Abstract
High-density lipoproteins (HDL) were examined as potential carriers of small peptides in plasma. HDL purified by density gradient centrifugation was delipidated and fractionated by size-exclusion chromatography under denaturing conditions. By hplc and mass spectrometry, more than 100 peptide components were found in the size range from 1,000 to 5,000 Da. By sequence analysis, peptides were identified as fragments of proteins such as apolipoproteins, fibrinogen, α1-proteinase inhibitor, and transthyretin. The results indicate that purified HDL bears a complex range of small peptides. It is unclear whether the peptides have any significant functional role as apolipopeptides, but they may represent a pathway for peptide delivery or scavenging and a significant reservoir of plasma peptides for diagnostic evaluation.
Keywords: High-density lipoprotein, Apolipoprotein, Peptidomics, Transthyretin, Apolipoprotein A-I, Proteomics
Abbreviations: HDL, high-density lipoprotein; MALDI TOF MS, matrix-assisted laser desorption time-of-flight mass spectrometry; m/z, mass/charge ratio
Lipoprotein particles in the blood circulation have multiple physiological roles as carrier molecules. In addition to the well recognized role of lipoproteins in cholesterol and triglyceride transport [1, 2], lipoprotein particles including high-density, low-density, and very low-density lipoproteins (HDL, LDL, and VLDL) serve as carriers for other lipids and for fat-soluble vitamins such as vitamins A, D, E, K, and carotenoids [3, 4]. A number of well-characterized proteins are major structural and functional components of HDL, LDL, and VLDL including apolipoproteins A-I, A-II, B, C-I, C-II, C-III, D, and E [1]. Other molecules without direct roles in lipoprotein metabolism such as endotoxin [5], tissue factor pathway inhibitor [6], paraoxonase [7], and ghrelin [8] also bind to HDL. The diversity of molecules binding to lipoprotein particles suggests that they can serve as a versatile adsorptive phase, analogous to stationary phases in reverse-phase chromatography. This characteristic led to a hypothesis that lipoproteins might serve as a reservoir for the hundreds of small peptides that circulate in human plasma [9, 10]. The complex pattern of plasma peptides contains large amounts of potential information, and some studies suggest this information may be probed for cancer detection or other diagnostic purposes [11, 12]. Initial promise of this approach has been slowed by reproducibility and standardization issues [13, 14]. Most of the small peptides in plasma or serum now are considered to circulate bound to albumin and subfractionation of binding components may improve analyses [15–17]. Here, we examine whether HDL serves as a major carrier of small peptides.
Materials and Methods
Calibrators and α-cyano-4-hydroxycinnamic acid and sinapinic acid were from Bruker Daltonics (Billerica, MA). Monisotopic masses of peptide calibrators were: angiotensin II, 1,046.54; angiotensin I 1,296.69; substance P, 1,347.74; bombesin, 1,619.82; ACTH 1–17, 2,093.09; ACTH 18–39, 2,465.20; somatostatin-28, 3,147.47. Average masses of protein calibrators were: bovine insulin, 5,734.61; ubiquitin, 8,565.89; equine cytochrome c, 12,361.20; equine myoglobin, 16,952.62. Samples were prepared and analyzed by MALDI TOF mass spectrometry as previously described [18] using a α-cyano-4-hydroxycinnamic acid matrix (10 mg/ml in 50% acetonitrile, 20% ethanol, 30% water, 0.1% trifluoroacetic acid) on a Bruker UltraFlex mass spectrometer in linear positive ion mode. Peptides were analyzed with 60 ns delayed extaction. Sequential Edman degradation of peptides purified by reverse-phase hplc was performed using a Procise 492 sequencer (Applied Biosystems, Foster City CA) as previously described [19].
HDL was prepared from 400 mL of blood freshly collected in citrate preservative from a healthy donor. Plasma was separated from cells by centrifugation and HDL was isolated from plasma by density gradient centrifugation in solutions of KBr [20]. Isolated HDL was dialyzed versus an ammonium bicarbonate solution, lyophilized, and delipidated by extraction with ether (1:1). The protein residue (approximately 100 mg protein) was dissolved in 6 M guanidine hydrochloride, 0.2 M tris(hydroxymethyl)aminomethane, pH 8.0 and chromatographed on a 3.2 X 190 cm column of Sephacryl S-200 eluted with the 3 M guanidine hydrochloride, 0.2 M tris, pH 8.0. Small aliquots of collected fractions were sampled with ZipTip C18 P10 (Millipore Corp, Billerica MA). After rinsing with 0.1% trifluoroacetic acid, peptides were eluted with 5 μL of sinapinic acid matrix. Two aliquots of 1 μL of eluate were dried on a target plate and analyzed by MALDI TOF mass spectrometry. Three pools containing peptides in the size approximate size range of 1,000–3,000 Da; 2,000–5,000 Da; and 3,000–7,000 were formed. Peptides from each pool of about 100 mL were captured on a Sep-Pak Vac RC 500 mg cartridge (Waters, Milford MA). After washing with 0.5% acetic acid, peptides were eluted from the cartridges with 4 mL of 70% acetonitrile. Specimens were concentrated to 800 μL under a stream of nitrogen.
Half of each peptide pool was analyzed by Waters HPLC system with 626 pump and 996 photodiode array detector. Separation was on a 3 X 250 mm BioBasic C18-silica column (Thermo Hypersil-Keystone, Bellefonte PA) eluted with a linear gradient over 120 minutes from 5–65% acetonitrile with 0.1% formic acid at a flow of 0.4 mL/min. Fractions of 0.4 mL were collected and 1 μL aliquots were analyzed by MALDI TOF mass spectrometry.
