Abstract
The recognition of microbial components by Toll-like receptors (TLRs) initiates signal transduction pathways, which trigger the expression of a series of target genes. It has been reported that TLR signaling is enhanced by cytokines such as IFN-γ, but the mechanisms underlying this enhancement remain unclear. The MyD88 adaptor, which is essential for signaling by many TLRs, recruits members of the IFN regulatory factor (IRF) family of transcription factors, such as IRF5 and IRF7, to evoke the activation of TLR target genes. In this study we demonstrate that IRF1, which is induced by IFN-γ, also interacts with and is activated by MyD88 upon TLR activation. We provide evidence that MyD88-associated IRF1 migrates into the nucleus more efficiently than non-MyD88-associated IRF1 and that this IRF1 selectively participates in the TLR-dependent gene induction program. The critical role of MyD88-dependent “IRF1 licensing” is underscored by the observation that the induction of a specific gene subset downstream of the TLR–MyD88 pathway, such as IFN-β, inducible NO synthase, and IL-12p35, are impaired in Irf1-deficient cells. Thus, our present study places IRF1 as an additional member participating in MyD88 signaling and provides a mechanistic insight into the enhancement of the TLR-dependent gene induction program by IFN-γ.
Keywords: inducible NO synthase, IL-12, NF-κB, synergy, Kaede
The IFN regulatory factor (IRF) family of transcription factors comprises nine members (1). The family members all share a homology in their N-terminal DNA-binding domain and recognize the consensus DNA sequence, termed the IFN-stimulated response element. The prevailing notion is that the IRF system governs a broad spectrum of cellular responses in immunity (1). Indeed, IRFs have recently gained much attention as key regulators of the Toll-like receptor (TLR)-dependent gene induction program. IRF3 is activated by TLR4 and TLR3 signaling and plays an essential role in IFN-β and chemokine gene induction (2). IRF5 and IRF7 directly interact with MyD88, an essential adaptor of TLRs, and regulate the TLR-dependent induction of proinflammatory cytokines and type I IFNs, respectively (3–6). IRF4 also interacts with MyD88 and acts as a negative regulator of MyD88–IRF5-mediated gene induction (7). IRF8 interacts with TNF-associated factor 6, a ubiquitin ligase involved in TLR signaling, and regulates the production of inflammatory mediators (8).
IRF1 was initially identified as a regulator of type I IFN gene transcription (9). Recently it has been shown that IRF1 activates the expression of a larger panel of genes, such as inducible NO synthase (iNOS) and IL-12p35, which are important for mounting an effective innate and adaptive immunity against pathogens (10, 11). In this context it is notable that IRF1 expression is efficiently induced by IFN-γ (12, 13) and IFN-γ enhances TLR signaling (13, 14), suggesting a function of IRF1 that links these two signaling events. Consistent with this notion are the observations that IRF1 gene-deficient mice (Irf1−/− mice) are susceptible to Listeria monocytogenes, Toxoplasma gondii, and Mycobacterium tuberculosis infections, and this susceptibility is similar to that of mice deficient in IFN-γ or TLR signaling molecules (10, 15–20).
In the present study we investigated how IFN-γ-induced IRF1 contributes to TLR-mediated signaling. We demonstrate that IRF1 forms a complex with MyD88, similar to the case of IRF4, IRF5, and IRF7. We also provide evidence that IRF1 induced by IFN-γ is activated by MyD88, which we refer to as “licensing,” and migrates rapidly into the nucleus to mediate an efficient induction of IFN-β, iNOS, and IL-12p35. Our study therefore revealed that IRF1 is a previously unidentified member of the multimolecular complex organized via MyD88 and that the IRF1 licensing by the TLR–MyD88 pathway constitutes a critical mechanism underlying the cooperation between IFN-γ and TLR signaling events.
Results
IRF1 Directly Interacts with MyD88.
We first examined the subcellular localization of IRF1 and MyD88. We expressed IRF1 tagged with YFP (IRF1YFP) together with MyD88 tagged with cyan fluorescent protein (CFP; MyD88CFP) in HEK293T cells and subjected these cells to microscopic analysis. As shown in Fig. 1a, IRF1YFP was predominantly expressed in the nucleus. Interestingly, however, a substantial fraction of IRF1YFP was expressed in the cytoplasm, and it showed a granular structure and colocalized with MyD88CFP (Fig. 1a). To examine the direct interaction between IRF1 and MyD88, these cells were also subjected to FRET analysis. As shown in Fig. 1 b and c, FRETC images revealed a strong energy transfer from MyD88CFP to IRF1YFP, similar to that from MyD88CFP to IRF5YFP or IRF7YFP. In contrast, FRET was not observed in IRF2YFP- and MyD88CFP-expressing cells (Fig. 1c). We also performed an immunoprecipitation assay using HEK293T cells expressing FLAG-tagged MyD88 and HA-tagged IRF1 (Fig. 1d). Consistent with the FRET analysis, IRF1, but not IRF2, was coimmunoprecipitated with MyD88 (Fig. 1d), indicating that MyD88 and IRF1 form a cytoplasmic complex and are in direct contact with each other.
