Abstract
The putative oxidation of hydroethidine (HE) has become a widely used fluorescent assay for the detection of superoxide in cultured cells. By covalently joining HE to a hexyl triphenylphosphonium cation (Mito-HE), the HE moiety can be targeted to mitochondria. However, the specificity of HE and Mito-HE for superoxide in vivo is limited by autooxidation as well as by nonsuperoxide-dependent cellular processes that can oxidize HE probes to ethidium (Etd). Recently, superoxide was shown to react with HE to generate 2-hydroxyethidium [Zhao, H., Kalivendi, S., Zhang, H., Joseph, J., Nithipatikom, K., Vasquez-Vivar, J. & Kalyanaraman, B. (2003) Free Radic. Biol. Med. 34, 1359–1368]. However, 2-hydroxyethidium is difficult to distinguish from Etd by conventional fluorescence techniques exciting at 510 nm. While investigating the oxidation of Mito-HE by superoxide, we found that the superoxide product of both HE and Mito-HE could be selectively excited at 396 nm with minimal interference from other nonspecific oxidation products. The oxidation of Mito-HE monitored at 396 nm by antimycin-stimulated mitochondria was 30% slower than at 510 nm, indicating that superoxide production may be overestimated at 510 nm by even a traditional superoxide-stimulating mitochondrial inhibitor. The rate-limiting step for oxidation by superoxide was 4 × 106 M−1·s−1, which is proposed to involve the formation of a radical from Mito-HE. The rapid reaction with a second superoxide anion through radical–radical coupling may explain how Mito-HE and HE can compete for superoxide in vivo with intracellular superoxide dismutases. Monitoring oxidation at both 396 and 510 nm of excitation wavelengths can facilitate the more selective detection of superoxide in vivo.
Keywords: detection, hydroethidine, MitoSOX, mitochondria, dihydroethidium
Oxidative stress resulting from mitochondrial dysfunction has been implicated in neurodegeneration, aging, cancer, and diabetes (1, 2). Oxidative damage to mitochondrial electron transport complexes and DNA can lead to mitochondrial DNA mutations, aberrant electron transport, disruption of calcium homeostasis, and activation of apoptosis (3). Mitochondria consume ≈85–95% of the oxygen inspired during respiration (1), most of which is reduced to water, but a small portion (estimates range from <0.1% to as high as 4%) of electrons leak from the respiratory chain to reduce oxygen to superoxide (O2•−) (4, 5). Complexes I and III are major sites of O2•− formation (3, 4). Both membrane potential and reduction state of respiratory chain carriers affect O2•− production (3).
Assessing the generation of O2•− in mitochondria is confounded by the lack of a sensitive and specific assay (6). Some O2•− sensors actually generate O2•− by either uncoupling the respiratory chain (e.g., luminol) or reacting with oxygen (e.g., luminol and nitroblue tetrazolium; refs. 7 and 8). Many O2•− sensors react with intracellular oxidoreductases and can artificially generate a “superoxide” signal [e.g., hydroethidine (HE), cytochrome c, nitroblue tetrazolium, epinephrine, lucigenin, and luminol; ref. 8]. To circumvent these technical problems, we report a technique to detect O2•− using HE and the recently developed probe, MitoSOX red, a mitochondrial superoxide indicator (Invitrogen, Mito-HE). Mito-HE is comprised of HE, a commonly used probe for O2•−, linked by a hexyl carbon chain to a triphenylphosphonium (TPP+) group. TPP+ cations target molecules to mitochondria, because the positive charge on the phosphonium is surrounded by three lipophilic phenyl groups, which facilitates movement across phospholipid bilayers and accumulation into the mitochondrial matrix in response to the negative membrane potential (9).
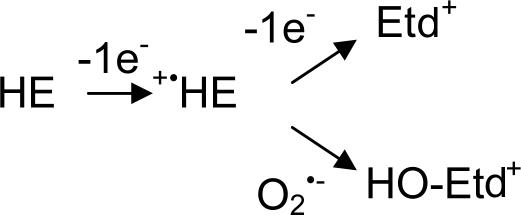
HE is the two-electron reduced form of ethidium (Etd+; 3,8-diamino-5-ethyl-6-phenylphenanthridinium). In 1984, Gallop et al. (10) reported that HE is readily taken up and internalized by live cells where HE can be oxidized to Etd+, which intercalates into nucleic acid, greatly enhancing its fluorescence when using 535-nm excitation and 610-nm emission wavelengths (11). Oxidation to Etd+ was originally attributed to the metabolic state of the cell and the cell's ability to dehydrogenate HE (11). However, in 1990, Rothe and Valet (12) showed in vitro that HE was oxidized by potassium superoxide to a red fluorescent product. HE has since been widely used to detect reactive oxygen species during the phagocytic respiratory burst (12, 13) and for the detection of intracellular O2•− (6, 14). However, Rothe and Valet also showed that HE was oxidized not only by O2•− but also by H2O2 plus peroxidase (12). HE may also be oxidized by other intracellular processes, involving oxidases or cytochromes, to yield Etd+ (11, 15–18). Consequently, increased Etd+ fluorescence is not necessarily proof of O2•− production. Swannell et al. (19) proposed HE could be oxidized to more than one red fluorescent product. In 2003, Zhao et al. (20) reported that HE is oxidized by O2•− to yield a hydroxylated product (HO-Etd+). The initial oxidation of HE had been proposed to involve the formation of a radical (15), implying the oxidation of HE by O2•− involves a two step mechanism (Scheme 1).
Scheme 1.