Approximately 1 μL of peptide pools from size-exclusion chromatography or pooled hplc fractions was analyzed on a ProteomeX liquid chromatography-ion trap mass spectrometry workstation (Thermo Finnigan Austin TX) using a 0.075 X 20 mm column of C18 silica as previously described [21]. During a 90 minute chromatography run, a scan of m/z over the range of 300–2,000 was performed every second. The three most abundant components of any scan are selected for analysis of fragment ion scans. Bioworks software is applied for interpretation of the partial sequence information and for protein identification.
Results and Discussion
Direct analysis of purified HDL and LDL from our laboratory or commercial sources by matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI TOF MS) showed multiple weak peaks (not shown) with a mass/charge (m/z) less than 6,630 which is the mass of apolipoprotein C-I, the smallest known apolipoprotein [1]. Patterns of peaks for HDL and LDL differed. However, it was not clear whether these weak peaks represented multiply charged forms of larger apolipoproteins, clusters of lipid molecules or other artifacts.
Purified HDL and LDL were delipidated to disrupt the structure of lipoproteins and to release peptide components of the particles. Soluble peptide components were analyzed directly by reverse-phase hplc to try to separate small peptide components from the major apolipoproteins, but efforts to isolate small peptide components by this approach and to identify them by sequential Edman degradation had limited success. Two peptides were isolated from LDL and identified by sequential Edman degradation for 10 cycles together with mass analysis. The peptides corresponded to residues 81- 103 of transthyretin (measured m/z = 2,451.8 versus 2,451.7 calculated) and residues 1–40 of apolipoprotein A-I (measured m/z = 4,496.2 versus 4,468.9). It is unclear whether a sequence polymorphism or peptide modification led to a difference between the measured and expected mass for the latter peptide. Finding of these peptides associated with LDL was somewhat surprising considering that their parent proteins are not recognized as components of LDL. Efforts to isolate peptides from LDL were hindered by gelling of delipidated specimens due to limited solubility of apolipoprotein B. Further analyses concentrated on analyses of HDL.
Multiple efforts to identify peaks from reverse-phase hplc by sequential Edman degradation were unsuccessful because two or more residues were identified for each cycle indicating that the specimen contained two or more peptides. Apparently, the high complexity of the mixture of peptides made it difficult to isolate single peptide components for sequence analysis. We concluded that analysis of the small peptide components required initial separation from the more abundant apolipoprotein components. Size fractionation of HDL polypeptides was accomplished by size-exclusion chromatography in a solution of guanidine hydrochloride. Fractions after the elution of major apolipoproteins were collected as 3 pools which contained peptides in the approximate size range from 3–6 kDa, 2–4 kDa, and 1–3 kDa. Analysis of each of these pools by reverse-phase hplc with monitoring at 220 nm revealed about 100 absorbance peaks for each pool (Fig. 1). The total absorbance at 220 nm for all peaks eluting between 10 and 75 min for all 3 pools was equivalent to the absorbance of about 1 mg of peptide, using insulin as a standard. Therefore, the recovered peptide components were estimated to represent about 1% of the total protein mass in the original HDL preparation. This is a minimal estimate, as there was no measure of losses during multiple steps in preparation, but it does suggest a minimal stoichiometry of about 1 small peptide molecule per each 5 apolipoprotein A-I molecules. Since the small peptides are a complex mixture of more than 100 components, none of the peptides individually are likely to represent major structural or functional elements of HDL. Analysis by MALDI TOF mass spectrometry of aliquots of each fraction collected during the hplc analysis indicated the presence of at least 174 unique components in the 3 pools in the m/z range from 1,200–6,400 (Fig. 2). This analysis was a conservative estimate of the number of components; extra peaks considered to arise from sodium or matrix adducts and doubly charged ions were edited out and a relatively high signal threshold was applied that excluded many low intensity peaks. Some components overlapped between 2 pools, but no components were in all 3 pools. The only apolipoprotein noted in the 3 pools was apolipoprotein C-I (two peaks at m/z = 6,630 and 6,432, eluting at about 78 min in hplc runs) in the 3–6 kDa pool. Similar complexity was observed for two separate HDL preparations.
Fig. 1.
