Abstract
Background
The role that human metapneumovirus (hMPV) plays in the etiology of upper respiratory tract infections (URIs) in children over a period of many years has not been evaluated previously.
Methods
By use of real-time reverse-transcriptase polymerase chain reaction, we retrospectively tested nasal wash (NW) specimens for hMPV that had been obtained from a cohort of 1532 infants and children with URIs who were prospectively followed for an average of 2.4 years during the period from 1982 to 2001. Virus genes were sequenced, and prospectively collected clinical data were analyzed.
Results
There were 2710 visits for URIs for which routine cultures did not reveal a viral etiology. Archival NW specimens from 2384 of these visits were available. hMPV RNA was detected in 118 (5%) of 2384 specimens. The mean age of the children with hMPV infection was 20 months, and 78% of illnesses occurred from December through May. Acute otitis media (AOM) was detected in 50% of these children. hMPV circulated each year, but the numbers of isolates detected varied by year. Reinfections with both homologous and heterologous strains occurred. Four distinct genetic lineages were present over the 20 years of surveillance, with several different lineages circulating during some seasons.
Conclusions
hMPV was detected in a substantial number of children with URIs and concomitant AOM.
Human metapneumovirus (hMPV) is a recently described paramyxovirus that is associated with acute respiratory tract disease [1–5]. Our previous work demonstrated that hMPV was a leading cause of lower respiratory tract infection (LRI) in previously healthy outpatient infants and children [6], and other reports have confirmed that hMPV infections are associated with hospitalizations of children [7–11]. However, long-term studies examining the frequency, seasonality, and clinical characteristics of hMPV infection in otherwise healthy children have not been reported. Using archival nasal wash (NW) specimens collected from a cohort of children presenting to our clinic for well and ill visits over a 20-year period, we used molecular diagnostic methods to determine rates of hMPV infection.
SUBJECTS, MATERIALS, AND METHODS
Study Design
The Vanderbilt Vaccine Clinic was established for the purpose of performing investigational vaccine trials and prospective surveillance for respiratory and enteric viruses in young children [6, 12–15]. Healthy full-term infants were enrolled at birth and followed until 5 years of age. Children with chronic diseases other than mild asthma were excluded from the clinic. Well and ill visits were conducted within the National Institutes of Health–supported General Clinical Research Center, with care provided by 4 pediatric infectious-diseases faculty and 2 nurse practitioners. During ill visits, signs and symptoms were recorded on a standardized clinical form and entered into a database. NW specimens were cultured for viral and bacterial respiratory pathogens.
Case Definitions
Upper respiratory tract infection (URI) included the following clinical syndromes: coryza, conjunctivitis, pharyngitis, otitis media (acute or serous), and stomatitis. NW specimens for viral culture were obtained from children with any of the following indications: URI accompanied by fever >38.3°C, acute otitis media (AOM), or evidence of LRI. Specimens were obtained by instilling sterile saline into one nostril and collecting the effluent. Specimens were kept on ice and processed for viral culture, as described elsewhere [6]. Aliquots of NW specimens were snap-frozen and stored at −70°C. Only specimens from patients who received a diagnosis of URI, without a diagnosis of LRI and with negative viral cultures for other pathogens, were selected for the present study. We chose to test only those specimens negative for other viruses, to investigate the role that hMPV plays as a sole pathogen in URI, since our previous work and studies by others have suggested that coinfection with hMPV in otherwise healthy children is uncommon [2, 4, 6, 7, 10, 11]. The 261 URI specimens tested in our previous study that focused on LRI [6] were not included in the present study.
Molecular Testing
RNA extraction
For the initial testing, RNA was extracted on a Qiagen BioRobot 9604 Workstation, using the QIAMP Viral RNA kit (Qiagen); 220 μL of NW specimen was used for extraction, and additional centrifugation steps were incorporated to remove all traces of ethanol. All specimens were thawed on ice and treated with Qiagen Protease by incubation for 30 min at 55°C before the addition of lysis buffer. Every 22 NW specimens were extracted concurrently with a positive hMPV control and a negative control (water). The eluted RNA was diluted 1:5 by adding 240 μL of RNA Storage Solution (Ambion) with 0.01 mg/mL yeast tRNA (Invitrogen). In comparison experiments in which RNA at varying dilutions was used, we found the dilution of 1:5 to be most sensitive for hMPV detection in this assay (data not shown). Subsequent re-extraction of NW specimens with a borderline hMPV-positive signal (defined below) was performed on an MWG Theonyx Liquid Handler, using the RNAqueous MAG-96 Viral RNA Isolation Kit (Ambion) to extract 100 μL of NW specimen.
Quantitative reverse-transcriptase polymerase chain reaction (RT-PCR)
The real-time RT-PCR assay was performed as described elsewhere [16]. The Stratagene Brilliant Plus Single-Step Quantitative RT-PCR Kit (Stratagene) was used for real-time RT-PCR analysis. A 50-μL reaction consisted of 5 μL of 10× RT-PCR buffer, 5 mmol/L magnesium chloride, dNTP mix, 0.3 μmol/L each forward and reverse primers, 0.2 μmol/L dual-labeled probe (FAM-BHQ3), 0.5 μL of 20 U/μL StrataScript reverse transcriptase, 0.5 μL of 5 U/μL SureStart Taq DNA polymerase, and 10 μL of template RNA. For assays run on the Stratagene M×4000 thermocycler (Stratagene), 0.15 μL of 10 μmol/L ROX reference dye was added to the core master mix. For assays run on the Applied Biosystems 7700 thermocycler, 1.5 μL of 10 μmol/L ROX reference dye was added.