HO-Etd+ can be separated from Etd+ by HPLC, providing a specific O2•− assay (20). However, detection of HO-Etd+ by fluorescence microscopy is confounded, because its emission spectrum strongly overlaps the emission of Etd+ (20, 21).
We investigated the oxidation of Mito-HE by O2•− and found Mito-HE was oxidized by O2•− in a manner similar to HE (Scheme 1). During the course of these investigations, we found that the O2•−-derived product of Mito-HE has a distinct excitation wavelength at 396 nm that was not present for other oxidation products. HO-Etd+ was also selectively excited at this wavelength. The ability to separate the fluorescence of the O2•−-derived product from other less-specific oxidation products can improve the detection and imaging of intracellular O2•− production.
Results
The Selective Oxidation of Mito-HE.
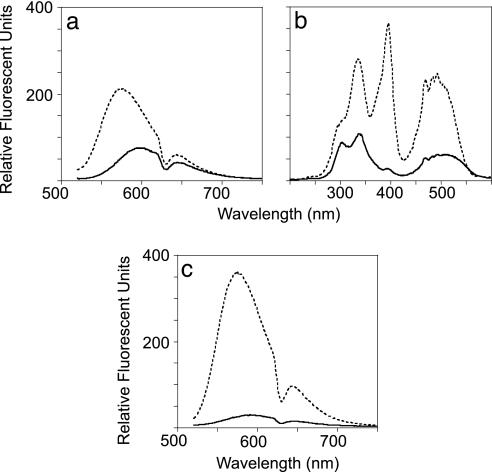
Superoxide generated by xanthine oxidase oxidized Mito-HE to give two closely eluting peaks by reverse-phase HPLC (Fig. 8, which is published as supporting information on the PNAS web site). The later peak was determined to be the two-electron-oxidized form of Mito-HE (3,8-diamino-5-hexyltriphenylphosphonium-6-phenylphenanthridinium, Mito-Etd+) by coelution with standards and mass spectrometry. The first peak contained one additional oxygen and was identified as the hydroxylated product, HO-Mito-Etd+. Oxidation of Mito-HE by hydrogen peroxide, peroxynitrite, hypochlorous acid, or a hydroxyl radical-generating system generated only a small percent of the fluorescence obtained from oxidation by O2•− (Table 1, which is published as supporting information on the PNAS web site). The fluorescence emission maximum for HO-Mito-Etd+ is blue-shifted ≈20 nm relative to Mito-Etd+ (Fig. 1a), similar to what was reported for the HE superoxide product, HO-Etd+ (20). Interestingly, the excitation spectrum of purified HO-Mito-Etd+ revealed an excitation maximum at 396 nm that was not present for Mito-Etd+ (Fig. 1b). Excitation at 396 nm (Fig. 1c) enhanced the fluorescence emission of HO-Mito-Etd+ by 70% and reduced the spectral overlap of Mito-Etd+ from 40% to 10%. The excitation spectrum of HO-Etd+ also had a distinct excitation at 396 nm that was not present for Etd+ (Fig. 9, which is published as supporting information on the PNAS web site). Therefore, excitation at 396 nm could serve as a more selective means for detecting O2•− using either HE or Mito-HE.
Fig. 1.
HO-Mito-Etd+ has a selective excitation wavelength at 396 nm. (a) The fluorescence emission of an equimolar concentration of Mito-Etd+ (—) overlapped 40% of the fluorescence emission of HO-Mito-Etd+ (− − −) exciting at 510 nm. (b) The excitation spectrum (emission at 579 nm) revealed HO-Mito-Etd+ (− − −) has a distinct excitation at 396 nm that is not present for Mito-Etd+ (—). (c) Excitation at 396 nm enhanced the fluorescence emission of HO-Mito-Etd+ (− − −) by 70% and reduced spectral overlap from Mito-Etd+ (—) to 10%, as compared with 510-nm excitation.
Detection of Mitochondrial Superoxide.
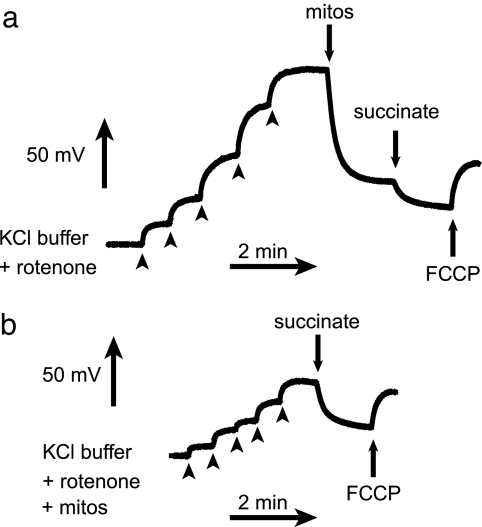
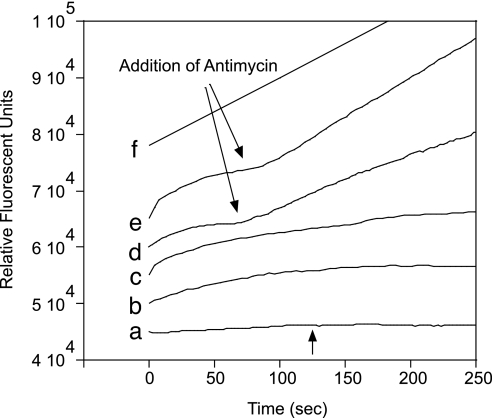
Mito-HE accumulated in isolated mitochondria in a manner that depended on the mitochondrial membrane potential, as well as by adsorption to membranes (Fig. 2). To test the relative ability of Mito-HE to detect O2•− using excitation wavelength (λex) = 396 nm, rat heart mitochondria respiring in State III were incubated with Mito-HE (Fig. 3). When oxygen concentrations were near the saturation limit for buffer (≈230 μM), mitochondria oxidized Mito-HE at a rate corresponding to 0.23 nmol O2•−·min−1·mg protein−1, while consuming 190 nmol O atm·min−1·mg protein−1. This corresponds to 0.1% of total electron flux resulting in O2•− formation. Because mitochondria consumed oxygen to concentrations <100 μM, endogenous superoxide formation was no longer observable. Antimycin stimulated O2•− production by Complex III to 0.81 nmol O2•−·min−1·mg protein−1 measured at λex = 396 nm (Fig. 3d). The rate of antimycin-stimulated fluorescence was 31% faster at λex = 510 nm (Fig. 3e), as compared with λex = 396 nm (Fig. 3d). Because HO-Mito-Etd+ is less fluorescent at λex = 510 nm, the 31% faster rate indicated that antimycin must have increased the formation of Mito-Etd+ in addition to HO-Mito-Etd+. The ratio of fluorescence emission from the two excitation wavelengths can be useful to assess nonspecific oxidation vs. oxidation mediated by O2•−.