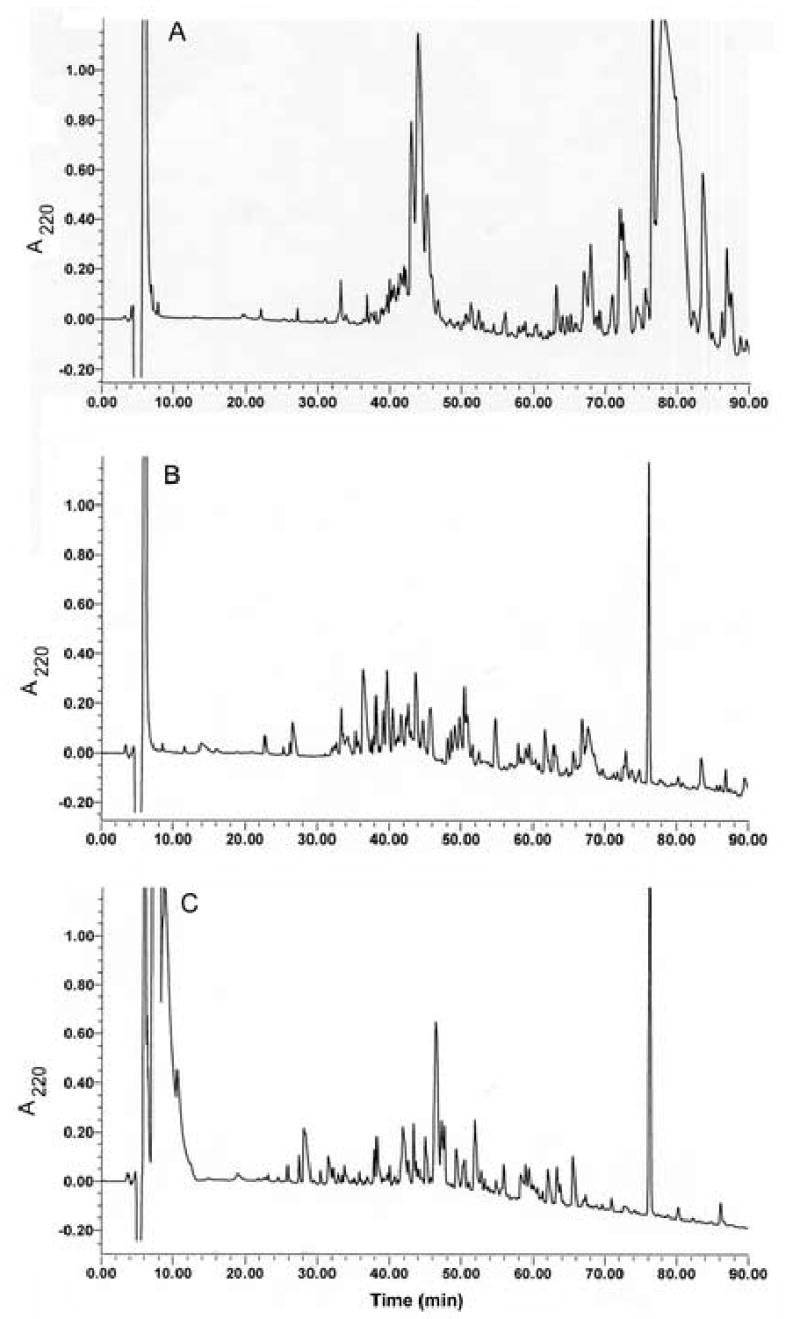
Analysis by hplc of 3 pools of HDL-associated peptides. The three pools were from size-exclusion chromatography of HDL-associated peptides with approximate size ranges of 3–6 kDa, panel A; 2–4 kDa, panel B; 1–3 kDa, panel C.
Fig. 2.
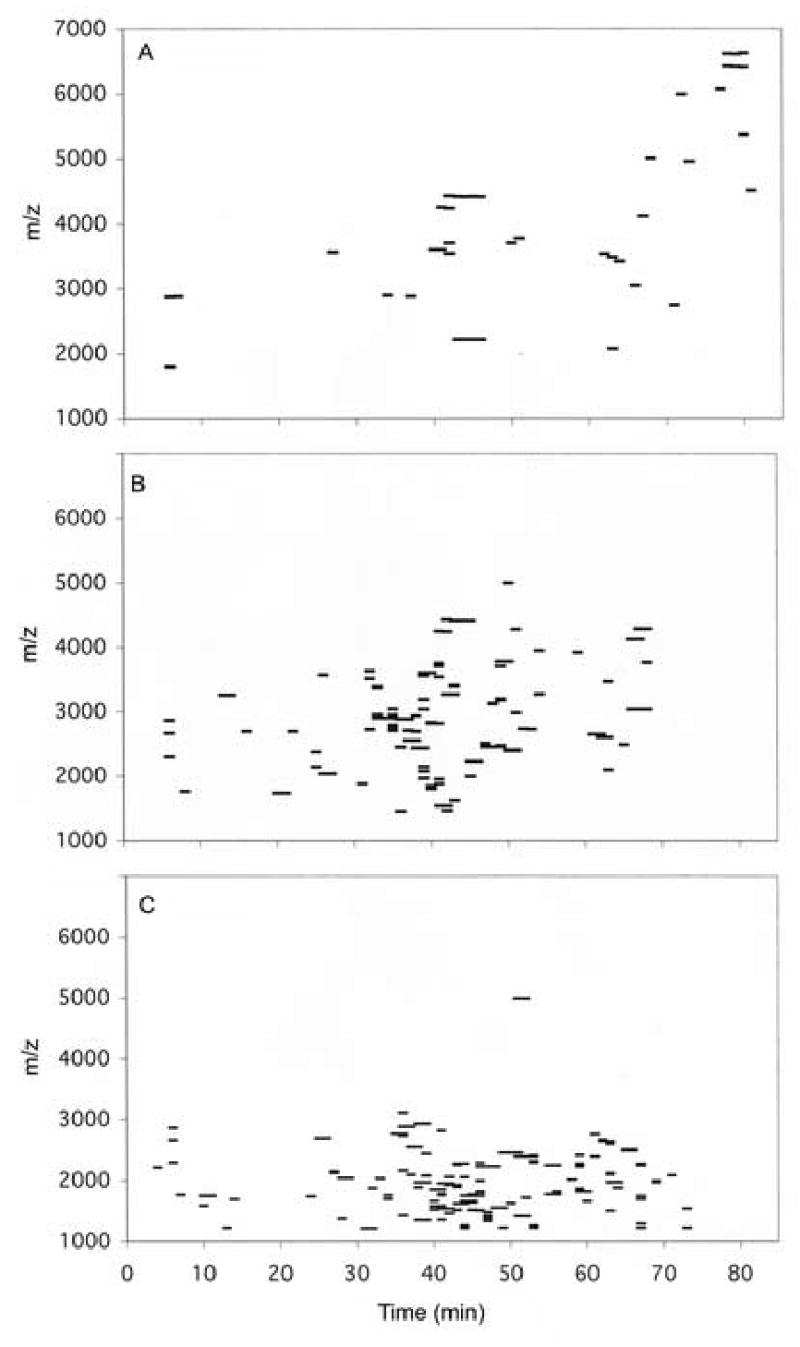
Analysis of hplc fractions by MALDI TOF mass spectrometry. Each fraction was collected for 1 min, and 1 μL out of a total of 400 μL for each fraction was analyzed as described in Methods.
