Abstract
Telomeres cap the ends of eukaryotic chromosomes and prevent them from being recognized as DNA breaks. We have shown that certain DNA damage responses induced during senescence and, at times of telomere uncapping, also can be induced by treatment of cells with small DNA oligonucleotides homologous to the telomere 3′ single-strand overhang (T-oligos), implicating this overhang in generation of these telomere-based damage responses. Here, we show that T-oligo-treated fibroblasts contain γH2AX foci and that these foci colocalize with telomeres. T-oligos with nuclease-resistant 3′ ends are inactive, suggesting that a nuclease initiates T-oligo responses. We therefore examined WRN, a 3′ → 5′ exonuclease and helicase mutated in Werner syndrome, a disorder characterized by aberrant telomere maintenance, premature aging, chromosomal rearrangements, and predisposition to malignancy. Normal fibroblasts and U20S osteosarcoma cells rendered deficient in WRN showed reduced phosphorylation of p53 and histone H2AX in response to T-oligo treatment. Together, these data demonstrate a role for WRN in processing of telomeric DNA and subsequent activation of DNA damage responses. The T-oligo model helps define the role of WRN in telomere maintenance and initiation of DNA damage responses after telomere disruption.
Keywords: exonuclease, γ-H2AX foci, Werner syndrome, senescence, oligonucleotide
Replication of mammalian cells ultimately is limited by a program of senescence, suggested to be a fundamental defense against cancer (1). Normal human fibroblasts in culture usually undergo 60 to 70 population doublings before entering proliferative arrest, termed “replicative senescence” (2). This state in fibroblasts largely depends on the p53 tumor suppressor pathway (3–7), and inactivation of this pathway can lead to escape from replicative senescence. In addition, some evidence suggests that the pRb pathway may present an additional barrier to proliferation in fibroblasts. Both the p53 and pRb pathways appear to cause senescence in human epithelial cells (3, 4). However, after an additional 20–30 population doublings, fibroblasts then enter another block to proliferation termed “crisis,” characterized by extensive cell death and massive chromosomal abnormalities (8, 9).
Senescence is controlled, in large part, by telomeres, tandem repeats of the DNA sequence TTAGGG that cap chromosome ends in mammalian cells (10). Telomere ends normally form a loop structure, with the 3′ single-strand overhang concealed within the proximal DNA duplex and stabilized by association with telomere repeat-binding factors (TRFs) (11). Telomeres shorten with each round of replication, and critical shortening of telomeres is associated with replicative senescence or cell death by apoptosis, depending on cell type (12, 13). Although the mechanism by which telomere shortening triggers senescence is not clear, it now is thought that features other than simply telomere length are important (14). For example, experimental disruption of the telomere loop structure by ectopic expression of a dominant-negative TRF leads to overhang loss, chromosomal fusions, and, ultimately, induction of DNA damage responses including senescence or apoptosis, mediated at least, in part, through the activation of the ATM kinase and p53 (15). Whether the 3′ overhang is lost or retained in senescent cells is controversial (16, 17). However, Celli et al. (18) recently reported that overhang loss is not necessary to generate telomere-associated DNA damage foci in mouse cells upon conditional depletion of TRF2.
Localized telomeric DNA damage responses may function other than to signal senescence or apoptosis. Recently, Verdun et al. (19) detected the recruitment of DNA damage response proteins to “unprotected” telomeres during the late S and G2 phases of the cell cycle in normal human fibroblasts and that inhibition of this process leads to telomere dysfunction. The authors conclude that localization of these DNA damage response proteins to the telomeres restores proper telomere structure and function after DNA replication.
We have shown that treatment of both normal and transformed cells with ssDNA oligonucleotides homologous to the telomere overhang (T-oligos) induces DNA damage responses (20–26). Oligonucleotides unrelated or complementary to the overhang are inactive (20–24, 27). T-oligos rapidly accumulate in the cell nucleus (20, 25) and induce and/or activate ATM, p53, p95/Nbs1, p16, pRb, and other DNA repair and cell cycle regulatory proteins (20–26). However, T-oligos induce these responses without disrupting the telomere structure and leave the endogenous telomere overhang intact (21), unlike experimental telomere disruption (28). Together, these data suggest that the telomere overhang plays a role in telomere-mediated DNA damage responses and that exogenously provided T-oligos mimic the endogenous telomere overhang. We propose that T-oligos within the nucleus are recognized at the telomere by telomere-associated proteins whose normal role is to monitor and affect telomere structure and function. In this case, T-oligos would have the potential to provide a novel and useful probe into the molecular mechanism of these telomere-associated responses. However, heretofore a telomeric site of action of T-oligos has not been demonstrated.