Fig. 2.
Mito-HE was taken up by isolated mitochondria in a manner that depended on mitochondrial membrane potential, as well as by adsorption to membranes. An ion-selective electrode was used to measure the concentration of Mito-HE in a mitochondrial suspension. Mito-HE (five sequential additions of 1 μM; arrowheads) was added to energized mitochondria, and the mitochondrial uptake of Mito-HE was measured as a decrease in free Mito-HE. The uncoupler trifluoromethoxyphenylhydrazone (0.5 μM) was added to induce mitochondrial uncoupling and Mito-HE release (measured as an increase in free Mito-HE). In a, Mito-HE was added to the chamber before the mitochondria, enabling an estimation of the extent of nonpotential dependent mitochondrial association. In b, mitochondria were present in the chamber before Mito-HE addition.
Fig. 3.
The fluorescence emission of isolated mitochondria incubated with Mito-HE. Each trace was evenly plotted above the previous one for clarity, and the rate of Mito-HE oxidation was calculated from the approximately linear slope of the first 60 sec. (a) Mito-HE (0.8 μM) incubated without mitochondria did not show an increase in fluorescence at λex = 396 nm, even upon the addition of 15 μM antimycin (arrow). (b) Fluorescence using λex = 396 nm of mitochondria respiring in State III with 0.8 μM Mito-HE. (c) Fluorescence using λex = 510 nm of mitochondria respiring in State III with 0.8 μM Mito-HE. (d) The same as b with λex = 396 nm with the addition of 15 μM antimycin (arrow). (e) The same as d, except λex = 510 nm. (f) Mito-HE (0.8 μM) without mitochondria was oxidized by xanthine/xanthine oxidase (generating 7.2 nM O2•− per sec) in the presence of 50 μg of DNA. The plotted curves represent 3–10 replicates of each experiment. Ethanol alone had no effect.
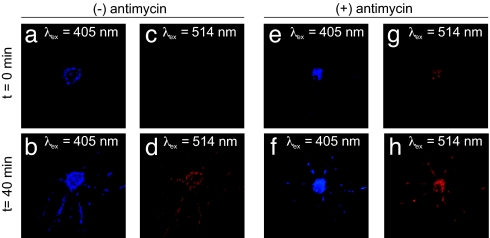
The two excitation wavelengths can be adapted to more selectively image O2•− in living cells. Standard lasers available on confocal microscopes can excite fluorophores at 405 and 514 nm, which are close enough to use in place of 396 and 510 nm. The fluorescence of cultured oligodendrocytes incubated with Mito-HE was more intense using 405-nm excitation yet qualitatively similar to 514-nm excitation (Fig. 4a–d). Fluorescence colocalized with mitochondria, as visualized with MitoTracker Deep Red (Invitrogen; data not shown). Antimycin treatment increased fluorescence at both excitation wavelengths, supporting dependency upon superoxide (Fig. 4 e–h). We have used the two excitations with confocal microscopy to observe increased mitochondrial localized fluorescence in five different types of primary cell cultures with the addition of superoxide-generating agents such as antimycin, paraquat, and menadione (not shown). This method can be adapted for widefield microscopy as well using a custom filter set (Fig. 10, which is published as supporting information on the PNAS web site).
Fig. 4.
Fluorescence from cultured oligodendrocytes incubated with Mito-HE using the two different excitation wavelengths, 405 (blue) and 514 nm (red). Instrument parameters were set and held constant to minimize autofluorescence at 405 nm, which is generally greater than at 514 nm. Oligodendrocyte after a 15-min incubation with 0.1 μM Mito-HE using λex = 405 nm (a) and then 40 min later (b). The same oligodendrocyte using λex = 514 nm (c and d). Fluorescence from oligodendrocytes was enhanced with 15 μM antimycin at λex = 405 nm (e) and 40 min later (f). The antimycin-stimulated increase in fluorescence was also observed by using λex = 514 nm (g and h).
Mechanism of Oxidation.
When Mito-HE was oxidized by xanthine oxidase, the fluorescence at 396/580 nm (λex/λem) was inhibited by Cu, Zn superoxide dismutase (SOD), but not by catalase (data not shown). The rate constant for Mito-HE oxidation by O2•− was calculated to be 3.9 ± 0.3 × 106 M−1·s−1 and was independent of whether fluorescence was measured at excitation/emission wavelength pairs of 396/580 nm (selective for HO-Mito-Etd+), 300/598 nm (selective for Mito-Etd+), or 510/580 nm (which excites both products). This is consistent with the rate-limiting step in the competition for O2•− being the initial oxidation of Mito-HE to a radical.