Every 22 NW RNA specimens were amplified with 1 hMPV-positive RNA isolation control, a sentinel control (water extraction), a no-template control, and 10-fold serially diluted hMPV sublineage B1 N gene transcript standards. The 10-fold serially diluted RNA transcript standards represented a range of 5 ×10 7–5 ×101 copies/reaction. For each assay, all specimens, standards, and controls were amplified in triplicate. Cycling conditions will be furnished on request. Specimens were defined as positive if they had a cycle threshold lower than that of the 5 ×101 copy dilution of the RNA control standard for all 3 replicates. Specimens positive in only 1 or 2 replicates were considered to be borderline positive and were re-extracted and retested, as described above.
hMPV sequencing and lineage assignment
Full-length F and G genes were amplified, by nested RT-PCR, from specimens positive in the real-time RT-PCR for N gene. Sequencing reactions were performed using the ABI PRISM BigDye Terminator Kit (Applied Biosystems). The end products were processed, by capillary electrophoresis, using a 3730 DNA Analyzer (Applied Biosystems) and were analyzed using DNA Sequencing Analysis (Applied Biosystems) and Sequencher (Gene Codes). Sequences were aligned with published hMPV sequences from Genbank, and phylogenetic analysis was performed using the neighbor-joining method from MacVector (version 7.1; Accelrys).
Statistical Analysis
χ2 or Fisher’s exact tests were used, as appropriate, for contingency table analyses of symptoms and signs associated with different viruses. Comparisons of the mean age and mean duration of symptoms for the different viral etiologies were calculated using a 2-tailed t test that assumed unequal variance. Analyses were performed using SPSS (version 11; SPSS). For analysis, parainfluenza virus 1, 2, and 3 were considered together as “PIV,” and influenza A and B viruses were considered together as “influenza virus.” Prospectively collected clinical data were compared for children with hMPV infection and those with URI due to respiratory syncytial virus (RSV), PIV, or influenza virus.
RESULTS
Demographics of study population
During the study period of January 1982 through December 2001, 1532 children were followed in the clinic for a mean of 2.4 years each, for a total of 3677 child-years. Half of the study population was male, 53% were white, and 44% were African American. Twelve percent of the child-years were represented by infants <6 months old, 15% by children 6–12 months old, 21% by children 1–2 years old, 15% by children 2–3 years old, and 36% by children >3 years old.
During the 20-year study period, there were 8045 visits that resulted in a diagnosis of URI. There were 4870 visits for URIs during which specimens were obtained from 850 distinct children. The mean age of the children with URI specimens was 18 months, with a male:female ratio of 1.2:1.0. Routine viral culture had previously identified 267 specimens with RSV, 344 with PIV, 285 with influenza virus, 532 with adenovirus, 554 with enteroviruses including rhinovirus, 122 with herpes virus, and 56 with rotavirus. There were 2710 URI visits with specimens collected that had previously been negative for virus, and2384 specimens were available for testing; 118 (5%) of these specimens tested positive for hMPV on both initial and confirmatory testing, and clinical data on these specimens were included in the analysis.
Seasonality and incidence of URI due to hMPV
Although hMPV was detected year-round, 78% of hMPV infections occurred from December through May, with 38% in March and April alone (figure 1). The peak number of hMPV infections overlapped with but occurred later than the peaks of both RSV and influenza virus infection in all study years. The prevalence of hMPV ranged from 1% to 5% of all URIs for a given year (figure 2), which is similar to the variability for RSV year to year (3%–11%), but varied less than did those for either influenza virus (0.3%–13%) or PIV (2%–14%). In some years, hMPV was associated with more URIs than was RSV.
Figure 1.
