Abstract
Hyaluronan synthase (HAS) utilizes UDP-GlcUA and UDP-GlcNAc in the presence of Mg2+ to form the GAG hyaluronan (HA). The purified HAS from Streptococcus equisimilis (seHAS) shows high fidelity in that it only polymerizes the native substrates, UDP-GlcNAc and UDP-GlcUA. However, other uridinyl nucleotides and UDP-sugars inhibited enzyme activity, including UDP-GalNAc, UDP-Glc, UDP-Gal, UDP-GalUA, UMP, UDP and UTP. Purified seHAS was ~40% more active in 25 mM, compared to 50 mM, PO4 in the presence of either 50 mM NaCl or KCl, and displayed a slight preference for KCl over NaCl. The pH profile was surprisingly broad, with an effective range of pH 6.5–11.5 and the optimum between pH 9 and 10. SeHAS displayed two apparent pKa values at pH 6.6 and 11.8. As the pH was increased from ~6.5, both Km and Vmax increased until pH ~10.5, above which the kinetic constants gradually declined. Nonetheless, the overall catalytic constant (120/sec) was essentially unchanged from pH 6.5 to pH 10.5. The enzyme is temperature labile, but more stable in the presence of substrate and cardiolipin. Purified seHAS requires exogenous cardiolipin for activity and is very sensitive to the fatty acyl composition of the phospholipid. The enzyme was inactive or highly activated by synthetic cardiolipins containing, respectively, C14:0 or C18:1(Δ9) fatty acids. The apparent Ea for HA synthesis is 40 kJ (9.5 kcal/mol) disaccharide. Increasing the viscosity by increasing concentrations of PEG, ethylene glycol, glycerol, or sucrose inhibited seHAS activity. For PEGs, the extent of inhibition was proportional to their molecular mass. PEGs with average masses of 2.7, 11.7, and 20 Kg/mol caused 50% inhibition of Vmax at 21, 6.5, and 3.5 mM, respectively. The apparent Ki values for ethylene glycol, glycerol, and sucrose were, respectively, 4.5, 3.3 and 1.2 mM.
Keywords: streptococcal, kinetics, pH, viscogens, temperature, divalent cations
ABBREVIATIONS: CLm cardiolipin; ECM, extracellular matrix; GAG, glycosaminoglycan; HA, hyaluronic acid, hyaluronate, hyaluronan; HAS, HA synthase; seHAS, Streptococcus equisimilis HAS; PBS, phosphate buffered saline; Tris, trishydroxymethylamino methane; TBS, tris-buffered saline; TBST, tris-buffered saline containing 0.05% Tween20
The glycosaminoglycan (GAG) hyaluronic acid (HA) was purified and characterized over 70 years ago (1). HA is found in some prokaryotes and is a ubiquitous extracellular component of vertebrates (2–4). HA is a linear heteropolysaccharide composed of the repeating disaccharide [→4)-β -D-glucuronic acid--β(1→3)-D-N-acetylglucosamine-(1→]. This glycosaminoglycan is a general constituent of extracellular matrices and is a major component in tissues such as cartilage and dermis, and in synovial and vitreous fluids. HA plays an important role during fertilization, embryogenesis, development, and differentiation (5–7). HA is also involved in a wide variety of cellular functions and behaviors, including cell migration, phagocytosis, and proteoglycan assembly (2,4,8). In addition, HA plays a role during wound healing, and is used as a drug delivery vehicle, a cosmetic ingredient, and as an analgesic (9–12).
In 1993, we cloned, sequenced, and functionally expressed the hasA gene encoding spHAS, the Group A S. pyogenes HAS (13,14). The similar Group C S. equisimilis HAS (seHAS) was reported four years later (15). Since the initial report, >20 HAS genes from many species have been identified and cloned. Two distinct types of HASs have been recognized (16): the unique Class II Pasteurella multocida HAS and the Class I family, which comprises all the other known HASs, including the streptococcal enzymes. The Class I enzymes produce HA in bacteria (17,18), virus-infected algae (19), amphibians (20,21) and mammals (22–25). The eukaryotic enzymes have been placed into three subfamilies, designated HAS1, HAS2 and HAS3, (26,27) each of which is ~30% identical with the three streptococcal HASs from pyogenes, equisimilis, and uberis. The Pasteurella HAS is in a category by itself as a Class II HAS, since it is very distinct from all the other synthases in its structure and mechanism of action (16,17).
Although the gene knockouts of HAS1 or HAS3 in mice showed no phenotype, the HAS2 knockout is lethal (28). The animals die during mid-gestation with critical defects in cardiac and vascular development. Surprisingly, the three HAS isozymes produce HA of different chain length in transfected cells in vitro (29–31). If this behavior occurs in vivo, then different HA product sizes may be important at different developmental stages, in wound healing, or other physiological situations. In the case of some prokaryotes (e.g. S. pyogenes and S. equisimilis), HA encapsulates the bacterium, thereby allowing evasion of the host’s immune system (32). Encapsulation is an effective pathogenic strategy because the bacterial and host HA, are identical in structure.
Cell-free HA biosynthesis was first achieved 45 years ago (33) and active HAS preparations were later obtained from detergent extracts of eukaryote (34,35) and prokaryote (36,37) membranes. Nonetheless, no active HAS was purified until Tlapak-Simmons et al. (38) reported the purification of recombinant spHAS and seHAS expressed in E. coli. The seHAS and spHAS enzymes were also characterized with respect to their kinetic constants (39), functional size (40), and their requirement for phospholipid, particularly CL, for full enzymatic activity (38).
The molecular mass of a streptococcal HAS is ~49 kDa, which is relatively small to mediate all the functions required for HA synthesis. The enzyme must bind the two precursors (UDP-GlcUA and UDP-GlcNAc) in the presence of MgCl2, catalyze two distinct glycosyltransferase reactions, bind to the growing HA chain, translocate the extended HA polymer through the enzyme (and through the lipid bilayer if the protein is in a membrane), and finally release the HA chain, typically after >10,000 monosaccharides (>2 × 106 Da) have been assembled. In this study we characterized purified seHAS by examining the effects of ionic strength, pH, osmolarity, temperature, lipids, synthetic lipids, nucleotides, sugars, and nucleotide-sugars on its activity.
METHODS AND MATERIALS
Materials, Strains and Plasmids
Reagents were supplied by Sigma unless stated otherwise. Media components were from Difco. Phosphate buffered saline (PBS) was formulated according to the GIBCO catalogue. The gene encoding HAS from S. equisimilis was inserted into the pKK223-3 vector (Amersham Pharmacia Biotech) and cloned into E. coli SURE™ cells (15). The pKK223-3 vector contains the strong tac promoter, which is regulated by the lac repressor and induced with isopropyl-β-D-thiogalactoside. To facilitate purification of seHAS (38), a C-terminal fusion of 6-His residues was introduced into the construct using synthetic oligonucleotides and standard polymerase chain reaction techniques. This modification does not significantly alter enzymatic activity (40). Synthetic cardiolipins containing only myristic acid (C14:0) or oleic acid (C18:1[Δ9]) were from Avanti.
Cell Growth and Membrane Preparation
E. coli SURE™ cells containing the HAS-encoding plasmids were grown at 32°C in Luria broth to an A600 of 1.5–1.6. The cells were then induced with 1 mM isopropyl-β-D-thiogalactoside and grown for an additional 3 h. The cells were harvested by centrifugation at 4°C for 30 min at 3000 × g, washed twice with PBS containing 1.3 M glycerol at 4°C, and then frozen at −80°C. Membranes from E. coli containing seHAS were obtained by modifications of a protoplast method reported previously (38). Cell pellets were thawed and resuspended to 1% of the original culture volume in 30 mM Tris (pH 8.2), 20% sucrose, 10 mM MgCl2, 1 mM dithiothreitol, 0.5 μg/ml leupeptin, and 0.7 μg/ml pepstatin. Lysozyme (20 mg/ml) in 0.1 M EDTA, pH 8 was added (0.1% of the initial culture volume) and the suspension was mixed until homogeneous and then incubated for 40 min on ice with constant mixing. Phenylmethylsulfonyl fluoride was added to a concentration of 46 μg/ml, and the suspension was sonicated on ice three times for 30 s each at 20 watts using a microtip probe (W-380; Heat Systems Ultrasonic, Inc, Farmingdale, NY). The lysate was diluted 2-fold in PBS containing 1.3 M glycerol, 1 mM dithiothreitol and the above protease inhibitors. MgCl2 (60 mM), DNase, and RNase (1 μg/ml each) were added to the indicated final concentrations. After 20 min on ice with constant mixing, debris was removed by centrifugation at 10,000 × g for 30 min at 4°C. The membranes were then harvested by ultracentrifugation at 100,000 × g for 1h. The membrane pellet was resuspended once with PBS containing 1.3 M glycerol and the above protease inhibitors, sonicated and recentrifuged at 100,000 × g for 1 h. The final pellets were stored at −80°C at ~300 μg protein/g cells.