Fig. 1.
Interaction of IRF1 with MyD88. (a) HEK293T cells were transfected with expression plasmids for IRF1YFP and MyD88CFP. Representative confocal images are shown. The arrows indicate colocalization of IRF1 with MyD88. (b and c) YFP and CFP images of HEK293T cells coexpressing IRFsYFP with MyD88CFP were obtained by using a fluorescence microscope. FRETC values were calculated and demonstrated by using a pseudocolor image (b) or FRETC/CFP values (c). (d) Lysates prepared from HEK293T cells transiently transfected with a combination of FLAG–MyD88 and HA–IRF1 or HA–IRF2 were immunoprecipitated (IP) with the anti-FLAG antibody and subjected to immunoblot (IB) analysis using the anti-HA or anti-FLAG antibody, as indicated. WCL, whole-cell extracts. (e) A schematic diagram of MyD88 truncated mutants is shown. Each FLAG–MyD88 mutant was coexpressed with HA–IRF1 in HEK293T cells and subjected to coimmunoprecipitation analysis.
We next examined the region of MyD88 that is responsible for its interaction with IRF1 using deletion mutants of MyD88, each tagged with FLAG (Fig. 1e) (3). Each FLAG–MyD88 mutant was coexpressed with HA–IRF1 in HEK293T cells and subjected to an immunoprecipitation assay. As shown in Fig. 1e, FLAG–MyD88(Δ173–296) and FLAG–MyD88(Δ1–151) interacted with HA–IRF1, but FLAG–MyD88(Δ134–296) and FLAG–MyD88(Δ60–296) failed to interact, suggesting that IRF1 interacts with the middle region of MyD88 (the intermediary domain and part of the Toll/IL-1 receptor domain), which is a similar interaction region for IRF5 (6, 7).
MyD88-Dependent Activation of IRF1.
The finding that MyD88 interacts with IRF1 prompted us to determine whether the function of IRF1 is influenced by MyD88 signaling. Therefore, we tested the ability of IRF1 to activate an IRF-binding-site-containing promoter-driven reporter gene (p125-luc) (21) in cells expressing MyD88. As shown in Fig. 2a, whereas the expression of IRF1 alone in HEK293T cells caused a marginal activation of the p125-luc reporter gene, the coexpression of IRF1 and MyD88 strongly activated the reporter gene. In contrast, the MyD88-dependent activation of NF-κB measured by using the pNF-κB-luc reporter gene was not affected by the coexpression of IRF1 (Fig. 2b).
Fig. 2.
MyD88-dependent activation of IRF1. (a and b) HEK293T cells were transfected with p125-luc (a) or pNF-κB-luc reporter (b) plasmids and expression vectors of the indicated combination of MyD88 (50 ng) and IRF1 (50 ng). After 24 h of transfection, cells were harvested and luciferase activity was measured. (c) Green (Kaede green), red (Kaede red), and blue (CFP) fluorescence images of HeLa cells expressing IRF1Kaede alone (Upper) or IRF1Kaede with MyD88CFP (Lower) were first obtained. A spot in the cytosolic portion (indicated by a red circle) of a HeLa cell was then pulsed with 405-nm light for 5 s, and red fluorescence images were collected every 10 s. N, nucleus. (d) Regions of interest encompassing the nucleus and the entire cell expressing IRF1Kaede alone (blue horizontal line) or IRF1Kaede with MyD88CFP (red horizontal line) were analyzed for total fluorescence intensity, and the percentage of IRF1Kaede residing in the nucleus was determined by using the formula described in Materials and Methods. Twelve cells were examined, and standard deviations are shown as vertical bars. (e) HEK293T cells were transfected with an expression vector of HA–IRF1 with or without FLAG–MyD88, after which HA–IRF1 was immunoprecipitated (IP) and subjected to two-dimensional gel electrophoresis and immunoblot (IB) analysis using the anti-HA antibody. IEF, isoelectric focusing.
To gain further insight into the mechanism underlying the MyD88-dependent activation of IRF1, we used a recently developed visualization technique using the Kaede protein (22). Briefly, Kaede is a recently cloned fluorescent protein that emits bright green fluorescence after its synthesis, which changes efficiently to a stable red fluorescence upon irradiation with 405-nm light, thereby providing a simple and powerful technique for the regional optical labeling of target molecules. We expressed IRF1Kaede with or without MyD88CFP in HeLa cells (Fig. 2 c and d). When a spot in the cytoplasmic portion (indicated by a red circle in Fig. 2c) of HeLa cells expressing IRF1Kaede alone was pulsed with 405-nm light, IRF1Kaede photoconverted to red was distributed mainly in the cytoplasm and slowly migrated into the nucleus (Fig. 2 c Upper and d, blue horizontal line). In contrast, IRF1Kaede bound to MyD88 showed a rapid migration to the nucleus after irradiation (Fig. 2c Lower and d, red horizontal line). These results further support the notion that IRF1 forms a complex with and is activated (or licensed) by MyD88 so as to undergo nuclear translocation.