Results and Discussion
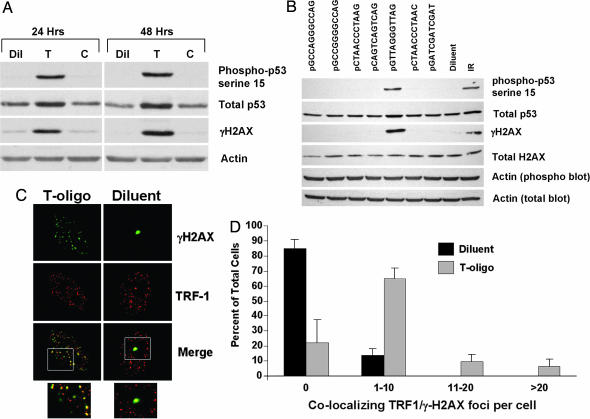
Phosphorylation of the histone variant H2AX, yielding γ-H2AX, is an early response to DNA damage (29) and has been shown to occur at short (30) and dysfunctional (31) telomeres, as well as at telomeres in cells serially passed to senescence (32). To determine whether T-oligo-treated cells contain γ-H2AX, normal human fibroblasts were treated with either an 11-base T-oligo, its complement, or diluent alone and then examined by Western blot. The T-oligo selectively and dramatically induced the phosphorylation of H2AX and p53 serine-15 as demonstrated in refs. 21 and 22) (Fig. 1A). To further exclude the possibility that these responses were triggered by G rich sequences independent of telomere homology or by sequences sharing only the same 3′ bases, other oligos with the same number and proportion (5 of 11) or more (6 of 11) G residues, as well as four sequences with the same 3′ termini (-AG) and two permutations of a repeat sequence with all four DNA bases, were compared with the 100% homolog (Fig. 1B). Again, only the telomere-homolog sequence was active, although other studies have shown that sequences with less-than-complete homology may be active so long as C residues are absent (27), suggesting that this feature, in addition to telomere homology, also is important. This specificity also was seen in another assay, the induction of apoptosis in MM-AN human melanoma cells (23), where only the T-oligo was active (Fig. 4, which is published as supporting information on the PNAS web site).
Fig. 1.
T-oligo treatment induces γ-H2AX foci at telomeres. (A) Normal human fibroblasts were treated with 40 μM T-oligo (T; GTTAGGGTTAG), a control, complementary oligo (C; CTAACCCTAAC), or an equal volume of diluent (Dil; water). After 24 and 48 h, the cells were collected and protein analyzed by Western blot. (B) Fibroblasts were treated with the indicated oligonucleotides at a concentration of 40 μM. As a control, cells were treated with an equal volume of diluent alone. Proteins were harvested for Western blot after 48 h. Two separate 10% gels were run with identical protein samples. One blot was probed for phospho-p53 serine-15, γ-H2AX, and actin (phospho blot actin); the other was probed for total p53, total H2AX, and actin (total blot actin). Both blots contained protein from IR (10 Gy)-treated fibroblasts as a positive control for the antibodies. (C) For immunofluorescence, fibroblasts were plated onto coverslips and treated with 40 μM T-oligo (GTTAGGGTTAG) or an equal volume of diluent (water). The cells were processed for immunofluorescence after 48 h. γ-H2AX was tagged with a FITC-linked secondary antibody (green) and TRF1 with a TRITC-linked secondary antibody (red). Cells containing γH2AX foci are shown from diluent- or T-oligo-treated cultures. In the case of diluent-treated cultures, only occasional cells contained any foci, but cells with at least one focus have been selected. The boxed areas seen on the merged images are shown enlarged below. (D) At least 100 cells from representative fields were scored for the number of γ-H2AX/TRF1 colocalizing foci. Values (mean ± SD) from three experiments (three donors) are shown.
Because both p53 and H2AX are known substrates for the ATM kinase (33, 34), these data are in agreement with our previous finding that T-oligo responses are mediated, at least in part, through ATM (21). Furthermore, γ-H2AX was present in foci in T-oligo-treated cells and colocalized with TRF1, a specific marker for telomeres (35), as demonstrated by immunostaining (Fig. 1C; see Fig. 5A, which is published as supporting information on the PNAS web site). Although occasional TRF1/γ-H2AX foci were found in ≈15% of diluent-treated cells (Figs. 1C and 4B), similar to those detected by Celli and de Lange (18) in mouse cells, T-oligo treatment greatly increased the percentage of colocalizing foci-containing cells to >80% (Fig. 1D). Moreover, in the majority of cases in both the occasional control cells with one or two foci and the plentiful T-oligo-treated cells with numerous colocalizing foci, the γ-H2AX foci fell between or immediately beside two TRF1 foci (Fig. 6A, which is published as supporting information on the PNAS web site). We now are investigating the possibility that these structures, far more frequent than expected by chance alone based on percent nuclear area occupied by these foci, represent telomeres on sister chromatids in proximity to phosphorylated histones surrounding telomeric and subtelomeric DNA (Fig. 6B). Localization of γ-H2AX to telomeres also was demonstrated by using the immuno-FISH technique (Fig. 7, which is published as supporting information on the PNAS web site), where telomeres are detected by hybridization to a telomere PNA probe. Together, these data demonstrate that treatment of normal cells with telomere-homologous DNA oligonucleotides stimulates phosphorylation of H2AX at telomeres.
