Abstract
The etiology of most chronic diseases involves interactions between environmental factors and genes that modulate important biological processes (Olden and Wilson, 2000). We are developing the publicly available Comparative Toxicogenomics Database (CTD) to promote understanding about the effects of environmental chemicals on human health. CTD identifies interactions between chemicals and genes and facilitates cross-species comparative studies of these genes. The use of diverse animal models and cross-species comparative sequence studies has been critical for understanding basic physiological mechanisms and gene and protein functions. Similarly, these approaches will be valuable for exploring the molecular mechanisms of action of environmental chemicals and the genetic basis of differential susceptibility.
INTRODUCTION
Environmental factors are implicated in many common conditions such as asthma, cancer, diabetes, hypertension, immune deficiency disorders, and Parkinson’s disease; however, the molecular mechanisms underlying these correlations are not well understood (Toscano and Oehlke, 2004). A paradigm used to explain this environment-disease relationship suggests that chemicals may be distributed or metabolized in cells and interact with, disrupt, or damage target genes. Disease may result when the affected genes regulate important biological processes such as DNA repair, cell cycle control, or differentiation. Understanding correlations between chemicals and diseases is made challenging, however, by the size of the human genome, the diversity of chemical combinations in the environment, the extent of genetic variability, and the specific circumstances of exposure.
The availability of abundant sequence data from diverse species offers new opportunities for understanding mechanisms of chemical actions. Cross-species comparative studies of sequences for toxicologically important genes and proteins can facilitate the identification of conserved and divergent regions to help elucidate the genetic basis of individual or species-specific responses to chemical exposure. We are developing CTD to support cross-species comparative studies of toxicologically important genes and proteins and their interactions with chemicals (Mattingly and others, ‘03; Mattingly and others, ‘04).
DATA INTEGRATION AND CURATION
A prototype version of CTD is available via the World Wide Web (http://ctd.mdibl.org; figure 1). The major data types in CTD are: 1) nucleotide and protein sequences, 2) reference publications, 3) curated genes, 4) Gene Sets (sets of curated genes), 5) a hierarchical vocabulary of chemicals, 6) Gene Ontology terms (hierarchical vocabulary of biological processes, cellular components, and molecular functions), and 7) organism taxonomy. To date, nucleotide and protein sequences are included for all vertebrates and invertebrates. Nucleotide sequences and annotations are acquired from the National Center for Biotechnology Information (NCBI). We include only Reference Sequences (RefSeqs) for H. sapiens, M. musculus, R. norvegicus, D. melanogaster, and C. elegans (Pruitt and others, ‘05). Amino acid sequences and annotations are acquired from the European Bioinformatics Institute’s UniProtKB/Swiss-Prot and TrEMBL databases (Bairoch and others, ‘05).
Fig. 1. CTD Home Page.
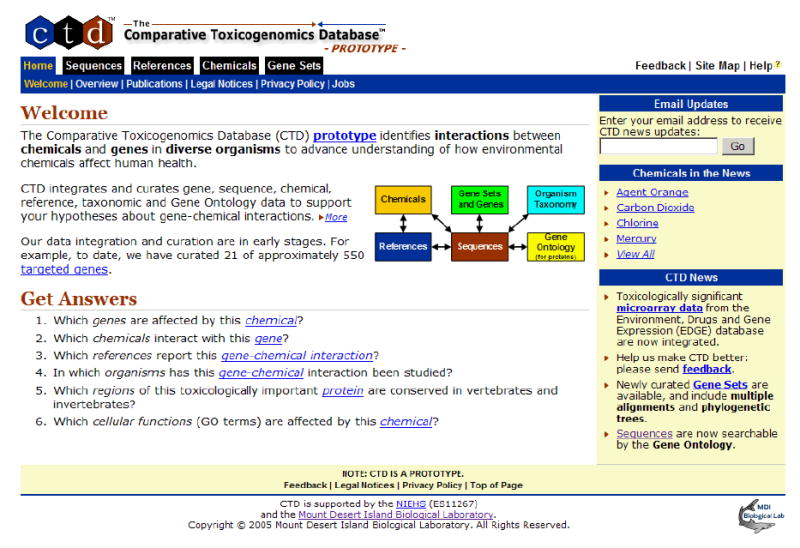
The CTD Home Page provides access to sequence, reference, chemical, and curated gene data. Information about CTD updates and mechanisms for joining the CTD email list and providing feedback are also presented.
References are acquired from PubMed and must contain information about chemical-gene interactions. This data set will continue to expand in scope and size as our curation capacity expands.
CTD integrates controlled, hierarchical vocabularies for organism taxonomy (NCBI; Wheeler and others, ‘02), chemicals (National Library of Medicine’s Medical Subject Headings and Supplementary Concepts (http://www.nlm.nih.gov/mesh/MBrowser.html), and Gene Ontology (GO; Harris and others, ‘04) to ensure consistency in data integration, annotation, access, and interpretation. These vocabularies enhance visitor query capabilities and enable us to link to related data in other biological resources. The chemical vocabulary has also been critical for establishing initial automated chemical-gene associations.