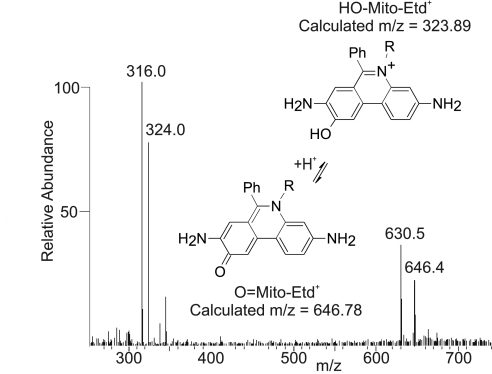
NMR spectra of purified O2•− oxidized Mito-HE (Fig. 11, which is published as supporting information on the PNAS web site) indicated the O2•− product had lost two protons, H11 and either H2 or H9, which was consistent for the proposed structure of HO-Mito-Etd+. Mass spectrometry of an O2•−-oxidized sample of Mito-HE revealed ions at m/z = 316.0 and 630.5 (Fig. 5), which were identified as Mito-Etd+ from comparison with the mass spectrum of a Mito-Etd+ standard (Fig. 12, which is published as supporting information on the PNAS web site). Mito-Etd+ can contaminate Mito-HE solutions, and a small percentage of Mito-Etd+ can also be generated upon oxidation by xanthine/xanthine oxidase (Fig. 8). Mass spectrometry supported the structure of HO-Mito-Etd+, revealing a doubly charged species at m/z = 324.0 (Fig. 5) consistent with the calculated mass of HO-Mito-Etd+, 647.79 atomic mass units. Because HO-Mito-Etd+ is doubly charged, it was surprising to observe a singly charged species at m/z = 646.4 (Fig. 5). This was resolved by attributing the m/z at 646.4 to the deprotonated form of HO-Mito-Etd+, O=Mito-Etd+ (Fig. 5). Collision-induced fragmentation of the parent ion at m/z 646.4 supported the proposed structure of O=Mito-Etd+ (Fig. 13, which is published as supporting information on the PNAS web site).
Fig. 5.
Mito-HE oxidized by xanthine/xanthine oxidase was analyzed by electrospray ionization and ion trap mass spectrometry. The ions at m/z = 316.0 and 630.5 were identified as Mito-Etd+. The doubly charged ion at m/z = 324.0 corresponded to the molecular weight of the hypothesized structure of HO-Mito-Etd+. The singly charged ion at m/z = 646.0 lead to the proposal of the carbonyl structure, O=Mito-Etd+.
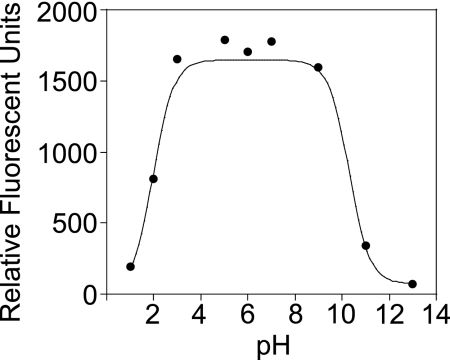
To determine the extent to which this deprotonation may occur in vivo, we determined the pKa of the HO-Mito-Etd+ hydroxyl group by measuring fluorescence using 396/579 nm (λex/λem) over a pH range from 1 to 12. The data were fit by using the Henderson–Hasselbalch equation, which revealed two pKas at 2.1 and 10.4 (Fig. 6). A third pKa was determined to be 0.65 by using fluorescence at λex = 396 nm and λem = 795 nm, which is maximal at acidic pH (data not shown). The pKas of 0.65 and 2.1 were assigned to the 8- and 3-ammonium groups of the phenanthridine ring, respectively, which are consistent with the previously determined pKas of Etd+, 0.8 and 2.0 (22). The pKa at 10.4 was consistent for an aromatic hydroxyl group. Moreover, the fluorescence emission of HO-Mito-Etd+ was maximal and independent of pH over the physiological range (Fig. 6).
Fig. 6.
The fluorescence emission of HO-Mito-Etd+ in the presence of 0.17 μg of DNA as a function of pH (λex = 396 nm; λem = 579 nm). This curve was fit to find two pKas at 2.1 and 10.5, and a third pKa was determined to be 0.65 by plotting the fluorescence emission at 795 nm with λex = 396 nm (not shown). Fluorescence of HO-Mito-Etd+ is maximal and independent of pH over the physiological range.
Discussion
Measurement of intracellular O2•− generation has been hampered by the lack of an assay sensitive enough to compete with endogenous SOD and yet selective for O2•−. Zhao et al. (20) have shown that the specificity for detecting O2•− with HE can be greatly improved if the hydroxylated product is distinguished from Etd+ by HPLC. However, the overlap in the emission spectra makes separating the two products difficult by fluorescence microscopy (20, 21). In the present study, we have uncovered a simple means to verify the selectivity of O2•− detection with either HE or Mito-HE by comparing fluorescence at two different excitation wavelengths. Excitation at 396 nm allows more selective imaging of the hydroxylated products produced by O2•− and has greater sensitivity than the current practice of 510 nm excitation (Fig. 1). HO-Mito-Etd+ was detected by using 396-nm excitation in both isolated mitochondria and cultured cells, and fluorescence was increased with agents that increase O2•− formation, such as antimycin. However, the rate of fluorescence generation from antimycin-stimulated mitochondria was 31% greater at 510- vs. 396-nm excitation, suggesting that Mito-Etd+ was being formed by nonsuperoxide-dependent pathways, and O2•− formation was overestimated by using 510-nm excitation. Thus, monitoring fluorescence using both excitation wavelengths can improve selectivity for distinguishing O2•− from other cellular oxidative processes.