HAS Extraction and Purification
Thawed membrane pellets (~1.3 mg of protein) were solubilized in 10 ml of extraction buffer containing 10 mM n-dodecyl-β-D-maltoside, 50 mM sodium and potassium phosphate (pH 7.0), 150 mM NaCl, 10 mM MgCl2, 1.0 mM β-mercaptoethanol, 2.7 mM glycerol, 0.5 μg/ml leupeptin, 0.7 μg/ml pepstatin, and 46 μg/ml phenylmethylsulfonyl fluoride for 2 h at 4°C with gentle mixing in a Micromixer E-36 (Taitec). The nonsolubilized membrane components were sedimented by centrifugation at 100,000 × g for 1 h at 4°C. Imidazole was then added to the supernatant (to a final concentration of 30 mM) to minimize nonspecific binding of E. coli proteins to the Ni2+-nitrilotriacetic acid (Ni-NTA) resin (QIAGEN Inc.). The extract (9.5 ml) was then applied directly to Ni2+-depleted NTA resin (0.3 ml in a mini-spin column; Bio-Rad) that had been equilibrated with extraction buffer, minus MgCl2, for 30 min at 4°C with constant mixing. The enzyme extract was removed and then incubated with 0.3 ml Ni2+-NTA resin for 2 h at 4°C with constant mixing. After incubation, the non-bound proteins were allowed to flow through the resin, which was then washed with 10 column volumes of extraction buffer to remove contaminating proteins. SeHAS was eluted in 0.3 ml of 25 mM sodium and potassium phosphate, pH 7.0, 50 mM NaCl, 1.0 mM dithiothreitol, 2.7 M glycerol, 1 mM n-dodecyl-β-D-maltoside, 0.5 μg/ml leupeptin, 46 μg/ml phenylmethylsulfonyl fluoride, 0.7 μg/ml pepstatin, and 200 mM histidine. The exposure to, or the presence of, histidine did not affect the activity of HAS. Protein concentrations were determined with the Coomassie protein assay reagent (Pierce) using bovine serum albumin as the standard (41). Purities of final seHAS preparations, assessed by SDS-PAGE (42), were >99% based on densitometric analysis of Coomassie-stained gels.
HAS Activity
HAS activity was normally determined in 100 μl of 25 mM sodium and potassium phosphate (pH 7.0), containing 50 mM NaCl, 20 mM MgCl2, 1.0 mM dithiothreitol, 1.0 mM EDTA, 2 M glycerol, 1 mM n-dodecyl-β-D-maltoside, 2 mM bovine CL, 1 mM UDP-GlcUA, 1.0 mM UDP-GlcNAc and 0.69 μM UDP-[14C]GlcUA (380 mCi/mmol; New England Nuclear). Purified seHAS (0.3–0.5 μg) was added to initiate the enzyme reaction and the mixture was gently agitated in a MicroMixer E-36 (Taitec) at 30°C for 1 h, or the indicated temperature and time. Modifications to the normal assay buffer for specific experiments are indicated in the figure legends. Reactions were terminated by the addition of sodium dodecylsulfate to 2% (w/v) final concentration at 22°C. The incorporation of [14C]GlcUA into high molecular mass HA was measured by descending paper chromatography using Whatman No. 3MM paper developed in 1 M ammonium acetate (pH 5.5), and ethanol (7:13). The origins were cut out, eluted overnight with 1 ml of distilled H2O in scintillation vials, 5 ml of Ultimagold scintillation fluid (Packard) was added and radioactivity was assessed using a Packard Model A2300 scintillation counter. For all the conditions reported, the assays used limiting amounts of seHAS protein and the kinetics were linear for ≥ 1 h at ≤ 30°C. Above 30°C, the kinetics were linear for ~45 min.
RESULTS
Effect of pH on Vmax and Km values
The effects of pH on the Michealis-Menten constants (Km) for both UDP-sugar substrates and the maximal velocity (Vmax) of purified seHAS at 30°C are shown in Fig. 1. The effective pH range for the enzyme was between 6.5–11.5, with the optimum between pH 9–10. Although the pH was varied from 4.5–12, the enzyme kinetics remained linear for 2 h. As the pH increased above pH ~5, the Vmax (Fig. 1A) and Km (Figs. 1B and 1C) values increased until pH ~10, above which the kinetic values then decreased. The synthase retained ~50% of its optimal activity at pH 6.5 or pH 11.5. The membrane-bound seHAS showed similar results (data not shown).
Figure 1. Effect of pH on the kinetic constants of purified seHAS.
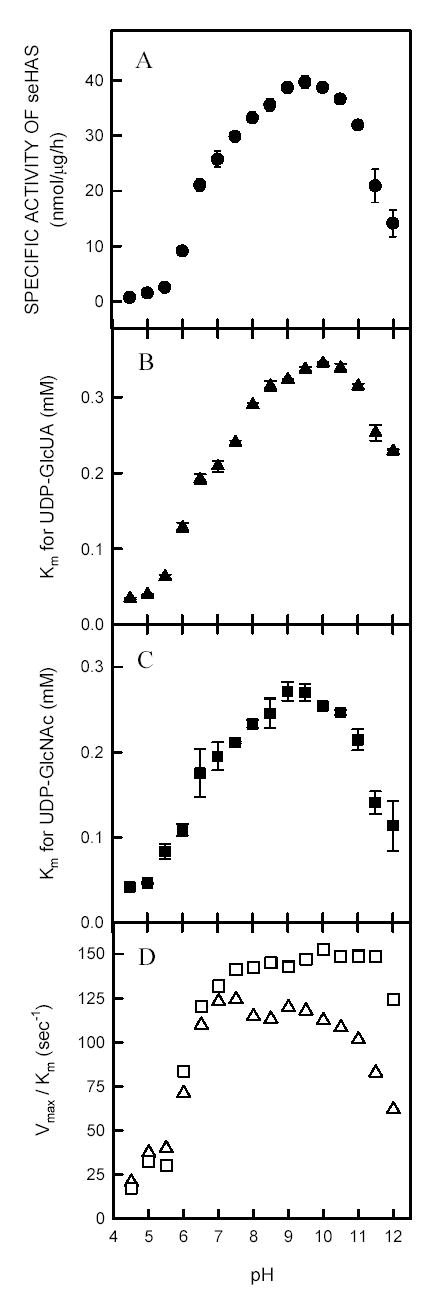
Rather than the normal sodium-potassium phosphate buffer, 25 mM solutions of potassium phosphate and phosphoric acid were mixed to achieve pH values ranging from 4.5–12. The maximal velocity of seHAS (Panel A) was determined at the indicated pH values using saturating concentrations of UDP-GlcUA and UDP-GlcNAc (1.0 mM each). The Km values for UDP-GlcUA (Panel B; KUDP-GlcUA) or UDP-GlcNAc (Panel C; KUDP-GlcNAc) were determined at the indicated pH values by varying the concentration of one substrate from 0.005 to 1.0 mM, while holding the other substrate concentration constant at 1.0 mM. The Km values were determined by Lineweaver-Burke analysis. Panel D shows the calculated catalytic constants at the indicated pH values, obtained by dividing the Vmax values by the KUDP-GlcUA (Δ) or the KUDP-GlcNAc values (○). The incorporation of substrate into HA was monitored as described in Materials and Methods.