To further assess the effect of interaction between MyD88 and IRF1 on the status of IRF1, we compared the mobility of IRF1 from MyD88-expressing HEK293T cells with that from non-MyD88-expressing HEK293T cells in a two-dimensional gel. As shown in Fig. 2e, the additional acidic charged subspecies of IRF1 were observed for cells coexpressing MyD88, presumably reflecting the MyD88 signaling-dependent phosphorylation of IRF1.
IRF1 Contributes to Ligand- and Cell-Type-Specific IFN-β Induction.
To further examine the function of IRF1 in the TLR–MyD88 signaling pathway we examined granulocyte/macrophage colony-stimulating factor-cultured bone marrow-derived dendritic cells (GM-DCs) from Irf1−/− mice for the induction of IFN-β mRNA by activating TLR9 using B- or K-type CpG-DNA (CpG-B) (23). As shown in Fig. 3a, IFN-β mRNA induction, constantly detected in this dendritic cell (DC) population albeit at much lower levels than that in plasmacytoid DCs (pDCs) stimulated with A- or D-type CpG-DNA (CpG-A) (24), was impaired in Irf1−/− GM-DCs. Interestingly, the induction was less affected in DCs from Irf3–Irf7 double-deficient (Irf3,7−/−) mice. In contrast, the induction of IFN-β mRNA in Irf1−/− GM-DCs upon stimulation with LPS or poly(I:C) was similar to that in wild-type GM-DCs, whereas it was severely impaired in Irf3,7−/− GM-DCs, indicating the utilization of distinct transcription factors by TLRs and their ligands in the induction of the same gene (Fig. 3a). Wild-type or mutant GM-DCs stimulated with LPS or CpG-B were also subjected to flow cytometric analysis of CD40 induction. Consistent with previous reports indicating that type I IFNs are essential for the maturation of DCs (25), the up-regulation of CD40 expression on Irf1−/− GM-DCs was suppressed in response to CpG-B as compared with wild-type cells, whereas it was normal in response to LPS (Fig. 3b). Conversely, in Irf3−/− GM-DCs the induction of CD40 was suppressed in response to LPS, whereas it was normal in response to CpG-B. These results indicate the important role of IRF1 in the MyD88-dependent signaling pathway induced by physiological stimuli.
Fig. 3.
IRF1-dependent IFN-β induction in GM-DCs. (a) GM-DCs derived from wild-type or mutant mice were stimulated with 0.3 μM CpG-B, 100 ng/ml LPS, or 100 μg/ml poly(I:C) for the indicated periods. Total RNA was prepared and subjected to real-time RT-PCR analysis. (b) Wild-type or mutant GM-DCs were unstimulated (gray) or stimulated with 100 ng/ml LPS (blue) or 0.3 μM CpG-B (red) for 18 h and analyzed for CD40 expression by flow cytometry. (c) Splenic pDCs from wild-type or mutant mice were stimulated with CpG-A (3 μM) and subjected to real-time RT-PCR analysis using IFN-α1 or IFN-β primers.
It is well established that pDCs are highly capable of producing type I IFNs upon stimulation with CpG-A (4). This type of induction depends completely on MyD88. To further assess the contribution of IRF1 to MyD88-dependent IFN induction, splenic CD11cintermediateB220+ pDCs were purified from wild-type, Irf1−/−, and Irf7−/− mice by using a cell sorter (>95% purity), stimulated with CpG-A, and analyzed for IFN-α and IFN-β induction by real-time RT-PCR. Consistent with a previous report (4), the induction of IFN-α and IFN-β mRNAs in response to stimulation with CpG-A was completely abolished in Irf7−/− pDCs, whereas it was normal in Irf1−/− pDCs (Fig. 3c). These results demonstrate further the cell-type-specific and ligand-specific involvement of IRF1 in the MyD88-dependent signaling pathway.
IRF1 Involved in Cooperation Between IFN-γ and TLR Signaling.