Our primary focus with curation is to construct cross-species toxicologically important genes and Gene Sets. Genes are defined in CTD by their constituent nucleotide and protein sequences from vertebrates and invertebrates and are constructed using sequence analysis methods in combination with literature review. Gene Sets group closely related curated genes, such as those that have undergone duplication events in specific species (e.g., CYP1A4, CYP1A5) or are members of large families (e.g., ABC transporters, G protein coupled receptors) and provide visitors with a broader perspective about their gene(s) of interest. To date we have curated seven Gene Sets that comprise 21 curated genes and 572 sequences from 84 unique species.
DATA ACCESS
From the CTD Home Page, visitors may access chemical, gene, and sequence data from a range of perspectives using sequence or reference query forms or by browsing the chemical vocabulary. Where possible, data are presented in a cross-species context.
For example, visitors may use the Comparative Sequence Query form to search for teleostei genes or sequences associated with dioxin (figure 2). Sequence results are organized by Gene Set and gene (where curated), organism, chemicals, sequence type (DNA, mRNA, protein), Gene Ontology annotation, and sequences. This presentation facilitates access to sequence data for toxicologically important genes and comparative studies that will provide insights into the role of specific genes in modulating chemical actions.
Fig. 2. Comparative Sequence Query Form and Results.
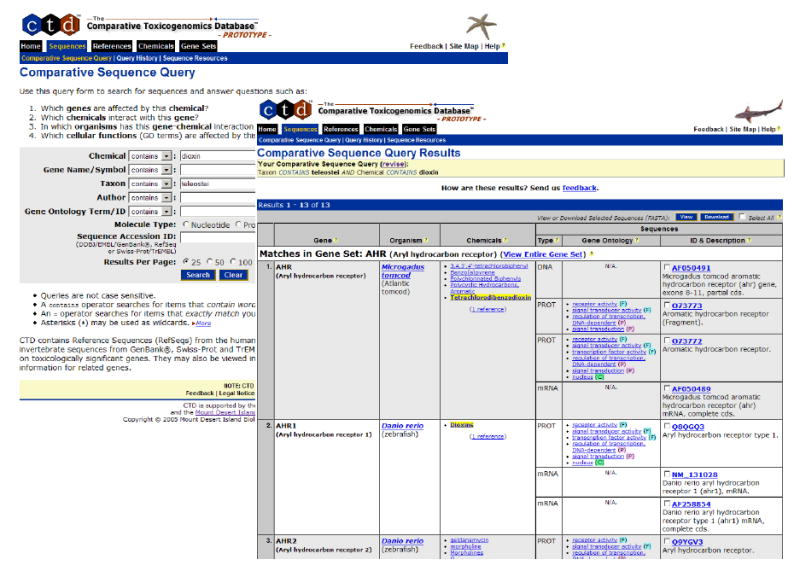
The Comparative Sequence Query form is used to retrieve molecular sequences. Comparative Sequence Query results are presented in a summary format, which includes the associated Gene Set and gene (where curated), source organism, chemicals, molecule type, Gene Ontology annotations, accession identifier, and description for each item. From this results page, one may access individual sequence detail pages which provide information about a selected sequence, or view or download selected FASTA-formatted sequences.
We aim to make CTD a community resource and encourage data contributions and feedback (ctd@mdibl.org).
Footnotes
This work is supported by NIEHS (R33 ES011267), NCRR (P20 RR-016463), and the Mount Desert Island Biological Laboratory.
References
- Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O’Donovan C, Redaschi N, Yeh LS. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33(Database issue):D154–159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L, Ringwald M, Balakrishnan R, Cherry JM, Christie KR, Costanzo MC, Dwight SS, Engel S, Fisk DG, Hirschman JE, Hong EL, Nash RS, Sethuraman A, Theesfeld CL, Botstein D, Dolinski K, Feierbach B, Berardini T, Mundodi S, Rhee SY, Apweiler R, Barrell D, Camon E, Dimmer E, Lee V, Chisholm R, Gaudet P, Kibbe W, Kishore R, Schwarz EM, Sternberg P, Gwinn M, Hannick L, Wortman J, Berriman M, Wood V, de la Cruz N, Tonellato P, Jaiswal P, Seigfried T, White R. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32(Database issue):D258–261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattingly CJ, Colby GT, Forrest JN, Boyer JL. The Comparative Toxicogenomics Database (CTD) Environ Health Perspect. 2003;111(6):793–795. doi: 10.1289/ehp.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattingly CJ, Colby GT, Rosenstein MC, Forrest JN, Jr, Boyer JL. Promoting comparative molecular studies in environmental health research: an overview of the comparative toxicogenomics database (CTD) Pharmacogenomics J. 2004;4(1):5–8. doi: 10.1038/sj.tpj.6500225. [DOI] [PubMed] [Google Scholar]
- Olden K, Wilson S. Environmental health and genomics: visions and implications. Nat Rev Genet. 2000;1(2):149–153. doi: 10.1038/35038586. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33(Database issue):D501–504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toscano WA, Oehlke KP. Systems Biology: New Approaches to Old Environmental Health Problems. Int J Environ Res Public Health. 2004;2:84–90. doi: 10.3390/ijerph2005010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Church DM, Lash AE, Leipe DD, Madden TL, Pontius JU, Schuler GD, Schriml LM, Tatusova TA, Wagner L, Rapp BA. Database resources of the National Center for Biotechnology Information: 2002 update. Nucleic Acids Res. 2002;30(1):13–16. doi: 10.1093/nar/30.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]