The three pH-activity profiles for the purified seHAS (Figs 1A–C) are bell-shaped curves, which typically indicate an enzyme mechanism that involves two protic residues. These data show two apparent pKa values; at pH 6.6 and pH 11.8. Maximum activity of the enzyme requires that the first residue, responsible for the inflection at pH 6.5, be protonated, but that the second residue, with an inflection at pH 11.8, be deprotonated. This indicates that the binding capacity for the substrates may decrease as the enzyme approaches its optimal Vmax. Noneteheless, the catalytic constants for seHAS (determined as the ratio of Vmax/Km ) remained very high and relatively constant over a broad pH range (Fig. 1D); ~120 sec−1 for UDP-GlcUA and ~150 sec−1 for UDP-GlcNAc. Presumbly, the overall kcat for HA biosynthesis would reflect the slower limiting value of 120 sec−1 for UDP-GlcUA. Thus, the conversion of substrates into products by seHAS is actually optimal over a ~5-log range, from pH ~6.5 to pH ~10.5. Theoretically, under these optimal conditions in vitro, purified seHAS could polymerize an HA chain of 107 Da (~25,000 disaccharide units) in just 3.5 min.
To assess the pH-stability of purified seHAS, the enzyme was pre-incubated at pH 5.5 or pH 11.5 in the absence of substrates at 30°C for one h and then assayed at the same pH or at pH 7, 8 or 9 (Fig. 2). When the enzyme was pre-incubated at pH 5.5 and then assayed at the same pH, it was 26% as active as the non pre-incubated control assayed at pH 5.5 (i.e. ~74% of the pH 5.5 activity was lost after the 1 h pretreatment). However, when the enzyme was pre-incubated at pH 5.5 and then assayed at pH 7, 8 or 9, the recovered activities were much greater at 127%, 178% and 221% of the pH 5.5 control values, respectively. Nonetheless, all three latter values at pH 7, 8 or 9 were substantially lower for seHAS pretreated at pH 5.5 (black bars) than for untreated seHAS (indicated by the gray bars). Pretreatment at pH 5.5 decreased the normal activity of seHAS at pH 7, 8 or 9 by about 85–90%.
Figure 2. Effect of pH on stability of purified seHAS.
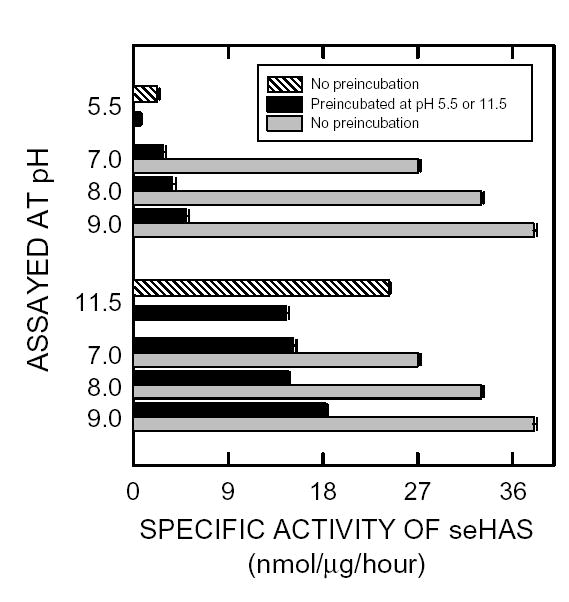
Purified seHAS was assayed at pH 5.5, 7, 8, 9 or 11.5 (Gray bars) for 1 hr at 30°C or preincubated at either pH 5.5 or 11.5 for 1 hr at 30°C and then assayed at pH 5.5, 7,8, 9 or 11.5 (Black bars) for 1 hr at 30°C as described in Methods.
In contrast, the enzyme preincubated at pH 11.5 and then assayed at pH 11.5, had ~60% of the control activity (Fig 2). SeHAS pre-incubated at pH 11.5 and then assayed at pH 7, 8 or 9, had 64%, 60% or 75% of the pH 11.5 control activity, respectively. Pretreatment of seHAS for one h at pH 11.5 resulted in a 40–50% loss in activity when the enzyme was then assayed at pH 7, 8 or 9, compared to the untreated controls. Thus, seHAS is considerably more stable at higher pH, where it is also intrinsically more active.
SeHAS activity depends on the fatty acyl composition of cardiolipin preparations
We reported previously that purified seHAS requires phospholipid for activity (38) and that the active functional unit of the enzyme in membranes is a protein monomer in complex with about 16 phospholipids (40). In both cases, the preferred phospholipid was cardiolipin. Since the majority of our studies with seHAS have been performed using CL preparations from bovine heart membranes, we compared the ability of several different commercial CL preparations to activate the purified enzyme (Fig. 3). Both CL preparations from natural membranes (bovine heart or E. coli) stimulated seHAS activity effectively in a dose-dependent manner, whereas the synthetic CL containing only short (14 carbons) saturated fatty acyl chains was completely ineffective in activating seHAS. In contrast, the synthetic CL containing only 18-carbon unsaturated oleic acids was the best activating cardiolipin for seHAS that we have found to date.
Figure 3. Differential activation of purified seHAS by different cardiolipin preparations.
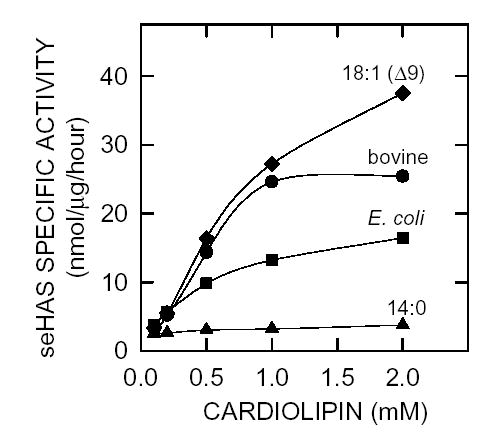
Recombinant seHAS was purified in the absence of phospholipids and then assayed in the presence of increasing amounts (0–2 mM) of commercial cardiolipin mixtures prepared by extraction from bovine heart (•) or E. coli membranes, (▪) or synthetic cardiolipin preparations containing only myristic (▴) or oleic (♦) fatty acids.
CL and UDP-GlcUA enhance the temperature stability of seHAS
Preincubation of purified seHAS at temperatures between 4°C and ~45°C without substrates or phospholipid for one hour decreased enzyme activity, indicating that the protein is temperature labile, even at 4°C (Fig. 4A). At higher temperatures, particularly above ~15°C, seHAS activity was lost in an almost linear manner with increasing temperature (Fig 4C). Only ~10% of the initial activity remained after 1 h at 45°C. Since the streptococcal HASs require CL or phosphatidylserine for activity, we assessed the effects of CL on the temperature stability of seHAS. If bovine CL and UDP-GlcUA were present, the stability of seHAS at higher temperatures was improved, with a significant decrease in HA biosynthesis occurring only above ~25°C (Fig. 4B and 4C). About 25% of the activity remained after pretreatment for one hour at 45°C. In the absence of UDP-GlcUA and bovine CL, the seHAS activity decreased by 25% for the 4°C sample (Fig. 4C). After pretreatment at 35–40°C, 3.3-fold more activity was recovered in the presence of CL and UDP-GlcUA. In other experiments, we observed a similar enhancement of seHAS stability at elevated temperatures in the presence of CL alone, but less of an effect with UDP-GlcUA alone. For example, preincubation of purified seHAS at 20°C for 1 hr with 1 mM UDP-GlcUA plus 2 mM CL, 1 mM UDP-GlcUA alone, 2 mM CL alone, or no additions gave relative activities of 100%, 34%, 85%, and 28%, respectively, when the samples were then assayed at 30°C for 1 h.
Figure 4. Effect of temperature, cardiolipin and substrate on the specific activity of purified seHAS.