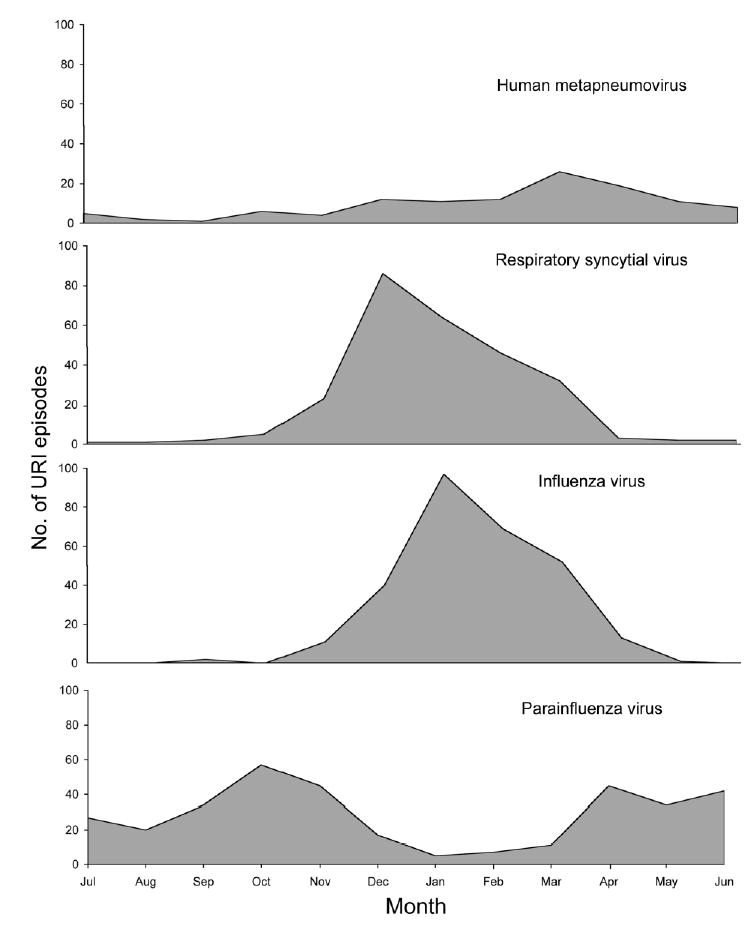
Epidemiologic pattern of upper respiratory tract infection (URI) with human metapneumovirus and other virus infections. Data are combined from 20 years of surveillance in the Vanderbilt Vaccine Clinic.
Figure 2.
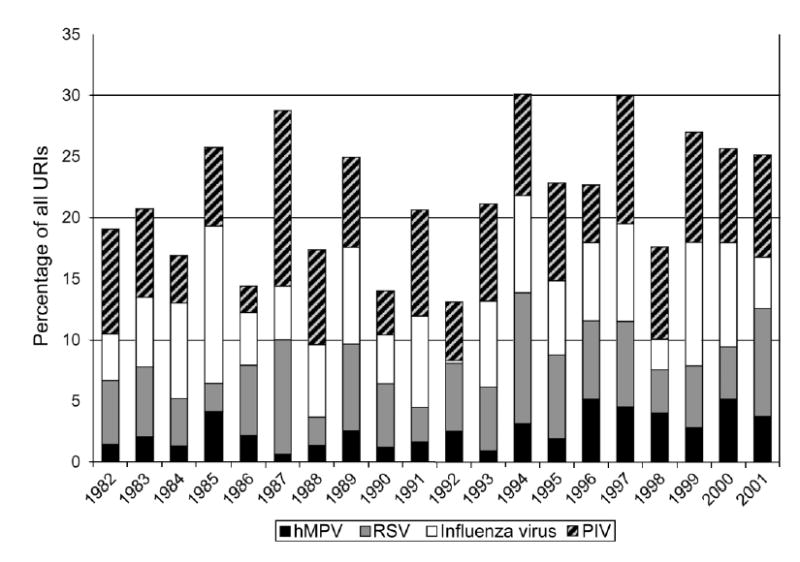
Annual rates of human metapneumovirus (hMPV) and other virus infections from 1962 to 2001 in the Vanderbilt Vaccine Clinic. The Y-axis shows the percentage of all upper respiratory tract infections (URIs) during the year accounted for by each virus. PIV, parainfluenza virus; RSV, respiratory syncytial virus.
Overall, during the 20-year study period, hMPV accounted for 5% of all URIs that occurred during March (figure 3). hMPV was present in warmer months, when little or no RSV and influenza virus was present. In the later months of the hMPV peak season, hMPV was associated with more disease than was RSV or influenza virus. During the peak months of February–May, hMPV accounted for 16% of all URIs over the 20-year period (figure 3).
Figure 3.
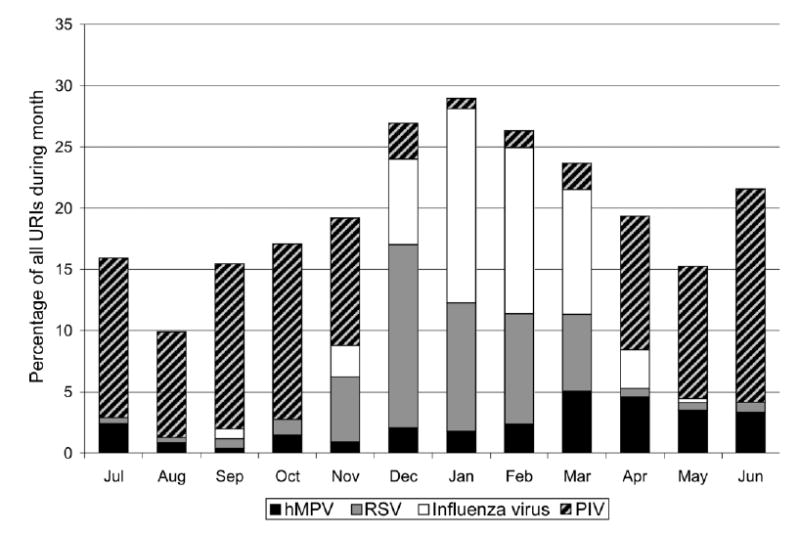
Cumulative monthly rates of human metapneumovirus (hMPV) and other virus infections from 1982 to 2001 in the Vanderbilt Vaccine Clinic. The Y-axis shows the percentage of all upper respiratory tract infections (URIs) during the month accounted for by each virus.