Sequence identification of peptides in pools was performed by hplc-ion trap mass spectrometry. Sequence database searches identified 68 potential peptide components in the 1–3 kDa and 2–4 kDa pools of peptide (Table 1). The third pool, with larger peptides was not analyzed, as most peptides were beyond the optimal size limit for sequence analysis by this technique. Table 1 summarizes data from 12 runs and includes peptides identified with a high confidence as indicated by high cross-correlation (XC) scores and achievement of a best match with a single sequence. Most of the peptides were identified in more than one run and in multiple spectral scans. This type of analysis does not identify all components; it has a limited mass range and will identify only components lacking post-translational modifications that affect interpretation versus the sequence database and that efficiently yield ions with a charge of +1, +2, or +3. For about half of the peptides identified by database searches, there was a component of appropriate mass by MALDI TOF mass spectrometry. Not all components are expected to show up equally with these two techniques, which differ in ionization and detection techniques. Of the 68 peptides identified, 19 were fragments derived from HDL-associated proteins including apolipoproteins A-I, A-IV, C-III and L and lipopolysaccharide binding protein; 49 peptides were fragments of proteins not considered to be components of HDL, proteins such as fibrinogen, transthyretin, α1-proteinase inhibitor, α1B-glycoprotein, and inter-α- trypsin inhibitor. Occurrence of peptides from proteins generally not associated with lipoproteins implies that these peptides associated with lipoprotein particles before HDL was isolated from plasma and that the peptides remained bound during the purification process and extensive dialysis. A notable aspect of the peptides recovered from HDL was that none were derived from the most abundant plasma proteins, albumin and immunoglobulin G or from apolipoprotein A-II which is the second most abundant apolipoprotein component of HDL. The limited number of proteins serving as sources for peptides suggests a relatively selective binding of peptides to HDL, a selective process for peptide generation, or both.
TABLE 1.
Summary of peptide sequences identified from 12 runs on capillary liquid chromatography-ion trap mass spectrometry. Adjoining residues are separated by a slash. A dash indicates the end of the native protein sequence. Abbreviations: XC, cross-correlation score; RHM, Relative Hydrophobic Moment; H, Hydrophobicity score; Fgn, fibrinogen; Apo, apolipoprotein, α1-PI, α1-Proteinase Inhibitor; LPS bp, lipopolysaccharide-binding protein; α1B-Glyco, α1B-Glycoprotein; Inter-α-TI, Inter-α-trypsin inhibitor,
| MH+ | z | XC | RHM | H | Peptide | Sequence |
|---|---|---|---|---|---|---|
| 1,077.53 | 2 | 2.3 | 0.25 | −1.22 | Fgn α-chain 6–16 | E/GDFLAEGGGVR/G |
| 1,120.48 | 2 | 3.2 | 0.07 | −3.32 | Fgn α-chain 573–582 | R/GDSTFESKSY/K |
| 1,168.54 | 2 | 3.3 | 0.14 | −0,96 | Apo C-III 1–11 | -/SEAEDASLLSF/M |
| 1,214.60 | 2 | 2.4 | 0.42 | −2,94 | Fgn α-chain 241–251 | M/ELERPGGNEIT/R |
| 1,309.55 | 1 | 3.5 | 0.22 | −1,91 | Fgn α-chain 2–15 | A/DSGEGDFLAEGGGV/R |
| 1,345.64 | 2 | 2.9 | 0.35 | −2.30 | Fgn α-chain 240–251 | R/MELERPGGNEIT/R |