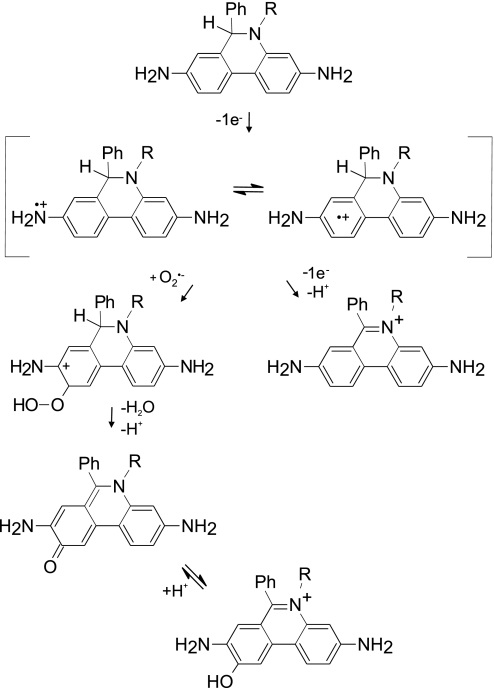
In 1934, Pauling and Neuman proposed the name superoxide based on the peculiar chemical bonding of KO2 (23, 24). However, superoxide more generally behaves as a mild reductant under physiological conditions rather than a “super”-oxidizing agent. The initial oxidation of the phenanthridine moiety by O2•− is recognized to generate a radical intermediate (14, 19, 20, 25) and is among the more rapid reactions of O2•− with organic molecules; the rate constant for HE was estimated here to be 2 × 106 M−1·s−1, and the oxidation of Mito-HE was twice as fast. This rate is ≈500 times slower than the rate of O2•− scavenging by SOD. However, the second reaction of the Mito-HE radical with O2•− would involve radical–radical coupling to produce a hydroperoxide intermediate and should approach the diffusion limit (>109 M−1·s−1). Hence, the Mito-HE radical could efficiently compete for O2•− with SOD in vivo to produce HO-Mito-Etd+. The hydroperoxide intermediate can spontaneously rearrange to lose a water molecule, yielding the hydroxylated product (Fig. 7). Hydroxylation appears to be relatively specific for O2•−, because other common biological oxidants generated only a small percent of the fluorescent signal generated by O2•− (Table 1). Although the hydroxyl radical in theory could add to the radical to give a hydroxylated product, the hydroxyl radical is a promiscuous oxidant that is far more likely to react with the multitude of other organic molecules in a cell (26).
Fig. 7.
Proposed scheme for the oxidation of Mito-HE [R = (CH2)6P+(Ph)3] or HE (R = CH2CH3) by O2•−. The radical intermediate is shown as a cation (HE·+), but it could alternatively be a neutral radical (Etd·).
Most fluorescent probes used to image oxidants in cells, including reduced forms of fluorescein, rhodamine, and Etd+, are susceptible to autooxidation through radical intermediates when illuminated (19). These probes can also be oxidized by a variety of intracellular peroxidases, oxidases, or cytochromes to yield radical intermediates that dismutate to give fluorescent products (12, 16–18). Therefore, the detection of the two-electron fluorescent product gives a rather nonselective assay of oxidative stress yet is commonly used as a superoxide indicator. Paradoxically, oxidation of HE by these alternative oxidative mechanisms could enhance O2•− detection by producing more HE radical, which increases the probability of trapping O2•− to form HO-Etd+.
The detection of HO-Etd+ is generally a semiquantitive assay for intracellular O2•−, because the relative fraction of O2•− reacting with SOD will be unknown. A second confounding variable can be the endogenous production of nitric oxide competing for O2•−, which may need to be inhibited by nitric oxide synthase inhibitors for O2•− to be detectable. On the other hand, we have found that the addition of an exogenous nitric oxide donor can inhibit the oxidation of Mito-HE (not shown) and can be used as a control of selectivity for O2•−.
The accumulation of Mito-HE depends upon the mitochondrial membrane potential with an ≈10-fold increase in uptake for every 60-mV increase in membrane potential (9). The mitochondrial membrane potential is typically in the range of −140 to −170 mV, which could concentrate Mito-HE in the mitochondria up to 1,000-fold relative to the medium. Such high intramitochondrial concentrations should allow Mito-HE to compete with manganese SOD for O2•−. However, excessive Mito-HE accumulation may stress mitochondria and enhance their sensitivity to inhibitors or other mitochondrial targeted molecules. Potentiometric probes may be added at the end of the incubation with Mito-HE to assess general mitochondrial integrity.
Careful optimization of conditions is necessary when using Etd+-based probes, and several caveats must be considered. Mitochondrial depolarization may reduce the efficiency of O2•− detection due to the decreased uptake of Mito-HE. Mito-HE itself, even at low (1- to 10-μM) concentrations, may disrupt membrane potential or increase mitochondrial permeability. We have observed a rapid loss of fluorescence from mitochondria and subsequent redistribution to the nucleus after incubation with as little as 2 μM Mito-HE. The optimal concentration of Mito-HE should be determined empirically in each cell type and varied in our experience from 0.1 to 2.5 μM. Because DNA is necessary to enhance the fluorescence of HO-Mito-Etd+, fluorescence might be limited by the amount of mitochondrial DNA, particularly in rho-naught cells or other pathological conditions where mitochondrial DNA has been deleted. Fluorescence intensity could artificially increase because of photooxidation. Photooxidation may be minimized by using the lowest possible concentration of Mito-HE and reducing the exposure to light throughout an experiment. Autofluorescence of cells will also be higher using 405-nm excitation and therefore should be controlled in microscopy experiments.