Clinical features of hMPV URI
The mean age of the children with URIs associated with hMPV was 20 months (range, 1–63 months), with a male:female ratio of 1.3:1.0. Neither of these differed significantly from those for URIs associated with other viral agents, except influenza virus (mean age, 27 months; P < .001). The mean duration of symptoms of hMPV infection before medical attention was sought was 2.7 days, which is similar to that of influenza virus infection (3.2 days; P = .2) but is shorter than that of RSV infection (4.3 days; P = .002) and PIV infection (3.8 days; P < .02). The signs and symptoms at presentation of children with hMPV infection are shown in table 1. Children with hMPV infection were less likely to be febrile than were children with influenza virus infection (54% vs. 85%; P < .001). Sixty-three percent of children with hMPV infection had some abnormality of their tympanic membranes, and 50% were prescribed antibiotics for concomitant AOM. The frequency of AOM associated with hMPV infection did not differ significantly from that associated with RSV, PIV, or influenza virus infection.
Table 1.
Clinical features at presentation of 118 children with human metapneumovirus upper respiratory tract infections.
| Symptom/sign | Children with symptom/sign, % |
|---|---|
| Fever | 54 |
| Coryza | 82 |
| Cough | 66 |
| Hoarseness | 8 |
| Otalgia | 31 |
| Rhinitis | 79 |
| Conjunctivitis | 3 |
| Pharyngitis | 44 |
| Abnormal tympanic membrane | 63 |
Circulation of hMPV lineages
Full-length F or G gene sequences were obtained for 76 isolates in the present study and were assigned to 1 of the proposed hMPV lineages (A1, A2, B1, or B2) on the basis of phylogenetic analysis. Of the 76 strains sequenced, 37 (49%) were of the B2 lineage, 21 (28%) were of the A2 lineage, 10 (13%) were of the B1 lineage, and 8 (11%) were of the A1 lineage. The occurrence of virus strains from different lineages during the study period is shown in figure 4. In some years, only a single lineage was present, whereas, in others, up to 3 different lineages circulated concurrently. The B2 lineage predominated in 1989–1991 and 1995–1996, whereas the A1 lineage was highly prevalent only in 1994, a year in which A2 and B1 lineages were also present. The B1 lineage was not detected before 1987 and was detected in a sporadic pattern. We did not detect differences in clinical presentation between viruses of different lineages.
Figure 4.
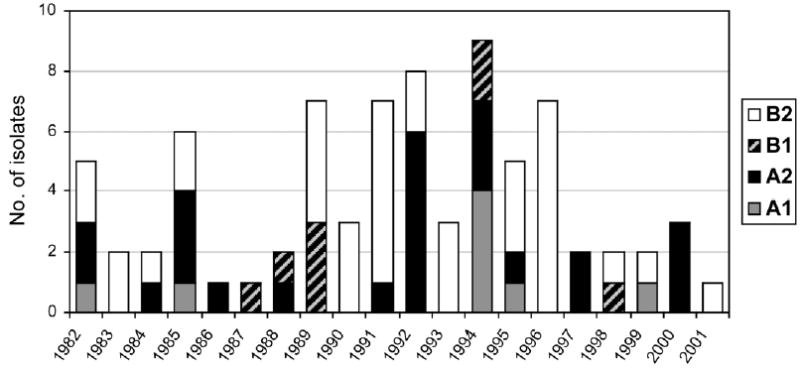
Rates, by year, of each genetic lineage of human metapneumovirus. Data are from 1982 to 2001 in the Vanderbilt Vaccine Clinic.
Recurrent infection with hMPV
We assessed the range of clinical presentations in individual children and the presence of recurrent hMPV infection within a population by combining data from the present study with data on patients with hMPV-associated LRIs in the same cohort from our previous study [6]. Since all of these children were followed for prolonged periods in the Vanderbilt Vaccine Clinic, it was possible to identify repeated infections in some distinct patients. Furthermore, we were able to determine the genetic lineage of viruses causing the initial and subsequent infections in some of these children, as is shown in table 2. One child (patient 1) had URI at 6 months of age caused by a B2 lineage virus and pneumonia at 33 months of age caused by an A2 lineage virus. Two other children (patients 2 and 3) had LRIs as infants and URIs at several years of age caused by viruses from the same major lineages (A or B). Patients with repeated URIs were infected with viruses from the same or different lineages.
Table 2.
Children in the Vanderbilt Vaccine Clinic for whom 2 human metapneumovirus (hMPV) infections were identified.
| Patient | Age, months | Diagnosis | hMPV lineage |
|---|---|---|---|
| 1 | 6 | URI | B2 |
| 33 | Pneumonia | A2 | |
| 2 | 4 | Bronchiolitis | A1 |
| 42 | URI | A2 | |
| 3 | 6 | Asthma | B2 |
| 38 | URI | B1 | |
| 4 | 6 | Bronchiolitis | B2 |
| 7 | URI | … | |
| 5 | 17 | URI | A2 |
| 42 | URI | A2 | |
| 6 | 12 | URI | B1 |
| 27 | URI | B1 | |
| 7 | 22 | URI | A2 |
| 30 | URI | B1 | |
| 8 | 38 | URI | B2 |
| 48 | URI | … | |
| 9 | 6 | URI | A1 |
| 14 | URI | … | |
| 10 | 26 | URI | … |
| 29 | URI | B1 | |
| 11 | 11 | URI | … |
| 30 | URI | A2 | |
| 12 | 12 | URI | … |
| 40 | URI | … | |
| 13 | 9 | URI | … |
| 24 | URI | … |
NOTE. …, Lineage could not be determined, because of inability to amplify F or G genes by reverse-transcriptase polymerase chain reaction; URI, upper respiratory tract infection.