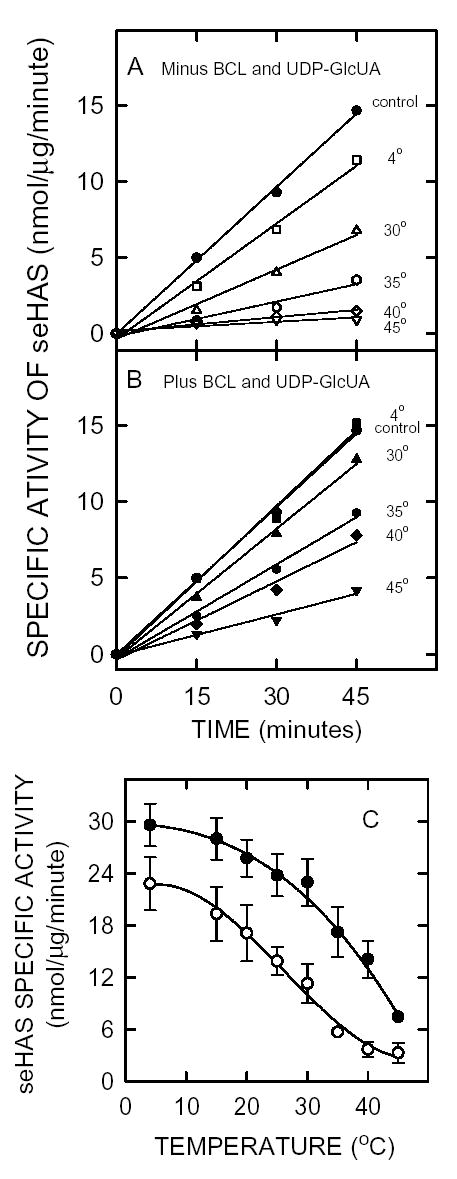
Purified seHAS was pre-incubated at pH 7.0 at temperatures ranging from 4°C to 45°C, as indicated, in the absence (Panel A, open symbols) or presence (Panel B, closed symbols) of 2 mM bovine CL plus 1 mM UDP-GlcUA for one hour. The control (•), which is shown for comparison in both A and B, was incubated at 4°C in the presence of bovine CL and UDP-GlcUA. The pre-incubated samples were then incubated with radiolabeled substrates and HAS activity was assessed at the indicated times at 30°C as described in Methods. Panel C shows the effect of CL plus UDP-GlcUA on the specific activity of purified seHAS at the indicated temperatures, as determined from the slopes of lines in A (○) or B (•).
Energy of activation
The kinetics of HA synthesis by the synthase (with CL present) remained linear for at least one hour at pH 7 (Fig. 5A) or pH 8.5 (not shown) between 4°C and 35°C. Although at pH 7 the enzyme was more active at 40°C than 35°C, the kinetics became slightly curvilinear, consistent with the thermal instability noted above. Activity was barely detected below 15°C and activity decreased above 40°C, e.g. at 45°C seHAS activity was similar to that at 25–30°C. At pH 8.5 the maximal activity occurred at ~35–40°C, whereas at pH 7 the maximal activity occurred at ~40°C. Standard activity assays are at 30°C, which represents a good compromise for linearity, high specific activity and thermal stability. An Arrhenius plot (Fig. 5B) of the Vmax values versus the reciprocal-temperature shows a linear increase in activity from 15–30°C, with a drop off between 30 and 35°C. The maximal velocity data at pH 7 and 8.5 from 15–35°C were used to calculate an apparent Eact of 9.5 kcal/mol (or 40 kJ/mol) for the biosynthesis of HA disaccharides.
Figure 5. Determination of Eact for seHAS.
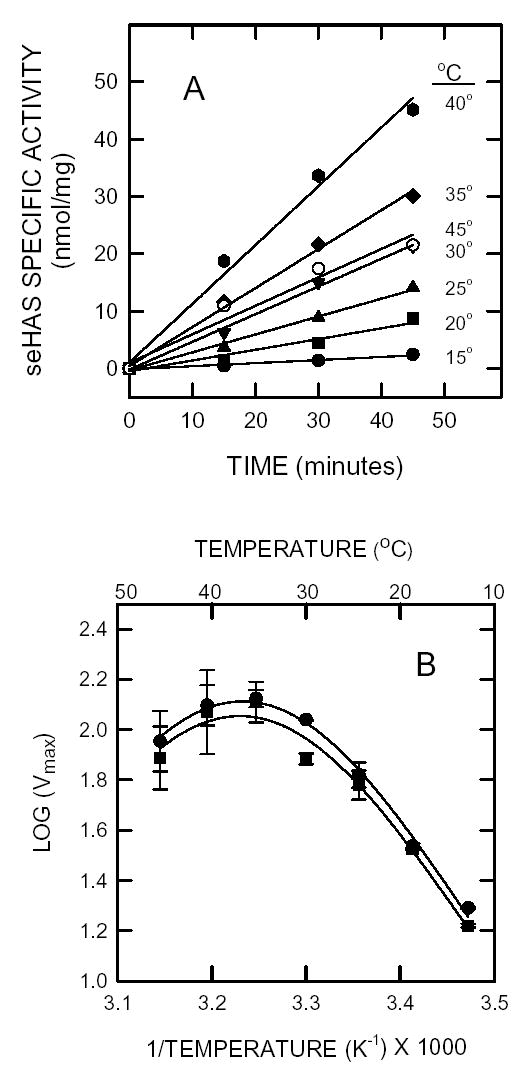
A. The purified enzyme was assayed at various temperatures, as indicated, between 15–45°C as in Fig. 4 at pH 7 and pH 8 (not shown). B. Arrhenius plot. The kinetic rate values (log Vmax), determined as in A, at pH 7.0 (▪) and pH 8 (•) are plotted versus 1/temperature.
Effect of ionic strength
The effect of increasing KCl or NaCl concentration on the HA biosynthesis reaction was examined in PBS at pH 7 (Fig. 6). The activity of purified seHAS was affected in a biphasic manner by increasing ionic strength and was also affected by the specific monovalent cation ion present. The enzyme had a higher specific activity in buffer containing KCl and lower phosphate concentration. In the absence of either KCl or NaCl, seHAS was twice as active in 25 mM versus 50 mM phosphate. In both cases there was slightly higher activity in KCl than NaCl. At 25 mM sodium or potassium phosphate, the highest seHAS activity with either KCl or NaCl occurred from 25–75 mM. However, as the concentration of either salt increased above 100 mM, the Vmax gradually decreased to essentially identical values at 1 M. For NaCl and KCl, these activities were ~10% and 20%, respectively, of the maximum activity. The enzyme is, therefore, sensitive to the nature and concentration of the monovalent cations present.
Figure 6. Effect of ionic strength on seHAS activity.
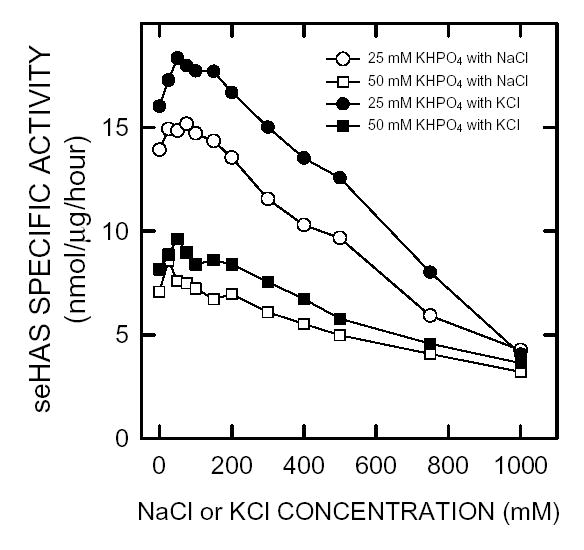
The effect of KCl and NaCl on purified seHAS was determined using the normal assay conditions to assess HAS activity described in Methods, except that two buffer concentrations were used: 25 mM KHPO4 (○,•) and 50 mM KHPO4 (□,▪) with varying NaCl (○,□) and KCl (•,▪) concentrations from 0–1.0 M. Kinetics were linear with respect to time and enzyme concentration under all conditions.
Inhibition of seHAS activity by nucleotides, sugars, and nucleotide-sugars
Using isolated membranes, we previously observed that uridine derivatives could inhibit seHAS activity, even though they were apparently not misincorporated into HA (39). To assess the specificity of this effect, purified seHAS was assayed using substrate concentrations below the Km values in the presence of a variety of monosaccarides, nucleotides or nucleotide-sugars (Fig. 7A). None of the monosaccarides tested inhibited seHAS activity, whereas the nucleotides and nucleotide-sugars caused variable inhibition. At a concentration of 2 mM the potency for the compounds tested, ordered from least to greatest inhibition (percent of control), was: ATP (10%), UDP-GalNAc (30%), UDP-Glc (30%), UDP-Gal (35%), UTP (45%), UDP-GalUA (45%), UMP (80%), and UDP (97%). Titrations of the more potent inhibitors revealed unexpected biphasic profiles (Fig. 8B), in which only partial inhibition occurred with most of these uridinyl compounds. In particular, the profiles for UDP-GalNAc, UDP-Gal, UDP-Glc, and UTP showed very abrupt transitions, essentially leveling off above 0.1 mM at 10–45% inhibition. Partial, biphasic inhibition by any of these uridinyl compounds is unexpected if their mode of action is to compete for the binding of either substrate, UDP-GlcNAc or UDP-GlcUA.