Because HE and Mito-HE accumulate to different extents within cells depending upon membrane potential, differences in fluorescence intensity between the two probes may not be directly compared to assess O2•− production between the two compartments. Higher concentrations of HE may be needed to achieve cytosolic concentrations similar to those of Mito-HE in mitochondria. Also, it is possible that HE could be oxidized by mitochondrially generated O2•−. Selective inhibitors would be needed to uncover the source of O2•−. Exact quantitation of products using fluorescence microscopy is fraught with difficulties, and therefore HPLC methods (20, 25) quantifying the relative amount of hydroxylated and nonhydroxylated products should be used in conjunction with fluorescence experiments when possible. If intracellular oxidation of Mito-HE to Mito-Etd+ should become 10- to 20-fold greater than HO-Mito-Etd+, a majority of fluorescence from 396-nm excitation could be due to Mito-Etd+. However, monitoring the oxidation at both 396 and 510 nm will reveal this potential artifact.
In conclusion, with judicious choice of conditions and an appreciation for the limitations of these probes, we have shown that the oxidation of HE to HO-Etd+ and Mito-HE to HO-Mito-Etd+ can be a sensitive indicator to monitor dynamic changes of endogenous O2•− generation.
Materials and Methods
MitoSOX Red mitochondrial superoxide indicator (Mito-HE), Mito-Etd+ HE, and ethidium were obtained from Invitrogen–Molecular Probes. The purity of Mito-HE was ascertained by HPLC (as described below), because Mito-Etd+ can be a contaminant in Mito-HE. If Mito-Etd+ contamination was ≥10%, Mito-HE was either purified by HPLC or reduced by an equimolar amount of NaBH4 (50 nmol in ethanol), with the reaction quenched by the addition of 120 nmol HCl. Recombinant human Cu, Zn SOD was expressed in Escherichia coli (27). Xanthine oxidase activity was assayed by the reduction of cytochrome c (Δε550 = 21 mM−1·cm−1) (28). Mito-HE was dissolved in DMSO to 5 mM and stored in the dark at −20°C for a maximum of 3 days. Mito-HE and all ethidium derivatives are carcinogenic and should be detoxified by reacting solutions diluted to ≤0.5 mg/ml ethidium with 0.2 volume of 5% hypophosphorous acid and 0.12 volume of fresh 0.5 M sodium nitrite, mixing carefully and venting the nitrogen gas evolved. DMSO penetrates gloves and skin and should also be used with caution.
Mito-HE and its oxidation products were separated by using C18 reverse-phase HPLC [Supelco (Bellefonte, PA) column, 15 cm × 4.6 mm, 5 μM] and a photodiode array detector. The mobile phase was H2O/CH3CN in 0.1% formic acid and 0.1% trifluoroacetic acid, using a linear gradient from 30% to 35% organic over 40 min. Fluorescence spectra of 1.0 μM HO-Mito-Etd+, Mito-Etd+, HO-Etd+, and Etd+ were obtained by using samples that were incubated for 15 min at 37°C with 1 mg/ml salmon sperm DNA, on a SLM Aminco (San Jose, CA) 8100c Fluorometer. The extinction coefficients for HO-Mito-Etd+ and Mito-Etd+ were determined to be ε478 = 9,400 M−1·cm−1 and ε488 = 5,800 M−1·cm−1, which were nearly identical to those reported for HO-Etd+ (29) and Etd+.
A SpectraMAX (San Jose, CA) Gemini fluorometer equipped with a 96-well plate reader was used for the SOD competition assays and also to compare fluorescence emission of Mito-HE oxidized by several oxidants. Assays were performed in 100 mM potassium phosphate buffer with 100 μM diethylenetriaminepentaacetic acid, pH = 7.4, 37°C. To determine rate constants, fluorescence emission was plotted as a function of the fractional inhibition (fi) by the addition of SOD, as described by Eq 1.
The rate of O2•− scavenging by SOD (ks) has been determined as 1.8 × 109 M−1·s−1 (30).
Superoxide was generated by xanthine oxidase, where the added volume of xanthine oxidase suspension was adjusted to generate 0.2 nM O2•− per sec with 0.5 mM xanthine, as measured by cytochrome c reduction. Alternatively, 10 μM Mito-HE was reacted with five 100-μM additions of peroxynitrite ± 25 mM ammonium bicarbonate with immediate vortexing, 100 μM hydrogen peroxide, or 100 μM hypochlorous acid. Hydroxyl radical was generated by using xanthine oxidase, 0.5 mM xanthine, and 10 μM Fe3+-EDTA. All assays were repeated at least three times and are reported ± standard deviation.
The uptake of Mito-HE into isolated mitochondria was determined by using an electrode selective for the triphenylphosphonium cation (31, 32). Rat livers were homogenized in 250 mM sucrose/5 mM Tris·HCl (pH 7.4)/1 mM EGTA, followed by differential centrifugation (33). The electrode was constructed as described (31), and the voltage was measured relative to an Ag/AgCl reference electrode. The electrode was calibrated by the sequential addition of 5 × 1 μM reduced Mito-HE to a stirred thermostated chamber at 30°C, containing 3 ml of KCl buffer (120 mM KCl/10 mM Hepes, pH 7.2/1 mM EGTA) and 4 μg/ml rotenone ± rat liver mitochondria (0.5 mg of protein per ml). Mitochondria were energized with succinate (10 mM), and the membrane potential was subsequently dissipated with 0.5 μM carbonylcyanide-p-trifluoromethoxyphenylhydrazone. Association of Mito-HE with mitochondria was measured as a decrease in the Mito-HE concentration in the medium.