DISCUSSION
We retrospectively tested specimens, by RT-PCR, that had been prospectively collected from otherwise healthy children with URI symptoms who were receiving comprehensive care in our clinic over a period of 20 years. We detected hMPV in 118 (5%) of 2384 specimens from children with URIs who had not received a prior virologic diagnosis, not including specimens from our previous study that focused on LRI [6]. These results suggest an overall prevalence of 3% in this cohort, which had 2710 URI episodes. This frequency was comparable to the prevalence in the same cohort of children with culture-positive URI episodes, for RSV (6%), influenza virus (6%), and PIV (7%). The role that hMPV plays in URI in otherwise healthy children appears to be less prominent than the role it plays in LRI in most pediatric populations studied, including, in our previous study, a population very similar to the cohort in the present study [2–11]. The decreased relative contribution of hMPV to URI is likely due, in part, to the high frequency of other viruses we identified in URI, such as adenovirus (11%) and picornaviruses including rhinovirus (10%). These viruses (which were not frequently associated with LRI in similar studies in this cohort) thus proportionally decreased the contribution of hMPV, as well as RSV, PIV, and influenza virus, which are also major causes of LRI in children. In addition, other respiratory viruses were identified shortly after the NW specimens were obtained, whereas hMPV was analyzed retrospectively—for some specimens, up to 20 years afterward. Hence, underestimation is likely due to the long-term storage at −70°C and the repeated freeze-thaw cycles of the NW specimens. Furthermore, we applied stringent criteria in the laboratory assays to define positive specimens; thus, we potentially underestimated the true prevalence of hMPV in URI. We elected to test only specimens that were previously negative for other viruses; thus, we potentially underestimated coinfections with hMPV and other viruses. However, we previously tested LRI specimens that were positive for other viruses and detected hMPV coinfection in only 4% of them [6]. Furthermore, other studies of hMPV in children have found low rates of coinfection [2, 4, 7, 10, 11], and hMPV is rarely detected in asymptomatic persons [4, 6].
We detected hMPV less frequently in URI specimens in the present study than in our previous study of hMPV-associated LRI, in which a much smaller number of URI specimens were tested [6]. There are several possible reasons for this discrepancy. In our previous study, all specimens were thawed and immediately extracted manually in small batches. The number of specimens tested in the present study required different methods for high-throughput testing. In addition, to protect patient confidentiality, specimens were thawed and realiquoted in batches of 72, to remove patient-specific identifiers before shipping to the diagnostic laboratory. Repeated freeze-thaw cycles have been shown to degrade viral RNA [17–20], and this may have led to decreased amounts of RNA in the specimens, although precautions were taken to preserve the integrity of the specimens at collection and during storage. In the present study, the RNA was extracted by automated methods. Some studies have suggested that automated nucleic acid–extraction methods are less efficient than manual methods [21–23], whereas others have reported similar sensitivities between the 2 methods [24, 25]. Although the URI specimens tested with the LRI specimens in our previous pilot-scale study were selected randomly (using computer methods) from all URI specimens, there were more specimens from recent years in that subset (data not shown), and this factor may have affected RNA quality as well.
The majority of hMPV infections occurred during the late winter and early spring months, overlapping the peak season for both RSV and influenza virus infection in the Nashville, Tennessee, area. However, the greatest number of hMPV infections occurred ~2 months after the peaks of RSV and influenza virus infection. This pattern is identical to the seasonality we observed in our earlier study of hMPV and LRI at this site [6], thus confirming the seasonal pattern of hMPV infections in children over an extended number of years. We detected hMPV year-round, similar to RSV in the Nashville area, as is shown in figure 3. Other investigators have also reported RSV infections during summer months in the southern United States [38–40], which is likely a regional phenomenon. During the seasonal peaks, hMPV accounted for as much as 16% of all URIs during the study period. Although hMPV causes annual community-wide epidemics of respiratory tract disease, the rates differ somewhat from year to year, from a minimum of 1% to a maximum of 5%. These findings are similar to the variability we reported for hMPV-associated LRI [6]. Other studies examining only a few seasons have also demonstrated variability [4, 7, 8, 10].
Children with URIs due to hMPV presented with typical symptoms, including rhinorrhea, cough, and fever. They were significantly less likely to have fever than were children with influenza virus infection. However, these findings could not be used to definitively identify the infectious agent, because of the large clinical overlap among the respiratory pathogens [2–4, 6–11]. Effective antiviral therapy is available for influenza viruses, and, thus, accurate diagnosis of these patients might alter patient management. Our data confirm, in a large prospective cohort, that rapid clinical diagnostic assays will be needed to distinguish hMPV infection from RSV, PIV, and influenza virus infections.