Although γ-H2AX foci have been shown to form at short (30) or dysfunctional (31) telomeres, treatment with T-oligo does not shorten telomeres (21, 24). We previously reported that T-oligos also have no effect on the length of the 3′ overhang after up to 7 days of treatment as determined by Southern blot (21, 24). However, to further document retention of the telomere overhang in cells made senescent by T-oligos, normal fibroblasts were treated once at time 0 with T-oligos and then challenged with provision of fresh serum-containing medium lacking T-oligos on days 7, 10, and 13 to document the expected (22) permanent growth arrest, senescence-associated β-gal positive staining, and induction of the senescence-associated proteins p53, p21, and p16 and activation of p53 and pRb (Fig. 8 A–C, which is published as supporting information on the PNAS web site). Paired cultures harvested on day 0 before T-oligo treatment and on day 14 after senescence were compared by telomeric-oligonucleotide ligation assay (16), an extremely sensitive assay for overhang length, and no loss was detected (Fig. 8D), confirming our previous results (22).
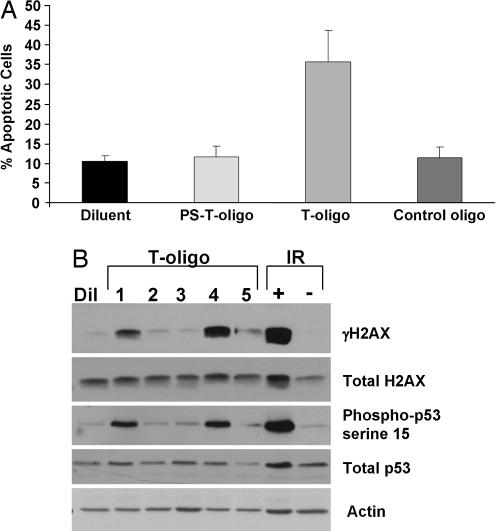
To study the features of T-oligos that affect their activity, we compared an 11-base T-oligo (GTTAGGGTTAG) synthesized with nonhydrolyzable phosphorothioate (PS) linkages to the physiologic phosphodiester-linked oligonucleotide. Hydrolyzable T-oligo readily induced apoptosis in MM-AN cells as expected (20, 23), but T-oligo with PS linkages was ineffective (Fig. 2A). To determine whether the effect of phosphorothioate linkages on T-oligo activity pertained generally to cells of multiple lineages, we examined normal fibroblasts and a well differentiated squamous cell carcinoma cell line (SCC12F). Similarly, whereas the physiologic hydrolyzable T-oligo induced an accumulation of cells within the S phase of the cell cycle as reported in refs. 20 and 22 in normal newborn fibroblasts and in the SCC12F cells, the PS-linked oligonucleotide did not (Fig. 9, which is published as supporting information on the PNAS web site). In addition, the phosphodiester-linked T-oligo also increased the sub-G0/G1 population of SCC12F cells, indicative of apoptosis. This effect was not seen in fibroblasts, likely reflecting the greater susceptibility of keratinocytes versus fibroblasts to apoptosis (36).
Fig. 2.
T-oligos with a blocked 3′ end are inactive. (A) Cultures of human MM-AN melanoma cells were treated once at time 0 with either 40 μM T-oligo (GTTAGGGTTAG) with phosphodiester linkages or with this oligonucleotide synthesized with phosphorothioate linkages (PS-T-oligo) to the control, complementary oligonucleotide, or diluent alone and harvested after 4 days. Apoptotic cells were identified as the sub-G0/G1 cells in FACS analysis. All experimental conditions were done in triplicate. The averages and SDs shown are calculated from two combined experiments. (B) Normal newborn fibroblasts were treated once at time 0 with either diluent alone or 40 μM of the following T-oligos: lane 1, all phosphodiester (no phosphorothioate) linkages (GTTAGGGTTAG); lane 2, two phosphorothioate linkages at both the 3′ and the 5′ ends (GsTsTAGGGTTsAsG); lane 3, all phosphorothioate linkages (GsTsTsAsGsGsGsTsTsAsG); lane 4, just the 5′ end blocked with 2 phosphorotioate linkages (GsTsTAGGGTTAG); or lane 5, just the 3′ end blocked with 2 phosphorothioate linkages (GTTAGGGTTsAsG). After 48 h, the cells were collected, and the proteins were analyzed by Western blot. Fibroblasts treated with 10 Gy ionizing radiation (IR+) or sham irradiated (IR−) were included as positive and negative controls.