| 1,350.63 | 2 | 3.4 | 0.15 | −2.03 | Fgn α-chain 3–16 | D/SGEGDFLAEGGGVR/G |
| 1,366.72 | 2 | 3.7 | 0.61 | −3,20 | Apo A-I 233–243 | A/LEEYTKKLNTQ/- |
| 1,380.59 | 1 | 3.6 | 0.19 | −1.85 | Fgn α-chain 1–15 | T/ADSGEGDFLAEGGGV/R |
| 1,384.61 | 2 | 3.5 | 0.22 | −2.73 | Fgn β-chain 3–14 | G/VNDNEEGFFSAR/G |
| 1,396.70 | 2 | 3.9 | 0.18 | −2.76 | Transthyretin 115–127 | Y/SYSTTAVVTNPKE/- |
| 1,465.66 | 2 | 4.8 | 0.14 | −2.45 | Fgn α-chain 2–16 | A/DSGEGDFLAEGGGVR/G |
| 1,484.66 | 1 | 3.0 | 0.21 | −1.02 | Apo C-III 1–14 | -/SEAEDASLLSFMQG/Y |
| 1,499.67 | 2 | 4.0 | 0.23 | −2.86 | Fgn α-chain 559–571 | S/SYSKQFTSSTSYN/R |
| 1,509.82 | 2 | 2.8 | 0.63 | −1.93 | Apo A-I 230–242 | F/LSALEEYTKKLNT/Q |
| 1,524.79 | 2 | 3.2 | 0.57 | −3.13 | Apo A-I 231–243 | L/SALEEYTKKLNTQ/- |
| 1,536.06 | 2 | 4.4 | 0.12 | −2.36 | Fgn α-chain 1–16 | T/ADSGEGDFLAEGGGVR/G |
| 1,572.68 | 2 | 3.7 | 0.07 | −3.9 | Fgn α-chain 253–268 | R/GGSTSYGTGSETESPR/N |
| 1,618.71 | 2 | 2.9 | 0.14 | −3.38 | Transthyretin 49–63 | K/TSESGELHGLTTEEE/F |
| 1,630.79 | 2 | 3.2 | 0.25 | −3.22 | Apo L 279–293 | R/VTEPISAESGEQVER/V |
| 1,637.88 | 2 | 3.0 | 0.56 | −2.22 | Apo A-I 230–243 | F/LSALEEYTKKLNTQ/- |
| 1,642.80 | 2 | 3.2 | 0.39 | −0.38 | Fgn α-chain 225–238 | K/VPPEWKALTDMPQM/R |
| 1,673.73 | 2 | 4.5 | 0.19 | −3.05 | Fgn α-chain 557–571 | K/SSSYSKQFTSSTSYN/R |
| 1,686.72 | 2 | 3.9 | 0.05 | −4.09 | Fgn α-chain 253–269 | R/GGSTSYGTGSETESPRN/P |
| 1,714.87 | 2 | 3.2 | 0.23 | −0.80 | α1-PI 62–77 | F/AMLSLGTKADTHDEIL/E |
| 1,744.67 | 2 | 3.8 | 0.11 | −4.04 | Fgn α-chain 584–600 | K/MADEAGSEADHEGTHST/K |
| 1,771.85 | 2 | 2.5 | 0.61 | −1.00 | Apo A-IV 264–279 | K/SLAELGGHLDQQVEEF/R |
| 1,797.87 | 2 | 5.4 | 0.44 | −1.73 | Apo A-IV 38–53 | L/FQDKLGEVNTYAGDLQ/K |
| 1,815.99 | 2 | 3.3 | 0.17 | +0.21 | α1-PI 283–299 | R/SASLHLPKLSITGTYDL/K |
| 1,842.99 | 2 | 2.7 | 0.60 | −0.95 | Apo A-I 227–242 | K/VSFLSALEEYTKKLNT/Q |
| 1,876.97 | 2 | 4.1 | 0.04 | −3.45 | Fgn α-chain 408–423 | R/EYHTEKLVTSKGDKEL/R |
| 1,877.96 | 2 | 2.5 | 0.35 | +0.09 | Transthyretin 104–120 | R/RYIAALLSPYSYSTTA/V |
| 1,951.00 | 2 | 3.9 | 0.19 | −1.82 | Fgn β-chain 23–41 | K/REEAPSLRPAPPPISGGGY/R |
| 1,970.01 | 2 | 3.0 | 0.30 | −1.03 | Transthyretin 110–127 | A/LLSPYSYSTTAVVTNPKE/- |
| 1,971.08 | 2 | 4.8 | 0.55 | −1.35 | Apo A-I 227–243 | K/VSFLSALEEYTKKLNTQ/- |
| 1,971.93 | 2 | 3.1 | 0.36 | −0.40 | α1B-Glyco 453–469 | R/SWVPHTFESELSDPVEL/L |
| 2,001.88 | 3 | 2.7 | 0.16 | −3.69 | Fgn α-chain 557–574 | K/SSSYSKQFTSSTSYNRGD/S |
| 2,033.07 | 2 | 4.5 | 0.13 | −3.84 | Fgn α-chain 407–423 | R/REYHTEKLVTSKGDKEL/R |
| 2,052.09 | 3 | 3.5 | 0.14 | −1.37 | Inter-α-TI H4 599–615 | K/YYLQGAKIPKPEASFSPR/R |
| 2,085.02 | 2 | 3.2 | 0.42 | +0.16 | α1B-Glyco 453–470 | R/SWVPHTFESELSDPVELL/V |
| 2,096.03 | 2 | 4.2 | 0.37 | −1.37 | Apo A-IV 35–53 | L/NALFQDKLGEVNTYAGDLQ/K |
| 2,099.14 | 2 | 5.1 | 0.44 | −1.73 | Apo A-I 226–243 | F/KVSFLSALEEYTKKLNTQ/- |
| 2,122.86 | 2 | 5.8 | 0.14 | −4.02 | Fgn α-chain 581–600 | K/SYKMADEAGSEADHEGTHST/K |
| 2,164.93 | 3 | 3.4 | 0.16 | −4.30 | α1-PI 6–24 | G/DAAQKTDTSHHDQDHPTFN/K |
| 2,165.91 | 3 | 3.6 | 0.17 | −4.42 | α1-PI 6–24 | G/DAAEKTDTSHHDQDHPTFN/K |
| 2,225.17 | 2 | 6.3 | 0.33 | −0.57 | Transthyretin 107–127 | T/IAALLSPYSYSTTAVVTNPKE/- |
| 2,238.15 | 2 | 3.9 | 0.48 | −1.45 | Apo A-IV 177–195 | R/LTPYADEFKVKIDQTVEEL/R |