Fifty percent of hMPV-infected children with URIs were concomitantly diagnosed with AOM and prescribed oral antibiotics. Whereas the presence of AOM was one of the criteria for obtaining a NW specimen and thus may have caused a selection bias for AOM, the frequency of AOM associated with hMPV did not differ from that for other viruses in the present study. These data show that hMPV—like RSV, PIV, and influenza virus—is frequently associated with AOM, which is the most common reason for the prescription of oral antibiotics to children [26, 30]. Studies have shown that influenza vaccine reduces the incidence of AOM in children [27–29]. The additional burden of disease due to hMPV-associated AOM should be considered in evaluation of the potential benefits of a vaccine.
There are 4 distinct genetic lineages of hMPV that have been described, which are provisionally designated as A1, A2, B1, and B2 lineages [31]. We have shown here that hMPV has circulated in the community for at least 20 years and that all 4 genetic lineages of hMPV have persisted during that time. More than 1 lineage was present concurrently during some seasons, whereas a single lineage dominated others. Interestingly, the hMPV B1 lineage was not detected in any specimen before 1987, although we tested specimens from 1982 onward. We cannot tell from these data whether the B1 lineage has arisen recently or circulates only every few years. That we did not detect B1 viruses between 1990 and 1993 or between 1995 and 1997 suggests the latter. hMPV strains from the B2 lineage were most common in the present study and accounted for almost one-half of the 76 isolates sequenced. This finding is in contrast with the findings of the majority of published hMPV epidemiology studies, in which overall A1 and A2 lineages account for approximately three-quarters of all disease [2–11]. However, many of these studies used RT-PCR primers that subsequently have been shown to be less sensitive for detection of hMPV B lineages, whereas the RT-PCR primers and probe used in the present study could detect all 4 lineages with equal sensitivity [16]. Furthermore, most published reports included only 1–3 years of study, which is not adequate for determination of long-term patterns of hMPV circulation. Our data show that both major lineages of hMPV are important in clinical disease. This finding may have important implications for the development of candidate vaccines or prophylactic monoclonal antibodies similar to the RSV antibody palivizumab.
There are limited data in animals, suggesting that there is a degree of cross-protective immunity between viruses representing major hMPV lineages, but only a few virus strains have been tested [31–34]. Our data suggest that previous hMPV infection does not protect against repeated URI with either homologous or heterologous strains. Although we were not able to identify the lineage for all strains, some patients had recurrent URIs due to hMPV within weeks or months. This pattern is consistent with studies of RSV infection in humans and animal models that showed that upper airway protection is limited and transient [35–37].
It is difficult to draw conclusions about the role that hMPV plays in LRI and subsequent URI. Of the 3 patients for whom both lineages of virus were determined, 1 was reinfected with a heterologous strain (patient 1), and 2 were reinfected with a virus of the same major lineage (patients 2 and 3). However, the prolonged period (>2 years) between the initial illness and subsequent infections could be long enough for the waning of protective immunity. Nonetheless, although most hMPV-associated LRIs occur during the first year of life [2, 3, 6–11], we detected URIs due to hMPV infection more frequently later during childhood. These data are consistent with the hypothesis that LRI was the primary presentation during infancy but that immunity induced by primary infection ameliorated subsequent infections and limited hMPV replication to the upper respiratory tract.
In summary, we have found that hMPV was associated with a substantial number of URI episodes in otherwise healthy outpatient children who were followed over a 20-year period. hMPV was detected during every year, although the prevalence varied season to season. All 4 genetic lineages were present, and often viruses from >1 lineage circulated during a single year. Children with URIs due to hMPV had clinical illnesses similar to those associated with other common viruses, including frequent AOM. Repeated infections of both the upper and lower respiratory tract occurred. These data provide a framework for further directed studies of strain-specific immunity and reinfection that have implications for development of vaccines and therapeutics. Furthermore, the substantial burden of disease due to hMPV suggests that a vaccine might have significant health and socioeconomic benefits.
Footnotes
Presented in part: Pediatric Infectious Disease Society/St. Jude Pediatric Microbial Research Conference, Memphis, 20 February 2004 (abstract 13); Pediatric Academic Societies Annual Meeting, San Francisco, 2 May 2004 (abstract 1899).
Potential conflicts of interest: J.V.W. received a onetime consulting fee from MedImmune in 2003 for attending a respiratory syncytial virus symposium and a onetime speaking fee from MedImmune for speaking at the Third Annual North Carolina Congress on Respiratory Viruses; K.M.E. receives grant support from MedImmune for influenza-vaccine studies; C.K.W., C.-F.Y., L.D.L., M.C., J.B.B., D.C., J.D.Q., and R.R.S. are employees of MedImmune; J.E.C. Jr. is a member of the Scientific Advisory Board of MedImmune and received research support for this study.
Financial support: National Institutes of Health (NIH; grant R03 AI054790 to J.V.W.); MedImmune (research grant to J.E.C.); National Institute of Allergy and Infectious Diseases/NIH Respiratory Pathogens Research Unit (grant NOI-AI-65298 to the Vanderbilt Vaccine Clinic); General Clinical Research Center (award RR 00095 to the Vanderbilt Vaccine Clinic).