The effects of phosphorothioate linkages were studied further by synthesizing T-oligos with no PS linkages, PS linkages throughout, two PS linkages at both the 3′ and 5′ ends, only the 5′ end, or only the 3′ end. The T-oligos containing PS linkages throughout or at both ends failed to induce phosphorylation of H2AX or p53 on serine-15, indicative of p53 activation (refs. 37 and 38; Fig. 2B), in contrast to fully hydrolyzable T-oligos, as observed in multiple cells types (21, 22) or T-oligos with an unblocked 3′ end (Fig. 2B). The same T-oligos that induced these phosphorylations also modestly induced total p53 protein (Fig. 2B), as also reported for the fully hydrolyzable 11-mer (21). The preferential activity of the phosphodiester-linked T-oligo is not due to preferential uptake and/or accumulation of this oligo in cultured cells (Fig. 10, which is published as supporting information on the PNAS web site). Indeed, intensity of nuclear staining after 4 h was far greater in the PS-linked T-oligos, presumably because of their very long half-life, in contrast to the estimated ≈4 h half-life for the readily hydrolyzable T-oligo (9). T-oligos containing a stabilizing 3′ phosphopropyl amine group (39) also poorly stimulated phosphorylation of p53 and H2AX, whereas a 5′ phosphopropyl amine group did not block the T-oligo activity (Fig. 11, which is published as supporting information on the PNAS web site). These data strongly suggest that 3′ → 5′ digestion of the T-oligo is required for its stimulation of DNA damage-like responses. Because both phosphorothioate linkages (40) and phosphopropyl amine groups (39) prevent exonucleolytic degradation of DNA, we hypothesize that the observed DNA damage responses are driven by the exonucleolytic degradation of the T-oligo within the nucleus.
The protein WRN, mutated in the autosomal recessive progeroid Werner syndrome (WS), recently has been shown to resolve telomeric loop structures in vitro when combined with TRF1 and TRF2 (41, 42). WRN contains both helicase and exonuclease domains (43) and localizes to telomeres in vivo (42). Recent work by Crabbe et al. (44) has shown that WRN helicase activity is necessary for proper replication of telomeres via lagging-strand DNA synthesis, possibly reflecting an ability of WRN to unwind G quadruplexes in the G rich telomere strand. To date, all mutations identified in WS are WRN truncations that eliminate the nuclear localization signal from the COOH end of the protein (45). Therefore, it is assumed that WRN mutations in WS generate a functional null phenotype by preventing the protein from reaching its site of action in the nucleus (46, 47). WS cells senesce prematurely compared with age-matched controls (48) and also demonstrate accelerated telomere shortening (49), characteristics cited in favor of this disease as an aging model (50). Cells from WS patients also show increased chromosomal deletions and translocations, both at baseline and after DNA damage (50), suggesting that WRN participates in DNA repair, replication and recombination, and maintenance of telomere length in addition to cell aging. However, the precise role of WRN in these pathways is poorly understood.
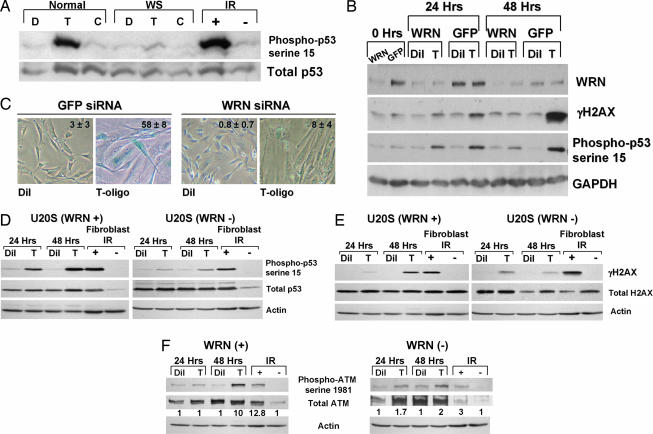
To determine whether WRN plays a role in T-oligo-induced DNA damage-like responses, fibroblasts from a WS patient and cells from an age-matched normal control were treated with T-oligos, and the DNA damage responses were measured by Western blot. Whereas normal fibroblasts phosphorylated p53 in response to T-oligo, the WS fibroblasts did not (Fig. 3A). Also, normal human fibroblasts made deficient in WRN by a single sequence siRNA were treated with T-oligo or diluent alone. Compared with control cells treated with siRNA-targeting GFP, WRN-depleted fibroblasts showed a reduction in T-oligo-induced p53 and H2AX phosphorylation, most strikingly at 48 h (Fig. 3B). Similar results also were obtained by using a mixture of WRN siRNAs (Smartpool by Dharmacon, Lafayette, CO) (Fig. 12, which is published as supporting information on the PNAS web site). Also, WRN-depleted fibroblasts failed to express senescence-associated β-gal activity after 6 days of T-oligo treatment (Fig. 3C), in contrast to WRN-expressing normal fibroblasts (Fig. 3C; refs. 22 and 24). WRN-deficient fibroblasts also have been shown to be resistant to hydrogen peroxide-induced senescence (51). Similarly, WRN depletion in U20S osteosarcoma cells by siRNA (Fig. 13, which is published as supporting information on the PNAS web site) greatly reduced the phosphorylation of p53 (Fig. 3D) and H2AX (Fig. 3E) after T-oligo treatment. Phosphorylation (activation) of ATM by T-oligo treatment also was reduced in the WRN-depleted U20S cells (Fig. 3F), consistent with a role for ATM in T-oligo responses.