| 2,246.21 | 3 | 4.0 | 0.45 | −1.12 | Apo A-I 225–243 | S/FKVSFLSALEEYTKKLNTQ/- |
| 2,293.99 | 3 | 3.1 | 0.14 | −4.36 | Fgn α-chain 584–605 | K/MADEAGSEADHEGTHSTKRGHA/K |
| 2,326.22 | 2 | 4.3 | 0.31 | −0.72 | Transthyretin 106–127 | Y/TIAALLSPYSYSTTAVVTNPKE/- |
| 2,451.21 | 3 | 3.9 | 0.26 | −1.03 | Transthyretin 81–103 | K/ALGISPFHEHAEVVFTANDSGPR/R |
| 2,462.28 | 3 | 3.7 | 0.45 | −1.10 | Apo A-I 223–243 | L/ESFKVSFLSALEEYTKKLNTQ/- |
| 2,553.10 | 2 | 4.4 | 0.13 | −3.35 | Fgn α-chain 557–579 | K/SSSYSKQFTSSTSYNRGDSTFES/K |
| 2,645.38 | 2 | 4.7 | 0.37 | −0.85 | Transthyretin 104–127 | R/RYIAALLSPYSYSTTAVVTNPKE/- |
| 2,692.11 | 3 | 3.2 | 0.09 | −4.55 | α1-PI 1–24 | -/EDPQGDAAEKTDTSHHDQDHPTFN/K |
| 2,730.31 | 2 | 3.7 | 0.12 | −2.15 | Apo L 279–303 | R/VTEPISAESGEQVERVNEPSILEMS/R |
| 2,816.32 | 3 | 3.0 | 0.19 | −2.47 | Fgn α-chain 529–555 | R/GSESGIFTNTKESSSHHPGIAEFPSRG/K |
| 2,861.33 | 3 | 3.9 | 0.14 | −4.31 | Fgn α-chain 584–610 | K/MADEAGSEADHEGTHSTKRGHAKSRPV/R |
| 2,931.29 | 3 | 2.9 | 0.13 | −3.46 | Fgn α-chain 557–582 | K/SSSYSKQFTSSTSYNRGDSTFESKSY/K |
| 2,955.56 | 3 | 2.8 | 0.32 | −1.23 | Transthyretin 101–127 | S/GPRRYIAALLSPYSYSTTAVVTNPKE/- |
| 2,974.23 | 3 | 4.6 | 0.08 | −3.51 | Fgn α-chain 573–600 | R/GDSTFESKSYKMADEAGSEADHEGTHST/K |
| 3,152.73 | 3 | 3.5 | 0.03 | −0.26 | LPS bp 1–30 | -/ANPGLVARITDKGLQYAAQEGLLALQSELL/R |
| 3,182.74 | 3 | 3.9 | 0.49 | −0.33 | Apo A-I 216–243 | R/QGLLPVLESFKVSFLSALEEYTKKLNTQ/- |
| 3,344.55 | 3 | 5.4 | 0.24 | −0.58 | Fgn α-chain 492–521 | R/HRHPDEAAFFDTASTGKTFPGFFSPMLGEF/V |
| 3,432.75 | 3 | 4.7 | 0.12 | −0.17 | Inter-α-TI H2 593–623 | R/SILQMSLDHHIVTPLTSLVIENEAGDERMLA/D |
| 3,604.69 | 3 | 3.6 | 0.15 | −2.98 | Fgn α-chain 522–555 | F/VSETESRGSESGIFTNTKESSSHHPGIAEFPSRG/K |
| 3,759.77 | 3 | 3.3 | 0.26 | −2.1 | α1-PI 6–38 | G/DAAEKTDTSHHDQDHPTFNKITPNLAEFAFSLY/R |
| 3,829.77 | 3 | 3.7 | 0.15 | −2.89 | Fgn α-chain 544–579 | H/HPGIAEFPSRGKSSSYSKQFTSSTSYNRGDSTFES/K |
| 3,923.77 | 3 | 3.7 | 0.14 | −2.92 | α1-PI 1–35 | -/EDPQGDAAEKTDTSHHDQDHPTFNKITPNLAEFAF/S |
Many of the peptides represented overlapping fragments such as 10 peptides from the C-terminal end of apolipoprotein A-I, 7 peptides from the C-terminal end of transthyretin, and 28 peptides representing groups of overlapping peptides in several regions of the α-chain of fibrinogen. The overlapping families of peptides suggest that most peptides are derived by cleavage following arginine or lysine residues and that both ends of peptides subsequently are trimmed by exopeptidases that remove one or two residues from one or both ends. This is not surprising considering the known occurrence of a variety of exopeptidases in blood; the loss of C-terminal arginine or lysine residues from peptides by carboxypeptidase N is a particularly well-characterized process of physiological importance [22]. A number of large fragments of apolipoprotein A-I have been observed previously by 2-dimensional electrophoresis suggesting susceptibility of this protein to degradation in vivo [23]. Shortening of apolipoprotein A-I significantly affects its distribution among lipoprotein particles and circulating half-life [24]. All of the protein fragments previously observed by 2-dimensional electophoresis are substantially larger than those described here.