Figure 7. Effect of nucleotides, nucleotide-sugars and sugars on seHAS activity.
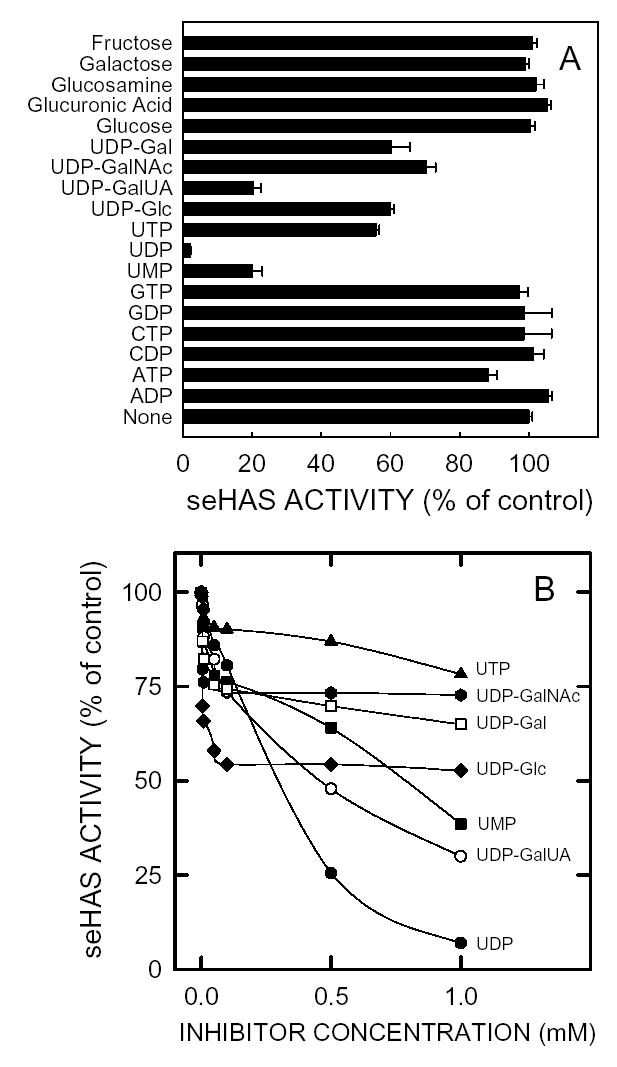
A. The activity of purified seHAS was determined using the normal activity assay, except the UDP-GlcUA and UDP-GlcNAc concentrations were, respectively, 0.02 mM and 0.035 mM and the indicated nucleotide, nucleotide-sugar or sugar was present at 2 mM. Values are presented as the mean ± SD for duplicate assays in three independent experiments (n=6), which were normalized to the no-addition control (set at 100 %). B. The two substrate concentrations were below their Km values, as in A and the nucleotide and sugar nucleotide concentrations ranged from 0–1 mM: UMP (▪), UDP (•), UTP (▴), UDP-Glc (♦), UDP-GalNAc (***), UDP-GalUA (○), and UDP-Gal (□).
Figure 8. Effect of different viscogens on seHAS activity.
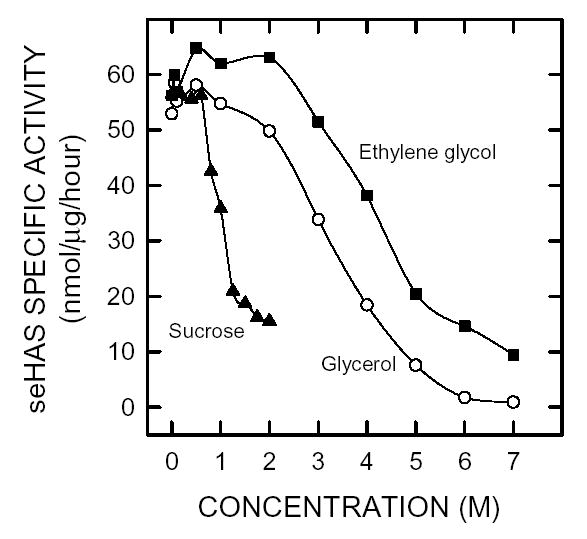
Purified seHAS was assayed using the normal conditions described in Methods, except that 2 M glycerol was omitted and ethylene glycol (▪), sucrose (▴) or glycerol (•) was included at the indicated concentration.
Effect of glycerol, ethylene glycol, or sucrose on HAS activity
The activity of seHAS might be sensitive to viscosity, since the enzyme remains attached to a very large polymer, whose viscosity can increase dramatically with increasing size or concentration. We, therefore, examined the effect of sucrose, glycerol, and ethylene glycol on the activity of purified seHAS (Fig. 8). SeHAS activity increased when the ethylene glycol concentration increased up to 2 M, whereas a similar increase was not seen in corresponding samples containing either glycerol or sucrose. All three agents inhibited seHAS activity with apparent Ki values of 4.5, 3.3, and 1.2 M for ethylene glycol, glycerol and sucrose, respectively. For example, a much higher concentration of ethylene glycol (6 M) was needed to achieve ~85% inhibition of seHAS activity compared to glycerol (4 M) or sucrose (2 M).
Effect of polyethylene glycol on HAS activity
To assess further the effect of viscosity on seHAS activity, we determined the effect of PEGs of various molecular masses on the maximal velocity of the purified enzyme (Fig. 9). As the molecular mass of the PEG increased, the Vmax of seHAS decreased; the concentration needed to inhibit seHAS activity was inversely proportional to the PEG molecular mass. Thus, a lower concentration was required to inactivate seHAS, when the PEG molecular mass was larger. The increasing order of PEG inhibitory ability (apparent Ki) was 21, 6.5, and 3.5 mM, respectively, for molecular masses of 2,751 g/mol, 11,756 g/mol and 20,000 g/mol.
Figure 9. Effect of PEGs of different molar masses on seHAS activity.
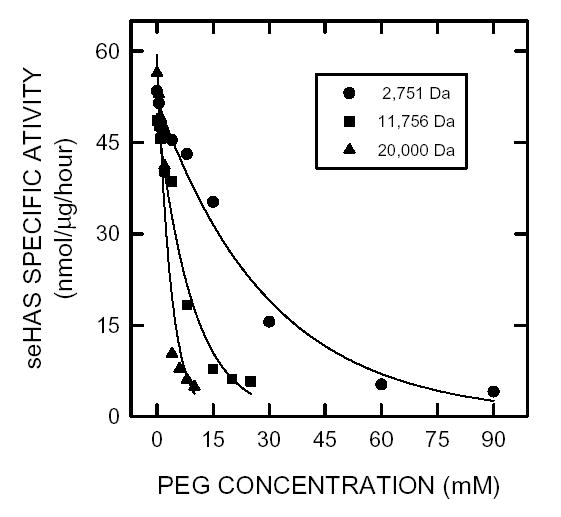
Purified seHAS was assayed using the normal conditions described in Methods, except that 2 M glycerol was omitted and PEGs of different average mass were added at the indicated concentration: 2751 g/mol (•), 11,756 g/mol (▪), and 20,000 g/mol (▴).
DISCUSSION
It has been difficult to detergent-solubilize and purify, with retention of activity, HA synthases from any source. The recombinant seHAS and spHAS have been the easiest Class I HA synthases to purify (39) and we have been able to study these enzymes more extensively. Recombinant mouse HAS1 is the only other HAS to be successfully purified and kinetically characterized (43). Thus, studies of the type described here have not been reported before. Based on the results from genetic (13,14), biochemical (44), and radiation inactivation studies (40,45), and studies with the purified enzymes (38,39,43), it is clear that a Class I HAS is the only protein necessary for HA biosynthesis – when the UDP-sugars are provided. There is no evidence that HAS requires a primer or any other proteins in order to synthesize HA. The kinetic behaviors of membrane-bound HASs from various sources are generally very similar. Typically, the UDP-GlcUA Km values are lower (e.g. 30–75 μM for the streptococcal and human HASs) than those for UDP-GlcNAc (e.g. 250–1000 μM for the three human enzymes). Vmax values for various HASs are more difficult to compare because different groups use different methods to normalize the amount of HA synthesis to the amount of HAS protein.