Fig. 3.
Loss of WRN decreases T-oligo effects. (A) Fibroblasts from a WS patient (WS) and from an age-matched normal donor (normal) were treated with either 40 μM T-oligo (T), control complementary oligo (C), or an equal volume of diluent (D) (water). After 48 h, proteins were collected and analyzed. Normal neonatal fibroblasts exposed to 10 Gy IR (IR+) or sham-irradiated (IR−) were collected 3 h after treatment and used as positive and negative controls, respectively. (B) Normal human fibroblasts were treated with custom siRNA for WRN (WRN) or for GFP as a negative control. One day after the last transfection (“0 h”) either diluent (Dil) or 40 μM T-oligo (T) was added to the cultures. Cells then were collected for Western blot analysis 24 or 48 h after the additions. (C) Normal human fibroblasts were transfected twice with either WRN or GFP siRNA as described. Cell cultures then were supplemented once with either T-oligo (T) or diluent (Dil), cultured for an additional 6 days and then stained for senescence-associated β-gal activity. The percent senescence-associated β-gal positive cells was calculated from counting positive cells in five representative fields from each condition and is presented as inserts in the photos. (D–F) U20S cells transfected with either a WRN siRNA or a scrambled siRNA were treated with either diluent (Dil) or 40 μM T-oligo (T) for 24 or 48 h, cells were collected, and compared against phospho p53 serine-15 (D), γ-H2AX (E), or phospho-ATM serine-1981 (F) by Western blot.
Together, these data suggest that WRN-mediated exonucleolytic degradation of telomere G rich DNA induces DNA damage responses. In this context, it is interesting that activation of the ATM kinase by the exonuclease/helicase complex Mre11/hRAD50/Nbs1, at least in response to double-stranded breaks, was shown recently to be stimulated by DNA (52). Moreover, p53 phosphorylation after T-oligo treatment is reduced in cells from ataxia telangiectasia patients (21), who are deficient in ATM function, suggesting that ATM mediates, at least in part, these phosphorylation events, which is consistent with the ATM phosphorylation (activation) demonstrated here (Fig. 3F). Whether WRN, like Mre11/hRad50/Nbs1 (52), also activates ATM directly in the presence of specific DNA substrates and whether exonucleolytic degradation of this substrate is required remains to be determined. Alternatively, because WRN has both helicase and exonuclease activities (43), we cannot rule out that the effect of WRN on T-oligos is derived from unwinding the G-quadruplex tetramer structures that could form between these G rich oligos (53). However, phosphorothioate linkages were shown not to inhibit G-quadruplex formation (53), although their effect on WRN helicase activity is not known, and the rate of G-quartet-mediated tetramer formation between small G-rich oligonucleotides at 40 μM would be extremely slow (54).
Loss of the 3′ overhang after telomere disruption by expression of a dominant-negative form of TRF2 has been attributed to the ERCC1/XPF endonuclease (55), and activation of DNA damage response pathways in mouse cells lacking TRF2 does not require loss of the overhang (18). However, these experimental conditions lacking functional TRF2 may not allow for normal functions of WRN at telomeres because of WRN′s known interactions with TRF2 (41, 42, 56). Hence, DNA damage signaling from telomeres in the absence of TRF2 may be qualitatively similar yet mechanistically different from that following physiologic insults and/or T-oligo treatment.
There is increasing evidence that WRN is involved in resolving D (displacement) loops such as those formed at telomeres. D loops are disrupted by the WRN helicase, and the invading 3′ strand is readily degraded by the WRN exonuclease (42, 57). Both TRF1 and TRF2 (41, 42) were found to regulate the WRN exonuclease activity on telomeric D loops, and WRN associates with telomeres during the S phase when replication of the telomere may require disruption of the T loop. It is tempting to speculate that WRN-mediated resolution of the telomere D loop, removing the 3′ overhang from the telomere duplex DNA, activates DNA damage responses and “marks” the telomere, facilitating repair, replication, and restoration of the loop structure, consistent with a role for WRN in telomere maintenance. This hypothesis is consistent with the recent findings of Verdun et al. (19) that DNA damage response proteins such as phospho-ATM and phospho-Nbs1 are recruited to telomeres in the late S and G2 phases of the cell cycle, a time when telomere ends have become exposed and are accessible to enzymatic modification. Inhibition of these DNA damage responses leads to telomere fusions (19), a hallmark of telomere dysfunction. These data suggest that the formation of foci of DNA damage proteins at telomeres during replication, initiated by WRN, promotes the restoration of proper telomere structure and function. It has been shown that WS cells readily senesce in response to critical telomere shortening and can be immortalized by telomerase expression that maintains telomere length (58, 59), and we propose that the WRN-mediated responses described here normally play a central role in telomere maintenance and that other secondary pathways capable of recognizing telomere dysfunction and inducing senescence remain intact in WS cells, leading to the Werner phenotype in vitro and in vivo.