To test the fluorescence emission of Mito-HE in isolated mitochondria, rat heart mitochondria were isolated as described (34, 35). Mitochondria were tested for coupled respiration with an oxygen electrode, and Mito-HE oxidation by mitochondria was followed with the SLM Aminco 8100c Fluorometer (λex = 396, 510 nm; λem = 579 nm) for 10 min. Mitochondria (600 μg of protein per ml) were incubated with 10 mM succinate/8.25 μM rotenone/2.5 mM ADP/0.8 μM Mito-HE with stirring at 30°C. The respiration buffer was 225 mM mannitol/75 mM sucrose/10 mM KCl/10 mM Tris·Cl/5 mM KH2PO4/0.1% fatty acid free BSA, pH = 7.2. Antimycin (15 μM) was added after 1 min to stimulate O2•− generation. Rates of fluorescence increase in the presence of mitochondria were converted to approximate rates of O2•− generation by comparison with a 0.8-μM Mito-HE standard oxidized by a known rate of superoxide (7.2 nM O2•− per sec generated with 130 μM xanthine and 2.5 μl of xanthine oxidase) in the presence of 50 μg of salmon sperm DNA.
Primary oligodendrocyte cultures were prepared from 1-day-old Sprague–Dawley rat pups, as reported (36). Cultured cells were transferred to the heated stage (37°C) of a Zeiss (Oberkochen, Germany) LSM510 confocal microscope with constant 5% CO2. Live cells were imaged with a 63× oil immersion objective by using either 405- or 514-nm laser excitation. A single field of view was used throughout an entire experiment and corrected for autofluorescence by collecting a 405-nm excitable signal before labeling with Mito-HE. Image acquisition conditions were kept constant for comparison between ±antimycin. Cells were incubated with 0.1 μM Mito-HE for 15 min, washed, and incubated in supplemented L15 media (36). Images were obtained immediately after Mito-HE addition and every 10 min thereafter. In some experiments, antimycin (15 μM) was added immediately after Mito-HE incubation. After confocal imaging of HO-Mito-Etd+, cells were incubated with 3.6 nM MitoTracker Deep Red (Invitrogen, Carlsbad, CA) to visualize mitochondrial colocalization and general integrity.
To determine the chemical structure of the oxidation product, samples were directly injected onto the electrospray interface of an LC-Q Classic ion trap mass spectrometer in positive ion mode. The mobile phase was 25% 10 mM ammonium acetate and 75% acetonitrile with 5 μl/min flow rate. The source voltage was 2.72 kV, and the capillary voltage was 32.7 V.
Supplementary Material
Acknowledgments
We appreciate the assistance of Brian Arbogast and Dr. Max Deinzer with mass spectrometry. K.M.R. was supported by a graduate fellowship from the Linus Pauling Institute, and M.F.R. was supported by the European Community's 6th Framework Program for Research, Contract LSHM-CT-2004-503116. This work was supported by Grants ES 00040, AG17141A, AT002034-02, TW006482-02, and PAT002034-01. We used the mass spectrometry and cell imaging facilities of the Environmental Health Sciences Center (ES00240) at Oregon State University in the performance of this research.
Abbreviations
- HE
hydroethidine
- Etd+
ethidium
- HO-Etd+
2-hydroxy ethidium
- Mito-HE
MitoSOX red mitochondrial superoxide indicator
- Mito-Etd+
3,8-diamino-5-hexyltriphenylphosphonium-6-phenylphenanthridinium
- HO-Mito-Etd+
hydroxylated Mito-HE
- SOD
superoxide dismutase
Footnotes
Conflict of interest statement: M.S.J. is an employee of Invitrogen, which donated reagents.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Shigenaga MK, Hagen TM, Ames BN. Proc Natl Acad Sci USA. 1994;91:10771–10778. doi: 10.1073/pnas.91.23.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green K, Brand MD, Murphy MP. Diabetes. 2004;53(Suppl 1):S110–S118. doi: 10.2337/diabetes.53.2007.s110. [DOI] [PubMed] [Google Scholar]
- 3.James AM, Murphy MP. J Biomed Sci. 2002;9:475–487. doi: 10.1159/000064721. [DOI] [PubMed] [Google Scholar]
- 4.Lenaz G, Bovina C, D'Aurelio M, Fato R, Formiggini G, Genova ML, Giuliano G, Merlo Pich M, Paolucci U, Parenti Castelli G, et al. Ann NY Acad Sci. 2002;959:199–213. doi: 10.1111/j.1749-6632.2002.tb02094.x. [DOI] [PubMed] [Google Scholar]
- 5.Fridovich I. Aging Cell. 2004;3:13–16. doi: 10.1046/j.1474-9728.2003.00075.x. [DOI] [PubMed] [Google Scholar]