Based on the predicted structural and physical properties of the peptides identified in Table 1, the ability to form amphipathic helices, the key structural motif that enables apolipoproteins to associate with lipids, may contribute to binding of the peptides to HDL. Interestingly, not only peptide fragments of apolipoproteins, but also several other peptides derived from other types of proteins, such as transthyretin and fibrinogen, were also predicted to have relatively high hydrophobic moments. A few peptides, such as α1-proteinase inhibitor (283–299) and Inter-α-trypsin inhibitor H2 (593–623), were not amphipathic but were highly enriched with hydrophobic amino acids, which could promote association with the lipid phase of HDL There were several peptides, however, with low propensity to form amphipathic helices or with relatively polar sequences, such as fibrinogen α-chain (584–600) and α1-proteinase inhibitor (1–24). The mechanism for association of small peptides with HDL is an interesting issue for further investigation.
Most small peptides such as bioactive peptides generated in blood, peptide hormones released from cells, or peptides administered pharmacologically are cleared rapidly from the blood circulation by filtration in the kidneys or by proteolytic degradation [25, 26]. Our findings suggest that HDL particles can bind a complex range of small peptides derived from the proteolytic degradation of major plasma proteins. Binding of peptides to lipoproteins or other protein carriers could serve as a mechanism to slow the renal clearance and proteolysis of peptides just as it does for small apolipoproteins [1] and the peptide hormone ghrelin [8]. Binding of peptides to liposomes or to albumin has been shown to offer protection from degradation and greatly extended circulating half-lives [25, 26]. The major reservoir of small bound peptides in plasma has been considered to be albumin [15–17]. However, our findings sugget that HDL particles also may represent significant reservoirs of small peptides in the circulation. Binding of peptide to HDL could serve as a mechanism for the trafficking of selected peptides in the circulation, and it may represent a subset of the plasma peptidome that will be of interest for diagnostic purposes.
Acknowledgments
John Stonik and Bonnie Meilinger provided technical assistance and Maureen Sampson prepared figures. Work was supported by the intramural program of the NIH.
References
- 1.Gotto A, Jr, Pownall HJ, Havel RJ. Introduction to the plasma lipoprteins. Meth Enzymol. 1986;128:3–41. doi: 10.1016/0076-6879(86)28061-1. [DOI] [PubMed] [Google Scholar]
- 2.Brewer HB, Jr, Remaley AT, Neufeld EB, Basso F, Joyce C. Regulation of plasma high-density lipoprotein levels by the ABCA1 transporter and the emerging role of high-density lipoprotein in the treatment of cardiovascular disease. Arterioscler Thromb Vasc Biol. 2004;24:1755–1760. doi: 10.1161/01.ATV.0000142804.27420.5b. [DOI] [PubMed] [Google Scholar]
- 3.Ribaya-Mercado JD, Ordovas JM, Russell RM. Effect of beta-carotene supplementation on the concentrations and distribution of carotenoids, vitamin E, vitamin A, and cholesterol in plasma lipoprotein and non-lipoprotein fractions in healthy older women. J Am Coll Nutr. 1995;14:614–620. doi: 10.1080/07315724.1995.10718550. [DOI] [PubMed] [Google Scholar]
- 4.Goulinet S, Chapman MJ. Plasma LDL and HDL subspecies are heterogenous in particle content of tocopherols and oxygenated and hydrocarbon carotenoids. Relevance to oxidative resistance and atherogenesis. Arterioscler Thromb Vasc Dis. 1997;17:786–796. doi: 10.1161/01.atv.17.4.786. [DOI] [PubMed] [Google Scholar]
- 5.Levels JH, Abraham PR, van den Ende A, van Deventer SJ. Distribution and kinetics of lipoprotein-bound endotoxin. Infect Immunol. 2001;69:2821–2828. doi: 10.1128/IAI.69.5.2821-2828.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broze GJ. Tissue pathway inhibitor and the current concept of blood coagulation. Blood Coag Fibrinol. 1995;6:S7–S13. doi: 10.1097/00001721-199506001-00002. [DOI] [PubMed] [Google Scholar]
- 7.Mackness MI, Mackness B, Durrington PN. Paraoxonase and coronary heart disease. Atherosclerosis Suppl. 2002;3:49–55. doi: 10.1016/s1567-5688(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 8.Beaumont NJ, Skinner VO, Tan TMM, Ramesh BS, Byrne DJ, MacColl T, Keen JN, Bouloux PM, Mikhailidis DP, Bruckdorfer KR, Vanderpump MP, Srai KS. Ghrelin can bind to a species of high density lipoprotein associated with paraoxonase. J Biol Chem. 2003;278:8877–8880. doi: 10.1074/jbc.C200575200. [DOI] [PubMed] [Google Scholar]