Our data strongly implicate the WRN-mediated displacement of the 3′ overhang, loop resolution, and subsequent activation of ATM and potentially other kinases as a mechanism for the formation of DNA damage-like foci. Both T-oligos and control oligos rapidly localize to the nucleus (ref. 20; Fig. 10), and we propose that this greatly increased concentration of single-stranded telomere overhang-specific DNA in the form of T-oligos provides a substrate to WRN in the vicinity of the telomere at the time of loop resolution and results in exaggerated DNA damage responses. Although the control oligonucleotides would be accessible equally to WRN, we propose that because of sequence and/or tertiary structural constraints, they do not serve as substrates for the exonuclease and, therefore, fail to activate the signaling cascade. These observations offer insight into the WS phenotype of altered telomere maintenance and accelerated aging. Further, they suggest that the physiologic critical telomere shortening after serial rounds of cell division or acute DNA damage, known to inhibit the binding of telomere-binding proteins TRF1 and TRF2 and increasing telomere erosion (60), may promote telomere loop disruption and WRN-mediated hydrolysis of the 3′ overhang that then initiates signaling through ATM, p53, and other effector proteins to safeguard genomic integrity.
Methods
Immunofluorescence.
Normal neonatal fibroblasts were cultured on glass coverslips and always refed 1 day before treating with either 40 μM T-oligo (GTTAGGGTTAG) or an equal volume of diluent (water). After 2 days, the cells were fixed and stained for γ-H2AX and TRF1 by using standard immunofluorescence protocols (61, 62). γ-H2AX was detected by using a mouse anti-γ-H2AX antibody and a FITC-conjugated goat anti-mouse IgG (code no. 115-095-146; Jackson Immunoresearch Laboratories, West Grove, PA). TRF1 was detected by using a rabbit anti-TRF1 antibody (catalog no. 581420; Calbiochem, San Diego, CA) and a Rhodamine Red-X-conjugated goat anti-rabbit IgG (code no. 111-295-144; Jackson Immunoresearch Laboratories). The cells were examined under ×1,000 magnification by using a Nikon Eclipse E400 microscope equipped with a RTke SPOT digital camera (Diagnostic Instruments, Sterling Heights, MI). FITC, TRITC, and DAPI images were overlapped by using the Advanced SPOT software.
siRNA Treatment.
All siRNAs were synthesized by Dharmacon. Normal neonatal fibroblasts were plated in p35 tissue culture dishes (Falcon tissue culture dish; Becton Dickinson Labware, Franklin Lakes, NJ) at a density of 200,000 cells per plate. The next day, the cells were transfected with 100 pmol per plate (2 ml of medium) siRNA directed against WRN (target sequence AGGCAUGUGUUCGGAAGAG or WRN smartpool siRNA), or GFP (catalog no. D-001300-01) as a negative control by using the protocol supplied by the manufacturer, for 6 h. The transfection protocol was repeated the following day. One day after the second transfection, cells were refed and supplemented with either diluent alone as a control or with 40 μM T-oligo (GTTAGGGTTAG). After the indicated times, cells were collected for protein analysis by Western blot.
For siRNA knockdown of WRN in U20S cells, an siRNA targeted against WRN mRNA (UGAAGAGCAAGUUACUUGCUU) was cloned into the pSilencer 3.1-H1 hygrovector (Ambion, Austin, TX). The pSilencer 3.1-H1 control hygrovector was used to express a scrambled siRNA with no significant homology to known human genes. Stable WRN knockdown and WT control cells were generated by transfection of U-20S cells and selection by growing in 200 μg/ml hydromycin B. Individual clones were selected and tested for WRN levels by Western blot and RT-PCR. The clone showing the greatest WRN knockdown and a control clone were selected for study.
Supporting Information.
Data on cells, oligonucleotides, antibodies, and chemicals used in this study are described as Supporting Methods, which is described as supporting information on the PNAS web site).
Supplementary Material
Acknowledgments
We thank Drs. Sheila Stewart and Ittai Ben-Porath for their insightful discussions and expert assistance with the telomeric-oligonuceotide ligation assay.
Abbreviations
- PS
phosphorothioate
- TRF
telomere repeat-binding factor
- WS
Werner syndrome.
Footnotes
Conflict of interest statement: Portions of the work reported in this article pertain to a patent application for which M.S.E. and B.A.G. are coinventors and, if awarded, will be assigned to the Trustees of Boston University (their employer) and then licensed to Semaco, Inc., a for-profit company created to commercialize intellectual property arising out of their laboratory. M.S.E. and B.A.G. both hold equity in Semaco, and B.A.G. is Semaco's Chief Scientific Officer.