References
- 1.Van Den Hoogen, DeJong JC, Groen J, et al. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–24. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peiris JS, Tang WH, Chan KH, et al. Children with respiratory disease associated with metapneumovirus in Hong Kong. Emerg Infect Dis. 2003;9:628–33. doi: 10.3201/eid0906.030009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boivin G, De Serres G, Cote S, et al. Human metapneumovirus infections in hospitalized children. Emerg Infect Dis. 2003;9:634–40. doi: 10.3201/eid0906.030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van den Hoogen BG, van Doornum GJ, Fockens JC, et al. Prevalence and clinical symptoms of human metapneumovirus infection in hospitalized patients. J Infect Dis. 2003;188:1571–7. doi: 10.1086/379200. [DOI] [PubMed] [Google Scholar]
- 5.Falsey AR, Erdman D, Anderson LJ, Walsh EE. Human metapneumovirus infections in young and elderly adults. J Infect Dis. 2003;187:785–90. doi: 10.1086/367901. [DOI] [PubMed] [Google Scholar]
- 6.Williams JV, Harris PA, Tollefson SJ, et al. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N Engl J Med. 2004;350:443–50. doi: 10.1056/NEJMoa025472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuevas LE, Nasser AM, Dove W, Gurgel RQ, Greensill J, Hart CA. Human metapneumovirus and respiratory syncytial virus, Brazil. Emerg Infect Dis. 2003;9:1626–8. doi: 10.3201/eid0912.030522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galiano M, Videla C, Puch SS, Martinez A, Echavarria M, Carballal G. Evidence of human metapneumovirus in children in Argentina. J Med Virol. 2004;72:299–303. doi: 10.1002/jmv.10536. [DOI] [PubMed] [Google Scholar]
- 9.Esper F, Martinello RA, Boucher D, et al. A 1-year experience with human metapneumovirus in children aged <5 years. J Infect Dis. 2004;189:1388–96. doi: 10.1086/382482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McAdam AJ, Hasenbein ME, Feldman HA, et al. Human metapneumovirus in children tested at a tertiary-care hospital. J Infect Dis. 2004;190:20–6. doi: 10.1086/421120. [DOI] [PubMed] [Google Scholar]
- 11.Mullins JA, Erdman DD, Weinberg GA, et al. Human metapneumovirus infection among children hospitalized with acute respiratory illness. Emerg Infect Dis. 2004;10:700–5. doi: 10.3201/eid1004.030555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards KM, Thompson J, Paolini J, Wright PF. Adenovirus infections in young children. Pediatrics. 1985;76:420–4. [PubMed] [Google Scholar]
- 13.Reed G, Jewett PH, Thompson J, Tollefson S, Wright PF. Epidemiology and clinical impact of parainfluenza virus infections in otherwise healthy infants and young children <5 years old. J Infect Dis. 1997;175:807–13. doi: 10.1086/513975. [DOI] [PubMed] [Google Scholar]
- 14.Fisher RG, Gruber WC, Edwards KM, et al. Twenty years of outpatient respiratory syncytial virus infection: a framework for vaccine efficacy trials. Pediatrics. 1997;99:E7. doi: 10.1542/peds.99.2.e7. [DOI] [PubMed] [Google Scholar]
- 15.Neuzil KM, Zhu Y, Griffin MR, et al. Burden of interpandemic influenza in children younger than 5 years: a 25-year prospective study. J Infect Dis. 2002;185:147–52. doi: 10.1086/338363. [DOI] [PubMed] [Google Scholar]
- 16.Maertzdorf J, Wang CK, Brown JB, et al. Real-time reverse transcriptase PCR assay for detection of human metapneumoviruses from all known genetic lineages. J Clin Microbiol. 2004;42:981–6. doi: 10.1128/JCM.42.3.981-986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glimaker M, Johansson B, Olcen P, Ehrnst A, Forsgren M. Detection of enteroviral RNA by polymerase chain reaction in cerebrospinal fluid from patients with aseptic meningitis. Scand J Infect Dis. 1993;25:547–57. doi: 10.3109/00365549309008542. [DOI] [PubMed] [Google Scholar]
- 18.Fong TL, Charboneau F, Valinluck B, Govindarajan S. The stability of serum hepatitis C viral RNA in various handling and storage conditions. Arch Pathol Lab Med. 1993;117:150–1. [PubMed] [Google Scholar]
- 19.Matsuo S, Sugiyama T, Okuyama T, et al. Preservation of pathological tissue specimens by freeze-drying for immunohistochemical staining and various molecular biological analyses. Pathol Int. 1999;49:383–90. doi: 10.1046/j.1440-1827.1999.00887.x. [DOI] [PubMed] [Google Scholar]
