protein related to the immunoglobulin E box-binding proteins. Mol Cell Biol. 1990;10:4384–4388. doi: 10.1128/mcb.10.8.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beckmann H, Kadesch T. The leucine zipper of TFE3 dictates helix-loop-helix dimerization specificity. Genes Dev. 1991;5:1057–1066. doi: 10.1101/gad.5.6.1057. [DOI] [PubMed] [Google Scholar]
- 47.Blackwell TK, Kretzner L, Blackwood EM, Eisenman RN, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;250:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- 48.Halazonetis TD, Kandil AN. Determination of the c-MYC DNA-binding site. Proc Natl Acad Sci USA. 1991;88:6162–6166. doi: 10.1073/pnas.88.14.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 50.Berberich S, Hyde-DeRuyscher N, Espenshade P, Cole M. Max encodes a sequence-specific DNA-binding protein and is not regulated by serum growth factors. Oncogene. 1992;7:775–779. [PubMed] [Google Scholar]
- 51.Ayer DE, Kretzner L, Eisenman RN. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- 52.Bendall AJ, Molloy PL. Base preferences for DNA binding by the bHLH-Zip protein USF: effects of MgCl2 on specificity and comparison with binding of Myc family members. Nucleic Acids Res. 1994;22:2801–2810. doi: 10.1093/nar/22.14.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morris JK, Richards JS. An E-box region within the prostaglandin endoperoxide synthase-2 (PGS-2) promoter is required for transcription in rat ovarian granulosa cells. J Biol Chem. 1996;271:16633–16643. doi: 10.1074/jbc.271.28.16633. [DOI] [PubMed] [Google Scholar]
- 54.Mantovani R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 1998;26:1135–1143. doi: 10.1093/nar/26.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dorn A, Bollekens J, Staub A, Benoist C, Mathis D. A multiplicity of CCAAT box-binding proteins. Cell. 1987;50:863–872. doi: 10.1016/0092-8674(87)90513-7. [DOI] [PubMed] [Google Scholar]
- 56.Ernst P, Smale ST. Combinatorial regulation of transcription I: general aspects of transcriptional control. Immunity. 1995;2:311–319. doi: 10.1016/1074-7613(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 57.Apt D, Watts RM, Suske G. High Sp1/Sp3 ratios in epithelial cells during epithelial differentiation correlate with the activation of the HPV-16 promoter. Virology. 1996;224:281–291. doi: 10.1006/viro.1996.0530. [DOI] [PubMed] [Google Scholar]
- 58.Heinemeyer T, Chen X, Karas H, Kel AE, Kel OV, Liebich I, Meinhardt T, Reuter I, Schacherer F, Wingender E. Expanding the TRANS-FAC database towards an expert system of regulatory molecular mechanisms. Nucleic Acids Res. 1999;27:318–322. doi: 10.1093/nar/27.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]