References
- 1.Campisi J. Cell. 1996;84:497–500. doi: 10.1016/s0092-8674(00)81023-5. [DOI] [PubMed] [Google Scholar]
- 2.Hayflick L, Moorhead PS. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 3.Rheinwald JG, Hahn WC, Ramsey MR, Wu JY, Guo Z, Tsao H, De Luca M, Catricala C, O'Toole KM. Mol Cell Biol. 2002;22:5157–5172. doi: 10.1128/MCB.22.14.5157-5172.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramirez RD, Herbert BS, Vaughan MB, Zou Y, Gandia K, Morales CP, Wright WE, Shay JW. Oncogene. 2003;22:433–444. doi: 10.1038/sj.onc.1206046. [DOI] [PubMed] [Google Scholar]
- 5.Shay JW, Pereira-Smith OM, Wright WE. Exp Cell Res. 1991;196:33–39. doi: 10.1016/0014-4827(91)90453-2. [DOI] [PubMed] [Google Scholar]
- 6.Beausejour CM, Krtolica A, Galimi F, Narita M, Lowe SW, Yaswen P, Campisi J. EMBO J. 2003;22:4212–4222. doi: 10.1093/emboj/cdg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bond JA, Wyllie FS, Wynford-Thomas D. Oncogene. 1994;9:1885–1889. [PubMed] [Google Scholar]
- 8.Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, Bacchetti S. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shay JW, Wright WE. Carcinogenesis. 2005;26:867–874. doi: 10.1093/carcin/bgh296. [DOI] [PubMed] [Google Scholar]
- 10.Greider CW. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 11.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 12.de Lange T. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- 13.Harrington L, Robinson MO. Oncogene. 2002;21:592–597. doi: 10.1038/sj.onc.1205084. [DOI] [PubMed] [Google Scholar]
- 14.Karlseder J, Smogorzewska A, de Lange T. Science. 2002;295:2446–2449. doi: 10.1126/science.1069523. [DOI] [PubMed] [Google Scholar]
- 15.Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- 16.Stewart SA, Ben-Porath I, Carey VJ, O'Connor BF, Hahn WC, Weinberg RA. Nat Genet. 2003;33:492–496. doi: 10.1038/ng1127. [DOI] [PubMed] [Google Scholar]
- 17.Chai W, Shay JW, Wright WE. Mol Cell Biol. 2005;25:2158–2168. doi: 10.1128/MCB.25.6.2158-2168.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Celli GB, de Lange T. Nat Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 19.Verdun RE, Crabbe L, Haggblom C, Karlseder J. Mol Cell. 2005;20:551–561. doi: 10.1016/j.molcel.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 20.Eller MS, Puri N, Hadshiew IM, Venna SS, Gilchrest BA. Exp Cell Res. 2002;276:185–193. doi: 10.1006/excr.2002.5531. [DOI] [PubMed] [Google Scholar]
- 21.Eller MS, Li GZ, Firoozabadi R, Puri N, Gilchrest BA. FASEB J. 2003;17:152–162. doi: 10.1096/fj.02-0197com. [DOI] [PubMed] [Google Scholar]
- 22.Li GZ, Eller MS, Firoozabadi R, Gilchrest BA. Proc Natl Acad Sci USA. 2003;100:527–531. doi: 10.1073/pnas.0235444100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puri N, Eller MS, Byers HR, Dykstra S, Kubera J, Gilchrest BA. FASEB J. 2004;18:1373–1381. doi: 10.1096/fj.04-1774com. [DOI] [PubMed] [Google Scholar]
- 24.Li GZ, Eller MS, Hanna K, Gilchrest BA. Exp Cell Res. 2004;301:189–200. doi: 10.1016/j.yexcr.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 25.Hadshiew IM, Eller MS, Gasparro FP, Gilchrest BA. J Dermatol Sci. 2001;25:127–138. doi: 10.1016/s0923-1811(00)00125-0. [DOI] [PubMed] [Google Scholar]
- 26.Goukassian DA, Helms E, van Steeg H, van Oostrom C, Bhawan J, Gilchrest BA. Proc Natl Acad Sci USA. 2004;101:3933–3938. doi: 10.1073/pnas.0306389101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohashi N, Yaar M, Eller MS, Truzzi F, Gilchrest BA. J Cell Phys. 2006 doi: 10.1002/jcp.20848. in press. [DOI] [PubMed] [Google Scholar]
- 28.van Steensel B, Smogorzewska A, de Lange T. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 29.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 30.Hao LY, Strong MA, Greider CW. J Biol Chem. 2004;279:45148–45154. doi: 10.1074/jbc.M403924200. [DOI] [PubMed] [Google Scholar]