TLR signaling cooperates with IFN-γ signaling for efficient induction of certain genes, including IL-12p35 and iNOS (11, 13, 14). In fact, the induction of these genes is likely mediated at least in part by the induction of IRF1 via IFN-γ produced upon TLR stimulation. We then examined the importance of IRF1–MyD88 interaction in the context of this cooperation (synergism). GM-DCs were pretreated with 200 units/ml IFN-γ for 4 h and subsequently stimulated with 0.3 μM CpG-B for the indicated periods. As shown in Fig. 4a, IRF1 mRNA was strongly induced by the pretreatment with IFN-γ, even in Myd88−/− GM-DCs. The subsequent CpG-B treatment only marginally increased IRF1 mRNA expression levels. TNF-α mRNA was induced upon stimulation with CpG-B alone, and this induction was not augmented by the pretreatment with IFN-γ and not affected by the Irf1 gene deficiency (Fig. 4a). In contrast, IFN-γ-pretreated GM-DCs, in which therefore IRF1 was up-regulated, exhibited an enhanced induction of IFN-β, IL-12p35, and iNOS mRNAs in response to stimulation with CpG-B (Fig. 4a). This enhancement was markedly suppressed in Irf1−/− or Myd88−/− GM-DCs (Fig. 4a). We also analyzed the effect of synergism between IFN-γ and MALP2, which activates the MyD88-dependent signaling pathway through TLR2 and TLR6 (26). Similar to the case of stimulation with CpG-B, the effect of synergism between MALP2 and IFN-γ was observed, and it depended on IRF1 and MyD88 (Fig. 4b). Collectively, these results support the notion that IRF1 induced by IFN-γ could be efficiently activated by MyD88 upon TLR stimulation, and this MyD88-dependent activation of IRF1 may likely be a mechanism underlying the cooperation/synergism between TLR and IFN-γ signaling.
Fig. 4.
IRF1 involved in IFN-γ and TLR synergy. GM-DCs from wild-type or mutant mice were pretreated with or without 200 units/ml IFN-γ for 4 h and then stimulated with 0.3 μM CpG-B (a) or MALP2 (b) for the indicated periods. Total RNA was prepared and subjected to real-time RT-PCR analysis of the indicated genes.
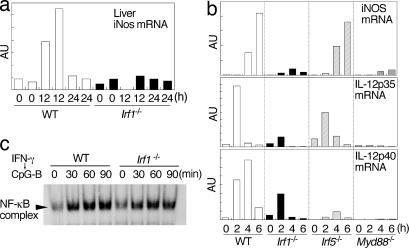
To examine the IRF1 dependence of the IFN-γ and TLR responses in vivo, we i.v. injected IFN-γ and then i.p. injected CpG-B into wild-type or Irf1−/− mice. All of the wild-type mice showed obvious evidence of systemic reactions such as reduced mobility and fur ruffling within a few hours. In contrast, Irf1−/− mice exhibited fewer signs of impairments than the control mice (data not shown). We collected liver samples from mice after injection and analyzed the expression of the iNOS gene. As shown in Fig. 5a, iNOS mRNA inductions were markedly suppressed in the Irf1−/− livers. These results indicate that IRF1 plays an essential role in the synergy between IFN-γ and TLR in vivo.
Fig. 5.
Role of IRF5 in cooperation between IFN-γ and TLR. (a) Wild-type or Irf1−/− mice were i.v. injected with IFN-γ (105 units) and then i.p. injected with CpG-B (100 μg) 1 h after. Liver samples were collected at the indicated periods (two mice for each time point) and subjected to real-time RT-PCR analysis of iNOS mRNA. (b) Peritoneal macrophages from wild-type or mutant mice were stimulated with 0.3 μM CpG-B and 30 units/ml IFN-γ for the indicated period. Real-time RT-PCR analysis for iNOS, IL-12p35, and IL-12p40 mRNAs was performed. (c) GM-DCs from wild-type and Irf1−/− mice were pretreated with 200 units/ml IFN-γ for 4 h and then stimulated with 0.3 μM CpG-B for the indicated periods. NF-κB activity was assessed by EMSA.
Role of IRF5 in Synergy Between IFN-γ and TLR.
We also analyzed the role of IRF5, another MyD88-interacting IRF family member (6), in TLR and IFN-γ synergy. The induction of iNOS and IL-12p35 mRNAs was again markedly suppressed in Irf1−/− or Myd88−/− peritoneal macrophages in response to IFN-γ CpG-B, whereas it was either unchanged or slightly suppressed in Irf5−/− cells compared with wild-type cells (Fig. 5b). In contrast, IL-12p40 mRNA induction was more markedly affected by the deficiency in Irf5 than by that in Irf1 (Fig. 5b). Taken together, IRF1 and IRF5 interact with a similar region of MyD88 (see Fig. 1e) but carry out their missions, which are distinct from each other.
Collaboration Between IRF1 and NF-κB.
It has been shown that the induction of IFN-β, IL-12p35, and iNOS genes is critically regulated by NF-κB (11, 27). Therefore, we next examined whether the loss of IRF1 affects the activation of NF-κB in response to stimulation with IFN-γ and CpG-B, and we found that the activation of NF-κB was normal in Irf1−/− GM-DCs (Fig. 5c). This result indicates that the activation of NF-κB is controlled independently of IRF1 in the pathway downstream of MyD88. It is likely that the synergy between IRF1 and NF-κB occurs at the gene promoter level.