- 6.Degli Esposti M. Methods. 2002;26:335–340. doi: 10.1016/S1046-2023(02)00039-7. [DOI] [PubMed] [Google Scholar]
- 7.Faulkner K, Fridovich I. Free Radic Biol Med. 1993;15:447–451. doi: 10.1016/0891-5849(93)90044-u. [DOI] [PubMed] [Google Scholar]
- 8.Tarpey MM, Fridovich I. Circ Res. 2001;89:224–236. doi: 10.1161/hh1501.094365. [DOI] [PubMed] [Google Scholar]
- 9.Ross MF, Kelso GF, Blaikie FH, James AM, Cocheme HM, Filipovska A, Da Ros T, Hurd TR, Smith RA, Murphy MP. Biochemistry (Moscow) 2005;70:222–230. doi: 10.1007/s10541-005-0104-5. [DOI] [PubMed] [Google Scholar]
- 10.Gallop PM, Paz MA, Henson E, Latt SA. BioTechniques. 1984;1:32–36. [Google Scholar]
- 11.Bucana C, Saiki I, Nayar R. J Histochem Cytochem. 1986;34:1109–1115. doi: 10.1177/34.9.2426339. [DOI] [PubMed] [Google Scholar]
- 12.Rothe G, Valet G. J Leukocyte Biol. 1990;47:440–448. [PubMed] [Google Scholar]
- 13.Perticarari S, Presani G, Banfi E. J Immunol Methods. 1994;170:117–124. doi: 10.1016/0022-1759(94)90251-8. [DOI] [PubMed] [Google Scholar]
- 14.Tarpey MM, Wink DA, Grisham MB. Am J Physiol Regul Integr Comp Physiol. 2004;286:R431–R444. doi: 10.1152/ajpregu.00361.2003. [DOI] [PubMed] [Google Scholar]
- 15.Benov L, Sztejnberg L, Fridovich I. Free Radic Biol Med. 1998;25:826–831. doi: 10.1016/s0891-5849(98)00163-4. [DOI] [PubMed] [Google Scholar]
- 16.LeBel CP, Ischiropoulos H, Bondy SC. Chem Res Toxicol. 1992;5:227–231. doi: 10.1021/tx00026a012. [DOI] [PubMed] [Google Scholar]
- 17.Henderson LM, Chappell JB. Eur J Biochem. 1993;217:973–980. doi: 10.1111/j.1432-1033.1993.tb18328.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhu H, Bannenberg GL, Moldeus P, Shertzer HG. Arch Toxicol. 1994;68:582–587. doi: 10.1007/s002040050118. [DOI] [PubMed] [Google Scholar]
- 19.Swannell RPJ, Caplin R, Nedwell DB, Williamson FA. FEMS Microbiol Ecol. 1992;101:173–182. [Google Scholar]
- 20.Zhao H, Kalivendi S, Zhang H, Joseph J, Nithipatikom K, Vasquez-Vivar J, Kalyanaraman B. Free Radic Biol Med. 2003;34:1359–1368. doi: 10.1016/s0891-5849(03)00142-4. [DOI] [PubMed] [Google Scholar]
- 21.Zhao H, Joseph J, Fales HM, Sokoloski EA, Levine RL, Vasquez-Vivar J, Kalyanaraman B. Proc Natl Acad Sci USA. 2005;102:5727–5732. doi: 10.1073/pnas.0501719102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luedtke NW, Liu Q, Tor Y. Chemistry. 2005;11:495–508. doi: 10.1002/chem.200400559. [DOI] [PubMed] [Google Scholar]
- 23.Pauling L. Trends Biochem Sci. 1979;4:N270–N271. [Google Scholar]
- 24.Neuman EW. J Chem Phys. 1934;2:31–33. [Google Scholar]
- 25.Fink B, Laude K, McCann L, Doughan A, Harrison DG, Dikalov S. Am J Physiol. 2004;287:C895–C902. doi: 10.1152/ajpcell.00028.2004. [DOI] [PubMed] [Google Scholar]
- 26.Beckman JS. Ann NY Acad Sci. 1994;738:69–75. doi: 10.1111/j.1749-6632.1994.tb21791.x. [DOI] [PubMed] [Google Scholar]
- 27.Leinweber B, Barofsky E, Barofsky DF, Ermilov V, Nylin K, Beckman JS. Free Radic Biol Med. 2004;36:911–918. doi: 10.1016/j.freeradbiomed.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 28.McCord JM, Fridovich I. J Biol Chem. 1968;243:5753–5760. [PubMed] [Google Scholar]
- 29.Zielonka J, Zhao H, Xu Y, Kalyanaraman B. Free Radic Biol Med. 2005;39:853–863. doi: 10.1016/j.freeradbiomed.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 30.Klug D, Rabani J, Fridovich I. J Biol Chem. 1972;247:4839–4842. [PubMed] [Google Scholar]
- 31.Kamo N, Muratsugu M, Hongoh R, Kobatake Y. J Membr Biol. 1979;49:105–121. doi: 10.1007/BF01868720. [DOI] [PubMed] [Google Scholar]
- 32.Davey GP, Tipton KF, Murphy MP. Biochem J. 1992;288:439–443. doi: 10.1042/bj2880439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chappell JB, Hansford RG. In: Subcellular Components: Preparation and Fractionation. Birnie GD, editor. London: Butterworths; 1972. [Google Scholar]
- 34.Palmer JW, Tandler B, Hoppel CL. J Biol Chem. 1977;252:8731–8739. [PubMed] [Google Scholar]
- 35.Duan JM, Karmazyn M. Mol Cell Biochem. 1989;90:47–56. doi: 10.1007/BF00225220. [DOI] [PubMed] [Google Scholar]
- 36.Martinez-Palma L, Pehar M, Cassina P, Peluffo H, Castellanos R, Anesetti G, Beckman JS, Barbeito L. Neurotox Res. 2003;5:399–406. doi: 10.1007/BF03033168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.