Purified seHAS and spHAS (data not included) had a surprisingly broad activity range between pH 6.5–11.5. Each bell shaped pH response curve for seHAS showed two apparent pKa inflections at pH ~6.6 and ~11.8, which suggest a two-base isomerization mechanism possibly involving histidine and arginine, respectively, at each pH. The pH optimum and range of high activity are much broader than the pH 7.1 optimum and narrow optimum range reported for broken Streptococcus pyogenes cell preparations (36). A neutral range pH optimum has also been reported for other membrane-bound HA synthases, e.g. 7.0–7.5 for rat fibrosarcoma HAS (46) and 7.0–8.1 for xlHAS (47). Although seHAS was labile under extreme low and high pH conditions, the presence of UDP-GlcUA and bovine CL enhanced its stability.
The temperature dependence seen in the Arrhenius plot (Fig. 4) shows that seHAS behaves within the Maxwell-Boltzman law only in the temperature range from 15–35°C. The activity of seHAS decreased above ~35°C (break temperature). The energy of activation (Eact) calculated in the temperature range between 15–35°C was 9.5 kcal/mol, which is approximately one-half of the value of 15 kcal/mol for the eukaryotic xlHAS (47) or 17 kcal/mol for the Streptococcus pneumoniae type 3 polysaccharide synthase (48). The lower Eact observed for seHAS could be because the enzyme was purified and did not have to extrude the HA product through a lipid bilayer, whereas the Eact values for the xlHAS and type 3 polysaccharide synthases were determined in membranes.
Many enzymes have a preference for either sodium or potassium and usually it is related to physiological situations in the organism. For example, rat liver phosphofructokinase is completely inactivated by NaCl, whereas KCl helps to stabilize the enzyme and increase its activity (49). In contrast, the purified seHAS was not very sensitive to the presence of KCl or NaCl; the enzyme was about 15–20% less active in the presence of NaCl compared to KCl. Enzyme activity decreased with increasing concentrations of either salt. Although activity of purified seHAS was inhibited at higher phosphate concentration, previous studies found that phosphate enhanced the stability and ability to store the purified enzyme (38).
Saccharide synthases from various organisms are very specific as to which sugar nucleotides are used to assemble the growing polymer chain. Membrane-bound native spHAS (33), recombinant spHAS (14,44) and recombinant seHAS (15) only incorporate UDP-GlcUA and UDP-GlcNAc. Nonetheless, these enzymes bind a variety of other nucleotides and nucleotide sugars, which are not utilized as substrates but which inhibit activity. Purified seHAS was inhibited substantially by UMP, UDP, UTP, UDP-Glc, UDP-GalNAc, UDP-GalUA, and UDP-Gal. The other nucleotides, nucleotide sugars, and sugars tested had no effect on enzyme activity. We also found similar results for purified spHAS (data not shown).
One interpretation of these above results is that uridine nucleotides or UDP-sugars can bind to the UDP-GlcNAc or UDP-GlcUA binding sites and serve as inhibitors. Consistent with this idea, we noted previously (39) that high concentrations of either UDP-sugar substrate caused inhibition, especially if the ratio of the two substrates was very different than 1:1. Although the other UDP-sugars or uridine nucleotides can likely occupy part of the substrate binding pockets, causing inhibition, most of them do not behave like typical competitive inhibitors. An unexpected finding (Fig. 7B) was that, with the exception of UDP, the inhibition curves for these compounds were biphasic and showed only partial inhibitory effects. For example, UDP-GalNAc and UDP-Glc only inhibited about 26% and 47%, respectively. This partial inhibitory behavior is inconsistent with their competition at the UDP-sugar substrate binding sites. Rather, the partial inhibition seems more consistent with an allosteric type of modulation at another binding site. Although an earlier study using isolated membranes found no evidence for such a site in seHAS, spHAS did display sigmoidal kinetic behaviour with increasing concentrations of UDP-GlcNAc (39). Allosteric regulation of spHAS, in S. pyogenes cells, which produces a very large HA capsule, might ensure that these growing cells conserve enough UDP-GlcNAc for cell wall biosynthesis. Although we did not detect it, allosteric regulation of seHAS may also occur, but may depend on maintenance of a particular cellular environment that is not present in the purified or membrane-bound enzyme.
Others have shown that membrane-bound synthases are inhibited by various nucleotides, nucleotide sugars, and sugars. For example, Stoolmiller and Dorfman (36) reported that spHAS is inhibited by UDP, GlcUA, and GalUA, although it is stimulated slightly by GlcNAc, and the type 3 capsular synthase is inhibited by UDP-Xylose (48). HA synthesis by membranes containing xlHAS was inhibited by ATP, CTP, GTP, UMP, UDP, UTP, UDP-Gal, UDP-GalUA, UDP-GalNAc, and UDP-Glc (45). Unknown HASs from teratocarcinoma cell membranes showed inhibition of HA production by UDP and UMP but stimulation of HA production by UTP, ATP, ADP, AMP, cAMP, ADP-Glc, and AMP (50). Stimulatory effects in crude membranes could be explained by some of the nucleotides serving as substrates for contaminating phosphatase, phosphodiesterase or other activities, thus protecting the precursors from degradation or metabolism. Examination of purified seHAS eliminates possible confounding results due to the metabolism or utilization of the various nucleotides by contaminating enzymes in crude membranes. Consequently, we did not observe stimulation with the purified seHAS that others may have noticed in membranes.
In general, the structural dynamics of a protein can play an important role in regulating its biochemical function (51). Since the interaction of proteins with the surrounding water molecules is linked with structural dynamics and Brownian motion, protein functions associated with (or dependent on) molecular motion are linked to solvent viscosity, which should thus affect a protein’s dynamics (52–57). In most cases, the rate constant for kinetic processes in solution is inversely proportional to the viscosity, and protein conformational changes that involve major surface changes show the most pronounced viscosity dependence.
PEGs of increasing molecular mass progressively inhibited seHAS, as the concentration increased. Similarly, increasing concentrations of glycerol, ethylene glycol, or sucrose inhibited seHAS activity. Interestingly, at lower concentrations the three latter viscogens had little effect, or actually stimulated seHAS activity slightly, followed by dramatic inhibition as the concentration increased. These co-solutes, which have much smaller molecular masses (sucrose, 342.3 g/mol; glycerol, 92.1 g/mol; and ethylene glycol, 62.4 g/mol) than the PEGs examined, required much greater concentrations to cause similar levels of inhibition, i.e. M rather than mM.
Interpretation of these effects on seHAS is hindered by the complexity of the multiple reactions the enzyme catalyzes in order to assemble an HA chain. Preliminary results using purified seHAS (58) have confirmed the earlier results of others, using crude membrane preparations (59,60), that HA synthesis occurs by addition of new sugars to the reducing end. The growing chain is, therefore, always activated by attachment to UDP, which is derived from the last sugar added (17). The Class I HASs do not release and then rebind HA chains as synthesis proceeds. Rather, the process is processive and chain release likely coincides with chain termination (16,17). The bond forming reactions proceed by an inversion mechanism to form β-glycoside bonds from the α-linked UDP-sugars, whereas these reactions are coupled to large stepwise movements, i.e. translocation, of the growing chain. Increasing solvent viscosity should more dramatically affect the latter reactions, which likely involve large movements of protein domains, compared to the substrate binding and bond forming reactions.
It is possible that the viscogens tested inhibit seHAS in a different manner or have additional effects not related to their viscosity. For example, they all contain -OH groups capable of interacting with the -OH rich HA. The PEGs or viscogens could potentially affect the dynamics of the enzyme-substrate interactions or hinder conformational changes in the enzyme needed to translocate the HA chain. The viscogens may have a greater effect on the rate of enzyme binding to substrate, whereas the PEGs may preferentially affect the HA translocation process. Unfortunately, since there are presently no assays available to assess the six individual, “partial” reactions in the overall synthase reaction, we cannot yet test these possibilities.