- 31.Takai H, Smogorzewska A, de Lange T. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 32.d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 33.Nakagawa K, Taya Y, Tamai K, Yamaizumi M. Mol Cell Biol. 1999;19:2828–2834. doi: 10.1128/mcb.19.4.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stiff T, O'Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA. Cancer Res. 2004;64:2390–2396. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 35.Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- 36.Marrot L, Belaidi JP, Jones C, Perez P, Riou L, Sarasin A, Meunier JR. J Invest Dermatol. 2003;121:596–606. doi: 10.1046/j.1523-1747.2003.12422.x. [DOI] [PubMed] [Google Scholar]
- 37.Caspari T. Curr Biol. 2000;10:R315–7. doi: 10.1016/s0960-9822(00)00439-5. [DOI] [PubMed] [Google Scholar]
- 38.Unger T, Sionov RV, Moallem E, Yee CL, Howley PM, Oren M, Haupt Y. Oncogene. 1999;18:3205–3212. doi: 10.1038/sj.onc.1202656. [DOI] [PubMed] [Google Scholar]
- 39.Zendegui JG, Vasquez KM, Tinsley JH, Kessler DJ, Hogan ME. Nucleic Acids Res. 1992;20:307–314. doi: 10.1093/nar/20.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Putney SD, Benkovic SJ, Schimmel PR. Proc Natl Acad Sci USA. 1981;78:7350–7354. doi: 10.1073/pnas.78.12.7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Machwe A, Xiao L, Orren DK. Oncogene. 2004;23:149–156. doi: 10.1038/sj.onc.1206906. [DOI] [PubMed] [Google Scholar]
- 42.Opresko PL, Otterlei M, Graakjaer J, Bruheim P, Dawut L, Kolvraa S, May A, Seidman MM, Bohr VA. Mol Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 43.von Kobbe C, Thoma NH, Czyzewski BK, Pavletich NP, Bohr VA. J Biol Chem. 2003;278:52997–53006. doi: 10.1074/jbc.M308338200. [DOI] [PubMed] [Google Scholar]
- 44.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 45.Oshima J. BioEssays. 2000;22:894–901. doi: 10.1002/1521-1878(200010)22:10<894::AID-BIES4>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 46.Marciniak RA, Lombard DB, Johnson FB, Guarente L. Proc Natl Acad Sci USA. 1998;95:6887–6892. doi: 10.1073/pnas.95.12.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsumoto T, Shimamoto A, Goto M, Furuichi Y. Nat Genet. 1997;16:335–336. doi: 10.1038/ng0897-335. [DOI] [PubMed] [Google Scholar]
- 48.Martin GM, Sprague CA, Epstein CJ. Lab Invest. 1970;23:86–92. [PubMed] [Google Scholar]
- 49.Schulz VP, Zakian VA, Ogburn CE, McKay J, Jarzebowicz AA, Edland SD, Martin GM. Hum Genet. 1996;97:750–754. doi: 10.1007/BF02346184. [DOI] [PubMed] [Google Scholar]
- 50.Opresko PL, Cheng WH, von Kobbe C, Harrigan JA, Bohr VA. Carcinogenesis. 2003;24:791–802. doi: 10.1093/carcin/bgg034. [DOI] [PubMed] [Google Scholar]
- 51.von Kobbe C, May A, Grandori C, Bohr V. FASEB J. 2004;18:1970–1972. doi: 10.1096/fj.04-1895fje. [DOI] [PubMed] [Google Scholar]
- 52.Lee JH, Paull TT. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 53.Sacca B, Lacroix L, Mergny JL. Nucleic Acids Res. 2005;33:1182–1192. doi: 10.1093/nar/gki257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wyatt JR, Davis PW, Freier SM. Biochemistry. 1996;35:8002–8008. doi: 10.1021/bi960124h. [DOI] [PubMed] [Google Scholar]
- 55.Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T. Mol Cell. 2003;12:1489–1498. doi: 10.1016/s1097-2765(03)00478-7. [DOI] [PubMed] [Google Scholar]
- 56.Opresko PL, von Kobbe C, Laine JP, Harrigan J, Hickson ID, Bohr VA. J Biol Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 57.Orren DK, Theodore S, Machwe A. Biochemistry. 2002;41:13483–13488. doi: 10.1021/bi0266986. [DOI] [PubMed] [Google Scholar]
- 58.Davis T, Singhrao SK, Wyllie FS, Haughton MF, Smith PJ, Wiltshire M, Wynford-Thomas D, Jones CJ, Faragher RG, Kipling D. J Cell Sci. 2003;116:1349–1357. doi: 10.1242/jcs.00331. [DOI] [PubMed] [Google Scholar]
- 59.Choi D, Whittier PS, Oshima J, Funk WD. FASEB J. 2001;15:1014–1020. doi: 10.1096/fj.00-0104com. [DOI] [PubMed] [Google Scholar]
- 60.Opresko PL, Fan J, Danzy S, Wilson DM, III, Bohr VA. Nucleic Acids Res. 2005;33:1230–1239. doi: 10.1093/nar/gki273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 62.Pichierri P, Rosselli F. EMBO J. 2004;23:1178–1187. doi: 10.1038/sj.emboj.7600113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.