To further analyze the relationship between IRF1 and NF-κB in IFN-γ and TLR synergy, we compared it with that in IFN-γ and TNF-α synergy, which is a classically known phenomenon (13, 28). As shown in Fig. 6, IRF1 expression was induced by pretreatment with IFN-γ and was slightly enhanced by the subsequent stimulation with CpG-B or TNF-α in GM-DCs. Consistent with reports (13, 28), the induction of IP-10 mRNA by stimulation with TNF-α was enhanced by the pretreatment with IFN-γ (Fig. 6); this enhancement did not depend on IRF1 (data not shown). In contrast, no effect of the synergy between IFN-γ and TNF-α for the induction of IFN-β, IL-12p35, and iNOS mRNAs was observed despite the efficient activation of NF-κB by stimulation with TNF-α, as revealed by the induction of an NF-κB-dependent gene such as IκBα (Fig. 6). Therefore, these results indicate that NF-κB activated by TNF-α cannot synergize with IRF1 that is simply induced by IFN-γ unless it is licensed by TLR–MyD88 signaling.
Fig. 6.
Effect of synergism between IFN-γ and TNF-α. GM-DCs from wild-type mice were pretreated with or without 200 units/ml IFN-γ for 4 h and then stimulated with 0.3 μM CpG-B or 10 ng/ml TNF-α for the indicated periods. Total RNA was prepared and subjected to real-time RT-PCR analysis of the indicated genes.
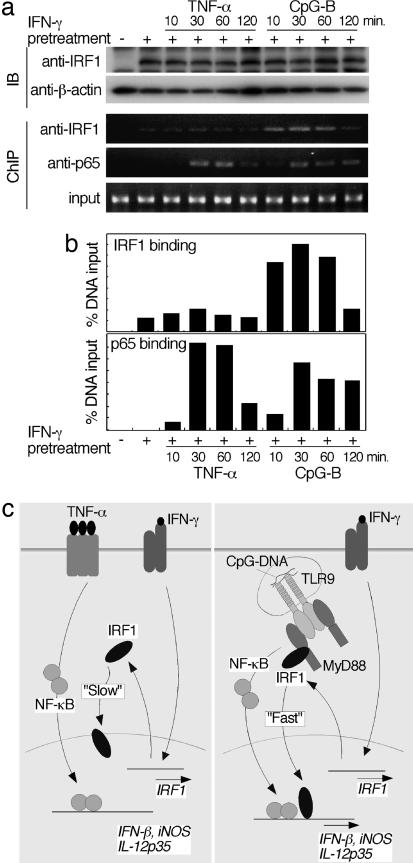
To further assess the activation status of IRF1 during stimulation with CpG-B or TNF-α we stimulated a macrophage cell line, RAW264.7, with IFN-γ and subsequently with CpG-B or TNF-α, and we performed a ChIP assay using the anti-IRF1 and anti-p65 antibodies as well as primers for the IL-12p35 promoter region containing an IFN-stimulated response element and an NF-κB-binding motif (11). RAW264.7 cells exhibited induction profiles of IFN-β, iNOS, and IL-12p35 mRNAs similar to those of GM-DCs after stimulation with IFN-γ and CpG-B or TNF-α (data not shown). As shown in Fig. 7a and b, a strong recruitment of the NF-κB component p65 to the IL-12p35 promoter was commonly observed for TNF-α- and CpG-B-stimulated cells. In contrast, IRF1 induced by IFN-γ was weakly recruited to the promoter region of IL-12p35, which was not enhanced by stimulation with TNF-α. A strong recruitment of IRF1 was observed only for CpG-B-stimulated cells. Interestingly, the kinetics of recruitment of p65 and IRF1 was similar during CpG-B stimulation (Fig. 7 a and b). Taken together, our results suggest that the interaction of IRF1 with MyD88 ensures the enhancement of the gene induction program for TLR9 signaling by IFN-γ, presumably in cooperation with NF-κB.
Fig. 7.
Relationship between IRF1 and NF-κB in IFN-γ and TLR synergy. (a) RAW264.7 cells were stimulated as in Fig. 6. The cells were subjected to immunoblotting (top two panels) and ChIP analysis (bottom three panels) using the indicated antibodies. (b) To quantify the results of ChIP analysis, the intensity of a band was measured by a densitometer. The intensity of the band corresponding to the factor binding to the promoter was compared with that corresponding to input DNA. (c) Schematic illustration of the role of IRF1 in the cross-talk of IFN-γ with TNF-α (Left) or CpG-DNA (Right). In both cases IRF1 induced by IFN-γ can migrate into the nucleus; however, the kinetics is significantly different between these two cases (see text).