An interesting result in the present study was that the phospholipid requirement for seHAS activity is very dependent on fatty acyl composition. Earlier results (38) showed that purified seHAS, which has little endogenous lipid, has very low activity, but the enzyme is activated by exogenous CL, phosphatidic acid or phosphatidylserine. This stimulatory effect was specific, since seven other lipids tested were either unable or poorly able to activate the enzyme. Here we tested the effect of two synthetic CLs, each made with a different type of fatty acyl chain. Purified seHAS was highly activated by CL containing only C18(Δ9) fatty acids (oleic acid), whereas it was essentially inactive in the presence of CL containing only C14 fatty acids (myristic acid). This intriguing observation indicates that the fatty acid composition of the associated lipids is critical for one or more seHAS functions. Future studies will be needed to determine the structural requirements of activating fatty acids and to identify the catalytic steps catalyzed by HAS that are lipid-dependent.
Acknowledgments
We thank Mandy Hayes and Jennifer Acuna for general technical assistance and for the preparation of membranes. This research was supported by National Institutes of Health Grant GM35978 from the National Institute of General Medical Sciences.
Footnotes
This research was supported by National Institute of General Medical Sciences grant GM35978 from the National Institutes of Health.
References
- 1.Meyer K, Palmer JW. The polysaccharide of the vitreous humor, . J Biol Chem. 1934;107:629–634. [Google Scholar]
- 2.Knudson CB, Knudson W. Hyaluronan-binding proteins in development, tissue homeostasis, and disease. FASEB J. 1993;7:1233–1242. [PubMed] [Google Scholar]
- 3.Fraser JR, Laurent TC, Laurent UB. Hyaluronan: its nature, distribution, functions and turnover, . J Intern Med. 1997;242:27–33. doi: 10.1046/j.1365-2796.1997.00170.x. [DOI] [PubMed] [Google Scholar]
- 4.Abatangelo G, Weigel PH. Elsevier Science Publishers B.V.; Amsterdam: 2000. New Frountiers in Medical Sciences: Redefining Hyaluronan. [Google Scholar]
- 5.Fenderson BA, Stamenkovic I, Aruffo A. Localization of hyaluronan in mouse embryos during implantation, gastrulation and organogenesis. Differentiation. 1993;54:85–98. doi: 10.1111/j.1432-0436.1993.tb00711.x. [DOI] [PubMed] [Google Scholar]
- 6.Lin Y, Mahan K, Lathrop WF, Myles DG, Primakoff P. A hyaluronidase activity of the sperm plasma membrane protein PH-20 enables sperm to penetrate the cumulus cell layer surrounding the egg, . J Cell Biol. 1994;125:1157–1163. doi: 10.1083/jcb.125.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toole BP. Hyaluronan in morphogenesis, . J Intern Med. 1997;242:35–40. doi: 10.1046/j.1365-2796.1997.00171.x. [DOI] [PubMed] [Google Scholar]
- 8.Turley EA, Bowman P, Kytryk MA. Effects of hyaluronate and hyaluronate binding proteins on cell motile and contact behaviour, . J Cell Sci. 1985;78:133–145. doi: 10.1242/jcs.78.1.133. [DOI] [PubMed] [Google Scholar]
- 9.Balazs EA, Denlinger JL. Viscosupplementation: a new concept in the treatment of osteoarthritis, . J Rheumatol Supp. 1993;39:3–9. [PubMed] [Google Scholar]
- 10.Goa KL, Benfield P. Hyaluronic acid. A review of its pharmacology and use as a surgical aid in ophthalmology, and its therapeutic potential in joint disease and wound healing. Drugs. 1994;47:536–566. doi: 10.2165/00003495-199447030-00009. [DOI] [PubMed] [Google Scholar]
- 11.Balazs EA. Analgesic effect of elastoviscous hyaluronan solutions and the treatment of arthritic pain. Cells Tissues Organs. 2003;174:49–62. doi: 10.1159/000070574. [DOI] [PubMed] [Google Scholar]
- 12.Surendrakumar K, Martyn GP, Hodgers EC, Jansen M, Blair JA. Sustained release of insulin from sodium hyaluronate based dry powder formulations after pulmonary delivery to beagle dogs, . J Control Release. 2003;91:385–94. doi: 10.1016/s0168-3659(03)00263-3. [DOI] [PubMed] [Google Scholar]
- 13.DeAngelis PL, Papaconstantinou J, Weigel PH. Isolation of a Streptococcus pyogenes gene locus that directs hyaluronan biosynthesis in acapsular mutants and in heterologous bacteria, . J Biol Chem. 1993a;268:14568–14571. [PubMed] [Google Scholar]
- 14.DeAngelis PL, Papaconstantinou J, Weigel PH. Molecular cloning, identification, and sequence of the hyaluronan synthase gene from Group A Streptococcus pyogenes, . J Biol Chem. 1993b;268:19181–19184. [PubMed] [Google Scholar]
- 15.Kumari K, Weigel PH. Molecular cloning, expression, and characterization of the authentic hyaluronan synthase from Group C Streptococcus equisimilis, . J Biol Chem. 1997;272:32539–32546. doi: 10.1074/jbc.272.51.32539. [DOI] [PubMed] [Google Scholar]
- 16.DeAngelis PL. Hyaluronan synthases: fascinating glycosyltransferases from vertebrates, bacterial pathogens, and algal viruses, . Cell Mol Life Sci. 1999;56:670–682. doi: 10.1007/s000180050461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weigel PH. Functional characteristics and catalytic mechanisms of the bacterial hyaluronan synthases. IUBMB Life. 2002;54:201–211. doi: 10.1080/15216540214931. [DOI] [PubMed] [Google Scholar]
- 18.Ward PN, Field TR, Ditcham WG, Maguim E, Leigh JA. Identification and disruption of two discrete loci encoding hyaluronic acid capsule biosynthesis genes hasA, hasB, and hasC in Streptococcus uberis, . Infect Immun. 2001;69:392–399. doi: 10.1128/IAI.69.1.392-399.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeAngelis PL, Jing W, Graves MV, Burbank DE, Van Etten JL. Hyaluronan synthase of chlorella virus PBCV-1. Science. 1997;278:1800–1803. doi: 10.1126/science.278.5344.1800. [DOI] [PubMed] [Google Scholar]
- 20.Meyer MF, Kreil G. Cells expressing the DG42 gene from early Xenopus embryos synthesize hyaluronan, . Proc Natl Acad Sci U S A. 1996;93:4543–7. doi: 10.1073/pnas.93.10.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeAngelis PL, Achyuthan AMJ. Yeast-derived recombinant DG42 protein of Xenopus can synthesize hyaluronan in vitro. J Biol Chem. 1996;271:23657–23660. doi: 10.1074/jbc.271.39.23657. [DOI] [PubMed] [Google Scholar]
- 22.Itano N, Kimata K. Mammalian hyaluronan synthases. IUBMB Life. 2002;54:195–199. doi: 10.1080/15216540214929. [DOI] [PubMed] [Google Scholar]
- 23.Spicer AP, Augustine ML, McDonald JA. Molecular cloning and characterization of a putative mouse hyaluronan synthase, . J Biol Chem. 1996;271:23400–23406. doi: 10.1074/jbc.271.38.23400. [DOI] [PubMed] [Google Scholar]
- 24.Shyjan AM, Heldin P, Butcher EC, Yoshino T, Briskin MJ. Functional cloning of the cDNA for a human hyaluronan synthase, . J Biol Chem. 1996;271:23395–23399. doi: 10.1074/jbc.271.38.23395. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe K, Yamaguchi Y. Molecular identification of a putative human hyaluronan synthase, . J Biol Chem. 1996;271:22945–22948. doi: 10.1074/jbc.271.38.22945. [DOI] [PubMed] [Google Scholar]
- 26.Weigel PH, Hascall VC, Tammi M. Hyaluronan synthases, . J Biol Chem. 1997;272:13997–14000. doi: 10.1074/jbc.272.22.13997. [DOI] [PubMed] [Google Scholar]
- 27.Spicer AP, McDonald JA. Characterization and molecular evolution of a vertebrate hyaluronan synthase gene family, . J Biol Chem. 1998;273:1923–1932. doi: 10.1074/jbc.273.4.1923. [DOI] [PubMed] [Google Scholar]