- 9.Raida M, Schulz-Knappe P, Heine G, Forssmann WG. Liquid chromatography and electrospray mass spectrometric mapping of peptides from human plasma filtrate. J Am Soc Mass Spectrom. 1999;10:45–54. doi: 10.1016/S1044-0305(98)00117-2. [DOI] [PubMed] [Google Scholar]
- 10.Schrader M, Schulz-Knappe P. Peptidomics technologies for human body fluids. Trends Biotechnol. 2001;19:S55–S60. doi: 10.1016/S0167-7799(01)01800-5. [DOI] [PubMed] [Google Scholar]
- 11.Bichsel VE, Liotta LA, Petricoin EF., III Cancer proteomics: from biomarker discovery to signal pathway profiling. Cancer J. 2001;7:69–78. [PubMed] [Google Scholar]
- 12.Rai AJ, Zhang Z, Rosenzweig J, Shih I, Pham T, Fung ET, Sokoll LJ, Chan DW. Proteomic approaches to tumor marker discovery. Identification of biomarkers for ovarian cancer. Arch Pathol Lab Med. 2002;126:1518–1526. doi: 10.5858/2002-126-1518-PATTMD. [DOI] [PubMed] [Google Scholar]
- 13.Semmes OJ, Feng Z, Adam BL, Banez LL, Bigbee WL, Campos D, Cazares LH, Chan DW, Grizzle WE, Izbicka E, Kagan J, Milik G, McLerran D, Moul JW, Partin A, Prasanna P, Rosenzweig J, Sokoll LJ, Srivastava S, Srivastava S, Thompson I, Welsh MJ, White N, Winget M, Yasui Y, Zhang Z, Zhu L. Evaluation of serum protein profiling by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry for the detection of prostate cancer: I. Assessment of platform reproducibility. Clin Chem. 2005;51:102–112. doi: 10.1373/clinchem.2004.038950. [DOI] [PubMed] [Google Scholar]
- 14.Hortin GL. Can mass spectrometric protein profiling meet desired standards of clinical laboratory practice? Clin Chem. 2005;51:3–5. doi: 10.1373/clinchem.2004.043281. [DOI] [PubMed] [Google Scholar]
- 15.Mehta AI, Ross S, Lowenthal MS, Fusaro V, Fishman DA, Petricoin EF, 3rd, Liotta LA. Biomarker amplification by serum carrier protein binding. Dis Markers. 2004;19:1–10. doi: 10.1155/2003/104879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowenthal MS, Mehta AI, Frogale K, Bandle RW, Araujo RP, Hood BL, Veenstra TD, Conrads TP, Goldsmith P, Fishman D, Petricoin EF, 3rd, Liotta LA. Analysis of albumin-associated peptides and proteins from ovarian cancer patients. Clin Chem. 2005;51:1933–1945. doi: 10.1373/clinchem.2005.052944. [DOI] [PubMed] [Google Scholar]
- 17.Lopez MF, Mikulskis A, Kuzdzal S, Bennett DA, Kelly J, Golenko E, DiCesare J, Denoyer E, Patton WF, Ediger R, Sapp L, Ziegert T, Lynch C, Kramer S, Whiteley GR, Wall MR, Mannion DP, Della Cioppa G, Rakitan JS, Wolfe GM. High-resolution serum proteomic profiling of Alzheimer disease samples reveals disease-specific, carrier-protein-bound mass signatures. Clin Chem. 2005;51:1946–54. doi: 10.1373/clinchem.2005.053090. [DOI] [PubMed] [Google Scholar]
- 18.Hortin GL, Meilinger B, Drake SK. Size-selective extraction of peptides from urine for mass spectrometric analysis. Clin Chem. 2004;50:1092–1095. doi: 10.1373/clinchem.2003.030742. [DOI] [PubMed] [Google Scholar]
- 19.Kaplan B, Cojocaru M, Unsworth E, Knecht A, Martin BM. Search for peptidic "middle molecules" in uremic sera: isolation and chemical identification of fibrinogen fragments. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;796:141–153. doi: 10.1016/j.jchromb.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 20.Brewer HB, Jr, Ronan R, Meng M, Bishop C. Isolation and characterization of apolipoproteins A-I, A-II, and A-IV. Methods Enzymol. 1986;128:223–246. doi: 10.1016/0076-6879(86)28070-2. [DOI] [PubMed] [Google Scholar]
- 21.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matthews KW, Mueller-Ortiz SL, Wetzel RA. Carboxypeptidase N: a pleiotropic regulator of inflammation. Mol Immunol. 2004;40:785–793. doi: 10.1016/j.molimm.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Golaz O, Hughes GJ, Frutiger S, Paquet N, Bairock A, Pasquali C, Sanchez JC, Tissot JD, Appel RD, Walzer C, Balant L, Hochstrasser DF. Plasma and red blood cell protein maps: Update 1993. Electrophoresis. 1993;14:1223–1231. doi: 10.1002/elps.11501401183. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt HHJ, Remaley AT, Stonik JA, Ronan R, Wellmann A, Thomas F, Zech LA, Brewer HB, Jr, Hoeg JM. Carboxyl-terminal domain truncation alters apolipoprotein A-I in vivo catabolism. J Biol Chem. 1995;270:5469–5475. doi: 10.1074/jbc.270.10.5469. [DOI] [PubMed] [Google Scholar]
- 25.Dennis MS, Zhang M, Meng YG, Kadkhodavan M, Kirchhofer D, Combs D, Damico LA. Albumin binding as a general strategy for improving the pharmacokinetics of proteins. J Biol Chem. 2002;277:35035–35043. doi: 10.1074/jbc.M205854200. [DOI] [PubMed] [Google Scholar]
- 26.Sethi V, Onvuksel H, Rubenstein I. Liposomal vasoactive intestinal peptide. Methods Enzymol. 2005;391:377–395. doi: 10.1016/S0076-6879(05)91021-5. [DOI] [PubMed] [Google Scholar]