Discussion
Among molecules that act downstream of MyD88, much attention has been recently focused on the role of IRF family members. IRF4, IRF5, and IRF7 all directly interact with MyD88 and regulate proinflammatory mediators and type I IFN induction (1, 3, 6, 7). In addition to these IRF family members, our present results support the notion that IRF1 also participates in the organization of a multimolecular complex via MyD88 and plays an essential role in the induction of IFN-β, iNOS, and IL-12p35 genes. IRF1 and IRF5 interact with a similar region in MyD88 (see Fig. 1e) but induce a distinct set of genes, providing a mechanistic basis for the diversity and complexity of the MyD88-dependent gene induction program. Our data indicate that IRF1, although expressed at low levels in unstimulated DCs and macrophages, is induced by IFN-γ in an autocrine or a paracrine manner, and the induced IRF1 binds to MyD88 and is modified by as yet unidentified signaling molecule(s) to migrate into the nucleus and induce genes. A fraction of IFN-γ-induced IRF1 could directly migrate into the nucleus and activate certain genes in a MyD88-independent manner (i.e., without licensing) (11, 13). However, our experimental data obtained by Kaede-tagged IRF1 or ChIP analysis indicate the operation of yet another mechanism by which IRF1, activated by MyD88 signaling, is selectively licensed to synchronize with the activation of NF-κB, leading to the cooperation between IRF1 and NF-κB, that is, the formation of a transcriptional complex (so-called “enhanceosome”) (29) for efficient activation of IFN-β, iNOS, and IL-12p35 gene transcription (Fig. 7c). We infer that the licensing of IRF1 involves its phosphorylation (Fig. 2e); however, the exact nature of IRF1 licensing and the mechanism by which the licensed IRF1 undergoes rapid nuclear translocation and cooperates with NF-κB need to be investigated further.
IRF1 was initially identified as an activator of type I IFN gene transcription (9). However, a series of gene disruption studies demonstrated that IRF3 and IRF7, rather than IRF1, are critical for type I IFN gene induction in virus-infected fibroblasts and pDCs (4, 21). In the present study we showed that IRF1 plays an essential role in type I IFN gene induction by CpG-B-mediated TLR9 activation in GM-DCs (Fig. 3). How can one reconcile previous and present data? Although no clear explanation can be offered at present, we infer that the IRF that participates in the gene induction is contingent on many elements such as cell type and the type of cellular compartment involved. In this context, CpG-B-mediated TLR9 signaling in GM-DCs, which presumably occurs in the lysosomal compartment (30), allows the activation of MyD88-associated IRF1, but not IRF3 or IRF7 (Fig. 3). On the other hand, the MyD88-associated IRF7 is activated in pDCs by the ingenious regulation of endosomal trafficking of CpG-A (30). It is worth noting that the IRF1-dependent type I IFN mRNA induction in GM-DCs is markedly weaker than those mediated by IRF3 and IRF7 in virus-infected fibroblasts and pDCs (Fig. 3) (4). Therefore, we also infer that, upon efficient activation of IRF3 and IRF7, the contribution of IRF1 may be overwhelmed by that of IRF3 and IRF7. During the course of our study, we learned that similar results on the role of IRF1 in TLR9-dependent type I IFN induction were also obtained by Hermann Wagner and colleagues (Institute for Medical Microbiology, Munich, Germany) (personal communication). Clearly, these issues will require further investigation.
The synergy between IFN-γ and TLR has been implicated in the host defense against pathogens (13, 14). IFN-γ produced by T cells and other cells is considered to enhance TLR signaling in DCs and macrophages for the efficient induction of inflammatory mediators to eliminate pathogens. Indeed, Myd88−/− as well as IRF1−/− mice are commonly susceptible to several pathogens, including L. monocytogenes, T. gondii, and M. tuberculosis (10, 15–19). Our present study provides insight into the mechanisms of the gene induction program during microbial infections, and the MyD88-dependent activation of IRF1 may explain the antimicrobial synergism between IFN-γ and TLR. On the other hand, as observed in IFN-γ- and CpG-B-injected mice, the host may suffer from detrimental effects of excessive MyD88–IRF1-mediated signaling elicited during acute infection. Thus, our study identifies IRF1 as an essential downstream regulator of the TLR–MyD88 signaling pathway and a potential target of therapeutic intervention to control beneficial as well as harmful immune responses.
Materials and Methods
Plasmid Construction.
Expression vectors of HA-tagged or YFP-tagged mouse IRFs and FLAG-tagged or CFP-tagged full-length mouse MyD88 and a series of deletion mutants of MyD88 were constructed as described previously (3). The Kaede-tagged IRF1 expression vector was constructed by using the pCAGGS-Kaede (22) (a gift from A. Miyawaki, RIKEN, Saitama, Japan).
Reagents.
The sequences of CpG-A (also called D19) and CpG-B (also called 1668) were described previously (4, 23, 24). poly(I:C) was purchased from Amersham Biosciences (Piscataway, NJ). LPS from Salmonella minnesota Re-595 and MALP2 (mycoplasmal macrophage-activating lipopeptide 2 kDa) were purchased from Sigma-Aldrich (St. Louis, MO) and Alexis (Lausen, Switzerland), respectively. Mouse IFN-γ, granulocyte/macrophage colony-stimulating factor, and TNF-α were purchased from PeproTech (Rocky Hill, NJ).
Fluorescence Microscopy.
Confocal microscopy, FRET analysis, and the calculation of corrected FRET (FRETC) were carried out as previously described (3). For the imaging of IRF1Kaede, stimulation and observation were performed by using an Olympus FV1000 system. Kaede green, Kaede red, and CFP were imaged by using excitation at 488, 543, and 457 nm, respectively. For the photoconversion of the Kaede protein, namely, from Kaede green to Kaede red conversion, a region of interest was pulsed with a 405-nm laser for 5 s. After pulsation, images were collected every 10 s. The following equation was used to determine the percentage of IRF1Kaede in the nucleus: (sum of intensities of total pixels in nucleus)/(sum of intensities of total pixels in cell) × 100%.