- 28.Camenisch TD, Spicer AP, Brehm-Gibson T, Biesterfeldt J, Augustine ML, Calabro A, Jr, Kubalak S, Klewer SE, McDonald JA. Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme, . J Clin Invest. 2000;106:349–360. doi: 10.1172/JCI10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brinck J, Heldin P. Expression of recombinant hyaluronan synthase (HAS) isoforms in CHO cells reduces cell migration and cell surface CD44, . Exp Cell Res. 1999;252:342–351. doi: 10.1006/excr.1999.4645. [DOI] [PubMed] [Google Scholar]
- 30.Itano N, Sawai T, Yoshida M, Lenas P, Yamada Y, Imagawa M, Shinomura T, Hamaguchi M, Yoshida Y, Ohnuki Y, Miyauchi S, Spicer AP, McDonald JA, Kimata K. Three isoforms of mammalian hyaluronan synthases have distinct enzymatic properties, . J Biol Chem. 1999;274:25085–25092. doi: 10.1074/jbc.274.35.25085. [DOI] [PubMed] [Google Scholar]
- 31.Koprunner M, Mulleggerl J, Lepperdinger G. Synthesis of hyaluronan of distinctly different chain length is regulated by differential expression of Xhas1 and 2 during early development of Xenopus laevis. Mech Dev. 2000;90:275–278. doi: 10.1016/s0925-4773(99)00238-5. [DOI] [PubMed] [Google Scholar]
- 32.Wessels MR, Bronze MS. Critical role of the Group A streptococcal capsule in pharyngeal colonization and infection in mice, . Proc Natl Acad Sci U S A. 1994;91:12238–42. doi: 10.1073/pnas.91.25.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markovitz A, Cifonelli JA, Dorfman A. The biosynthesis of hyaluronic acid by Group A streptococcus. VI. Biosynthesis from uridine nucleotides in cell-free extracts, . J Biol Chem. 1959;234:2343–2350. [PubMed] [Google Scholar]
- 34.Mian N. Characterization of a high-Mr plasma-membrane-bound protein and assessment of its role as a constituent of hyaluronate synthase complex, . Biochem J. 1986;237:343–357. doi: 10.1042/bj2370343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng KF, Schwartz NB. Solubilization and partial purification of hyaluronate synthetase from oligodendroglioma cells, . J Biol Chem. 1989;264:11776–11783. [PubMed] [Google Scholar]
- 36.Stoolmiller AC, Dorfman A. The biosynthesis of hyaluronic acid by Streptococcus, . J Biol Chem. 1969;244:236–246. [PubMed] [Google Scholar]
- 37.Triscott MX, van de Rijn I. Solubilization of hyaluronic acid synthetic activity from streptococci and its activation with phospholipids, . J Biol Chem. 1986;261:6004–6009. [PubMed] [Google Scholar]
- 38.Tlapak-Simmons VL, Baggenstoss BA, Clyne T, Weigel PH. Purification and lipid dependence of the recombinant hyaluronan synthases from Streptococcus pyogenes and Streptococcus equisimilis, . J Biol Chem. 1999a;274:4239–4245. doi: 10.1074/jbc.274.7.4239. [DOI] [PubMed] [Google Scholar]
- 39.Tlapak-Simmons VL, Baggenstoss BA, Kumari K, Weigel PH. Kinetic characterization of the recombinant hyaluronan synthases from Streptococcus pyogenes and Streptococcus equisimilis, . J Biol Chem. 1999b;274:4246–4253. doi: 10.1074/jbc.274.7.4246. [DOI] [PubMed] [Google Scholar]
- 40.Tlapak-Simmons VL, Kempner ES, Baggenstoss BA, Weigel PH. The active streptococcal hyaluronan synthases (HASs) contain a single HA synthase monomer and multiple cardiolipin molecules, . J Biol Chem. 1998;273:26100–26109. doi: 10.1074/jbc.273.40.26100. [DOI] [PubMed] [Google Scholar]
- 41.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding, . Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 42.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 43.Yoshida M, Itano N, Yamada Y, Kimata K. In vitro synthesis of hyaluronan by a single protein derived from mouse HAS1 gene and characterization of amino acid residues essential for the activity. . J Biol Chem. 2000;275:497–506. doi: 10.1074/jbc.275.1.497. [DOI] [PubMed] [Google Scholar]
- 44.DeAngelis PL, Weigel PH. Immunochemical confirmation of the primary structure of streptococcal hyaluronan synthase and synthesis of high molecular weight product by the recombinant enzyme. Biochemistry. 1994;33:9033–9039. doi: 10.1021/bi00197a001. [DOI] [PubMed] [Google Scholar]
- 45.Pummill PE, Kempner ES, DeAngelis PL. Functional molecular mass of a vertebrate hyaluronan synthase as determined by radiation inactivation analysis, . J Biol Chem. 2001;276:39832–39835. doi: 10.1074/jbc.M105489200. [DOI] [PubMed] [Google Scholar]
- 46.Hopwood JJ, Fitch FW, Dorfman A. Hyaluronic acid synthesis in a cell-free system from rat fibrosarcoma, . Biochem Biophys Commun. 1974;61:583–590. doi: 10.1016/0006-291x(74)90997-8. [DOI] [PubMed] [Google Scholar]
- 47.Pummill PE, Achyuthan AM, DeAngelis PL. Enzymological characterization of recombinant Xenopus DG42, a vertebrate hyaluronan synthase, . J Biol Chem. 1998;273:4976–81. doi: 10.1074/jbc.273.9.4976. [DOI] [PubMed] [Google Scholar]
- 48.Forsee WT, Cartee RT, Yother J. Biosynthesis of type 3 capsular polysaccharide in Streptococcus pneumoniae. Enzymatic chain release by an abortive translocation process, . J Biol Chem. 2000;275:25972–25978. doi: 10.1074/jbc.M002613200. [DOI] [PubMed] [Google Scholar]
- 49.Reinhart GD. Influence of pH on the regulatory kinetics of rat liver phosphofructokinase: a thermodynamic linked-function analysis. Biochemistry. 1985;24:7166–7172. doi: 10.1021/bi00346a022. [DOI] [PubMed] [Google Scholar]
- 50.Prehem P. Synthesis of hyaluronate in differentiated teratocarcinoma cells. Characterization of the synthase, . Biochem J. 1983a;211:181–189. doi: 10.1042/bj2110181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Falke JJ. Enzymology. A moving story. Science . 2002;295:1480–1481. doi: 10.1126/science.1069823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gavish B, Werber MM. Viscosity-dependent structural fluctuations in enzyme catalysis. Biochemistry. 1979;18:1269–1275. doi: 10.1021/bi00574a023. [DOI] [PubMed] [Google Scholar]
- 53.Ansari A, Jones CM, Henry ER, Hofrichtner J, Eaton WA. The role of solvent viscosity in the dynamics of protein conformational changes. Science. 1992;256:1796–1798. doi: 10.1126/science.1615323. [DOI] [PubMed] [Google Scholar]
- 54.Skamnaki VT, Owen DJ, Nobel MEM, Lowe ED, Lowe G, Oikonomakas NG, Johnson LN. Catalytic mechanism of phosphorylase kinase probed by mutational studies. Biochemistry. 1999;38:14718–14730. doi: 10.1021/bi991454f. [DOI] [PubMed] [Google Scholar]
- 55.Goldbeck RA, Paquette SJ, Kliger DS. The effect of water on the rate of conformational change in protein allostery, . Biophys J. 2001;81:2919–2934. doi: 10.1016/S0006-3495(01)75932-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walser R, van Gunsteren WF. Viscosity dependence of protein dynamics. . Proteins: Struct, Funct, Genet. 2001;42:414–421. [PubMed] [Google Scholar]
- 57.Fenimore PW, Frauenfelder H, McMahon BH, Parak FG. Slaving: solvent fluctuations dominate protein dynamics and functions, . Proc Natl Acad Sci U S A. 2002;99:16047–51. doi: 10.1073/pnas.212637899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tlapak-Simmons VL, Weigel PH. Hyaluronan synthesis by the purified Class I hyaluronan synthase from S. pyogenes and S. equisimilis occurs by addition to the reducing end, . Glycobiol. 2002;12:708. [Google Scholar]
- 59.Prehem P. Synthesis of hyaluronate in differentiated teratocarcinoma cells. Mechanism of chain growth, . Biochem J. 1983b;211:191–198. doi: 10.1042/bj2110191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Asplund T, Brinck J, Suzuki M, Briskin MJ, Heldin P. Characterization of hyaluronan synthase from a human glioma cell line, . Biochim Biophys Acta. 1998;8:377–388. doi: 10.1016/s0304-4165(98)00010-5. [DOI] [PubMed] [Google Scholar]