- 20.Ward CL, Dempsey MH, Ring CJ, et al. Design and performance testing of quantitative real time PCR assays for influenza A and B viral load measurement. J Clin Virol. 2004;29:179–88. doi: 10.1016/S1386-6532(03)00122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knepp JH, Geahr MA, Forman MS, Valsamakis A. Comparison of automated and manual nucleic acid extraction methods for detection of enterovirus RNA. J Clin Microbiol. 2003;41:3532–6. doi: 10.1128/JCM.41.8.3532-3536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forman MS, Valsamakis A. Verification of an assay for quantification of hepatitis C virus RNA by use of an analyte-specific reagent and two different extraction methods. J Clin Microbiol. 2004;42:3581–8. doi: 10.1128/JCM.42.8.3581-3588.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller Z, Stelzl E, Bozic M, Haas J, Marth E, Kessler HH. Evaluation of automated sample preparation and quantitative PCR LCx assay for determination of human immunodeficiency virus type 1 RNA. J Clin Microbiol. 2004;42:1439–43. doi: 10.1128/JCM.42.4.1439-1443.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyachi H, Masukawa A, Asai S, et al. Quantitative assay of hepatitis C virus RNA using an automated extraction system for specific capture with probes and paramagnetic particle separation. J Clin Microbiol. 2003;41:572–5. doi: 10.1128/JCM.41.2.572-575.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi PY, Kauffman EB, Ren P, et al. High-throughput detection of West Nile virus RNA. J Clin Microbiol. 2001;39:1264–71. doi: 10.1128/JCM.39.4.1264-1271.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halasa NB, Griffin MR, Zhu Y, Edwards KM. Differences in antibiotic prescribing patterns for children younger than five years in the three major outpatient settings. J Pediatr. 2004;144:200–5. doi: 10.1016/j.jpeds.2003.10.053. [DOI] [PubMed] [Google Scholar]
- 27.Clements DA, Langdon L, Bland C, Walter E. Influenza A vaccine decreases the incidence of otitis media in 6- to 30-month-old children in day care. Arch Pediatr Adolesc Med. 1995;149:1113–7. doi: 10.1001/archpedi.1995.02170230067009. [DOI] [PubMed] [Google Scholar]
- 28.Heikkinen T, Ruuskanen O, Waris M, et al. Influenza vaccination in the prevention of acute otitis media in children. Am J Dis Child. 1991;145:445–8. doi: 10.1001/archpedi.1991.02160040103017. [DOI] [PubMed] [Google Scholar]
- 29.Belshe RB, Mendelman PM, Treanor J, et al. The efficacy of live attenuated, cold-adapted, trivalent, intranasal influenza virus vaccine in children. N Engl J Med. 1998;338:1405–12. doi: 10.1056/NEJM199805143382002. [DOI] [PubMed] [Google Scholar]
- 30.Heikkinen T, Thint M, Chonmaitree T. Prevalence of various respiratory viruses in the middle ear during acute otitis media. N Engl J Med. 1999;340:260–4. doi: 10.1056/NEJM199901283400402. [DOI] [PubMed] [Google Scholar]
- 31.van den Hoogen BG, Herfst S, Sprong L, et al. Antigenic and genetic variability of human metapneumoviruses. Emerg Infect Dis. 2004;10:658–66. doi: 10.3201/eid1004.030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacPhail M, Schickli JH, Tang RS, et al. Identification of small-animal and primate models for evaluation of vaccine candidates for human metapneumovirus (hMPV) and implications for hMPV vaccine design. J Gen Virol. 2004;85:1655–63. doi: 10.1099/vir.0.79805-0. [DOI] [PubMed] [Google Scholar]
- 33.Skiadopoulos MH, Biacchesi S, Buchholz UJ, et al. The two major human metapneumovirus genetic lineages are highly related antigenically, and the fusion (F) protein is a major contributor to this antigenic relatedness. J Virol. 2004;78:6927–37. doi: 10.1128/JVI.78.13.6927-6937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang RS, Mahmood K, Macphail M, et al. A host-range restricted parainfluenza virus type 3 (PIV3) expressing the human metapneumovirus (hMPV) fusion protein elicits protective immunity in African green monkeys. Vaccine. 2005;23:1657–67. doi: 10.1016/j.vaccine.2004.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hall CB, Walsh EE, Long CE, Schnabel KC. Immunity to and frequency of reinfection with respiratory syncytial virus. J Infect Dis. 1991;163:693–8. doi: 10.1093/infdis/163.4.693. [DOI] [PubMed] [Google Scholar]
- 36.Prince GA, Horswood RL, Chanock RM. Quantitative aspects of passive immunity to respiratory syncytial virus infection in infant cotton rats. J Virol. 1985;55:517–20. doi: 10.1128/jvi.55.3.517-520.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sami IR, Piazza FM, Johnson SA, et al. Systemic immunoprophylaxis of nasal respiratory syncytial virus infection in cotton rats. J Infect Dis. 1995;171:440–3. doi: 10.1093/infdis/171.2.440. [DOI] [PubMed] [Google Scholar]
- 38.Washburne JF, Bocchini JA, Jr, Jamison RM. Summertime respiratory syncytial virus infection: epidemiology and clinical manifestations. South Med J. 1992;85:579–83. doi: 10.1097/00007611-199206000-00004. [DOI] [PubMed] [Google Scholar]
- 39.Halstead DC, Jenkins SG. Continuous non-seasonal epidemic of respiratory syncytial virus infection in the southeast United States. South Med J. 1998;91:433–6. doi: 10.1097/00007611-199805000-00004. [DOI] [PubMed] [Google Scholar]
- 40.Centers for Disease Control and Prevention. Respiratory syncytial virus activity—United States, 2003–2004. MMWR Morb Mortal Wkly Rep. 2004;53:1159–60. [PubMed] [Google Scholar]