Reporter Assay.
HEK293T cells were transiently cotransfected with 100 ng of the reporter plasmid (p125-luc or pNF-κB-luc) together with expression plasmids for IRF1 and MyD88, and luciferase activity was measured as described (3, 21).
Immunoprecipitation and Immunoblotting.
Immunoprecipitation and immunoblotting were carried out as described (3).
Two-Dimensional Gel Electrophoresis.
HEK293T cells were transfected with HA–IRF1 in combination with or without FLAG–MyD88. Cell lysates were immunoprecipitated with the anti-HA antibody and then subjected to isoelectric focusing by using a 24-cm Immobiline DryStrip (pH 4–7) and an IPGphor isoelectric focusing unit (GE Healthcare, Piscataway, NJ). For separation in the second dimension, 10% SDS/PAGE was used. Gels were transferred to a nitrocellulose membrane and then subjected to immunoblot analysis by using the anti-HA antibody.
Mice.
The generation of Irf1−/−, Irf3−/−, Irf5−/−, Irf7−/−, and Irf3/7−/− mice was described (4, 6, 21, 31), and Myd88−/− mice were provided by S. Akira (Osaka University, Osaka, Japan).
DC Generation and Isolation.
GM-DCs, resident peritoneal macrophages, and splenic pDCs (B220+CD11cintermediate cells) were prepared as described (4, 7).
RNA Analysis.
Quantitative real-time RT-PCR analysis was performed by using a LightCycler system (Roche, Mannheim, Germany). Data were normalized by the level of β-actin expression in each sample. The primers used for IFN-α1, IFN-β, TNF-α, IP-10, IL-12p40, IκBα, and β-actin were described (2, 4, 6, 7). The following primers were also used: iNOS, 5′-CACCTTGGAGTTCACCCAGT-3′ and 5′-ACGACTCGTACTTGGGATGC-3′; IL-12p35, 5′-CTTAGCCAGTCCCGAAACC-3′ and 5′-GCTCCCTCTTGTTGTGGAAG-3′.
EMSA and Flow Cytometry.
EMSA and flow cytometry were performed as described (4).
ChIP Assay.
The ChIP assay was performed by using a Quick ChIP kit (IMGENEX, San Diego, CA), a Phusion high-fidelity PCR kit (Daiichi Pure Chemicals, Tokyo, Japan), and anti-IRF1 and anti-p65 polyclonal antibodies (Santa Cruz Biotechnology, Santa Cruz, CA). The following primers for the IL-12p35 promoter region were used: 5′-TTGCTTTCGCTCTGAGTGTG-3′ and 5′-GCTGACCTTGGGAGACACAT-3′.
Acknowledgments
We thank M. Shishido, T. Koga, and S. Harumiya for technical assistance and A. Miyawaki for Kaede and YFP (also known as Venus) expression plasmids. This work was supported by a Kakenhi Grant-in-Aid for Scientific Research on Priority Areas (“Integrative Research Toward the Conquest of Cancer”) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and the Uehara Memorial Foundation.
Abbreviations
- TLR
Toll-like receptor
- IRF
IFN regulatory factor
- iNOS
inducible NO synthase
- DC
dendritic cell
- GM-DC
granulocyte/macrophage colony-stimulating factor-cultured bone marrow-derived DC
- pDC
plasmacytoid DC
- CFP
cyan fluorescent protein
Footnotes
The authors declare no conflict of interest.
References
- 1.Honda K, Taniguchi T. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 2.Sakaguchi S, Negishi H, Asagiri M, Nakajima C, Mizutani T, Takaoka A, Honda K, Taniguchi T. Biochem Biophys Res Commun. 2003;306:860–866. doi: 10.1016/s0006-291x(03)01049-0. [DOI] [PubMed] [Google Scholar]
- 3.Honda K, Yanai H, Mizutani T, Negishi H, Shimada N, Suzuki N, Ohba Y, Takaoka A, Yeh WC, Taniguchi T. Proc Natl Acad Sci USA. 2004;101:15416–21. doi: 10.1073/pnas.0406933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 5.Kawai T, Sato S, Ishii KJ, Coban C, Hemmi H, Yamamoto M, Terai K, Matsuda M, Inoue J, Uematsu S, et al. Nat Immunol. 2004;5:1061–1068. doi: 10.1038/ni1118. [DOI] [PubMed] [Google Scholar]
- 6.Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, Kano S, Honda K, Ohba Y, Mak TW, Taniguchi T. Nature. 2005;434:243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 7.Negishi H, Ohba Y, Yanai H, Takaoka A, Honma K, Yui K, Matsuyama T, Taniguchi T, Honda K. Proc Natl Acad Sci USA. 2005;102:15989–15994. doi: 10.1073/pnas.0508327102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J, Kong HJ, Li H, Huang B, Yang M, Zhu C, Bogunovic M, Zheng F, Mayer L, Ozato K, et al. J Biol Chem. 2006;281:10073–10080. doi: 10.1074/jbc.M507788200. [DOI] [PubMed] [Google Scholar]
- 9.Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y, Miyata T, Taniguchi T. Cell. 1988;54:903–913. doi: 10.1016/s0092-8674(88)91307-4. [DOI] [PubMed] [Google Scholar]
- 10.Kamijo R, Harada H, Matsuyama T, Bosland M, Gerecitano J, Shapiro D, Le J, Koh SI, Kimura T, Green SJ, et al. Science. 1994;263:1612–1615. doi: 10.1126/science.7510419. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Cao S, Herman LM, Ma X. J Exp Med. 2003;198:1265–1276. doi: 10.1084/jem.20030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimura T, Nakayama K, Penninger J, Kitagawa M, Harada H, Matsuyama T, Tanaka N, Kamijo R, Vilcek J, Mak TW, et al. Science. 1994;264:1921–1924. doi: 10.1126/science.8009222. [DOI] [PubMed] [Google Scholar]
- 13.Schroder K, Hertzog PJ, Ravasi T, Hume DA. J Leukocyte Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 14.Shi S, Nathan C, Schnappinger D, Drenkow J, Fuortes M, Block E, Ding A, Gingeras TR, Schoolnik G, Akira S, et al. J Exp Med. 2003;198:987–997. doi: 10.1084/jem.20030603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fremond CM, Yeremeev V, Nicolle DM, Jacobs M, Quesniaux VF, Ryffel B. J Clin Invest. 2004;114:1790–1799. doi: 10.1172/JCI21027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan IA, Matsuura T, Fonseka S, Kasper LH. J Immunol. 1996;156:636–643. [PubMed] [Google Scholar]
- 17.Scanga CA, Aliberti J, Jankovic D, Tilloy F, Bennouna S, Denkers EY, Medzhitov R, Sher A. J Immunol. 2002;168:5997–6001. doi: 10.4049/jimmunol.168.12.5997. [DOI] [PubMed] [Google Scholar]
- 18.Taki S, Sato T, Ogasawara K, Fukuda T, Sato M, Hida S, Suzuki G, Mitsuyama M, Shin EH, Kojima S, et al. Immunity. 1997;6:673–679. doi: 10.1016/s1074-7613(00)80443-4. [DOI] [PubMed] [Google Scholar]
- 19.Seki E, Tsutsui H, Tsuji NM, Hayashi N, Adachi K, Nakano H, Futatsugi-Yumikura S, Takeuchi O, Hoshino K, Akira S, et al. J Immunol. 2002;169:3863–3868. doi: 10.4049/jimmunol.169.7.3863. [DOI] [PubMed] [Google Scholar]
- 20.Dalton DK, Pitts-Meek S, Keshav S, Figari IS, Bradley A, Stewart TA. Science. 1993;259:1739–1742. doi: 10.1126/science.8456300. [DOI] [PubMed] [Google Scholar]
- 21.Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 22.Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. Proc Natl Acad Sci USA. 2002;99:12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krieg AM, Yi AK, Matson S, Waldschmidt TJ, Bishop GA, Teasdale R, Koretzky GA, Klinman DM. Nature. 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 24.Verthelyi D, Ishii KJ, Gursel M, Takeshita F, Klinman DM. J Immunol. 2001;166:2372–2377. doi: 10.4049/jimmunol.166.4.2372. [DOI] [PubMed] [Google Scholar]
- 25.Hoebe K, Janssen EM, Kim SO, Alexopoulou L, Flavell RA, Han J, Beutler B. Nat Immunol. 2003;4:1223–1229. doi: 10.1038/ni1010. [DOI] [PubMed] [Google Scholar]
- 26.Takeuchi O, Kaufmann A, Grote K, Kawai T, Hoshino K, Morr M, Muhlradt PF, Akira S. J Immunol. 2000;164:554–557. doi: 10.4049/jimmunol.164.2.554. [DOI] [PubMed] [Google Scholar]
- 27.Ghosh S, May MJ, Kopp EB. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 28.Sun D, Ding A. Nat Immunol. 2006;7:375–381. doi: 10.1038/ni1308. [DOI] [PubMed] [Google Scholar]
- 29.Carey M. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 30.Honda K, Ohba Y, Yanai H, Negishi H, Mizutani T, Takaoka A, Taya C, Taniguchi T. Nature. 2005;434:1035–1040. doi: 10.1038/nature03547. [DOI] [PubMed] [Google Scholar]
- 31.Matsuyama T, Kimura T, Kitagawa M, Pfeffer K, Kawakami T, Watanabe N, Kundig TM, Amakawa R, Kishihara K, Wakeham A, et al. Cell. 1993;75:83–97. [PubMed] [Google Scholar]