Abstract
Truncation mutations of the GLI3 zinc finger transcription factor can cause Greig cephalopolysyndactyly syndrome (GCPS), Pallister–Hall syndrome (PHS), and postaxial polydactyly type A (PAP-A). GLI3 is homologous to Drosophila Cubitus interruptus (Ci), which regulates the patched (ptc), gooseberry (gsb), and decapentaplegic (dpp) genes. Ci is sequestered in the cytoplasm and is subject to posttranslational processing whereby the full-length transcriptional activator form (Ci155) can be cleaved to a repressor form (Ci75). Under hedgehog signaling, the Ci155 form translocates to the nucleus whereas in the absence of hedgehog, the Ci75 form translocates to the nucleus. Based on the correlation of GLI3 truncation mutations and the human phenotypes, we hypothesized that GLI3 shows transcriptional activation or repression activity and subcellular localization similar to Ci. Here we show that full-length GLI3 localizes to the cytoplasm and activates PTCH1 expression, which is similar to full-length Ci155. PHS mutant protein (GLI3-PHS) localizes to the nucleus and represses GLI3-activated PTCH1 expression, which is similar to Ci75. The GCPS mutant protein has no effect on GLI3-activated PTCH1 transcription, consistent with the role of haploinsufficiency in this disorder. The PAP-A mutant protein (GLI3-PAP-A) showed less specific subcellular localization but still inhibited GLI3-activated PTCH1 transcription, suggesting it may be a weaker allele than the GLI3-PHS mutation. These data show that GLI3 mutations in humans mimic functional effects of the Drosophila ci gene and correlate with the distinct effects of these mutations on human development.
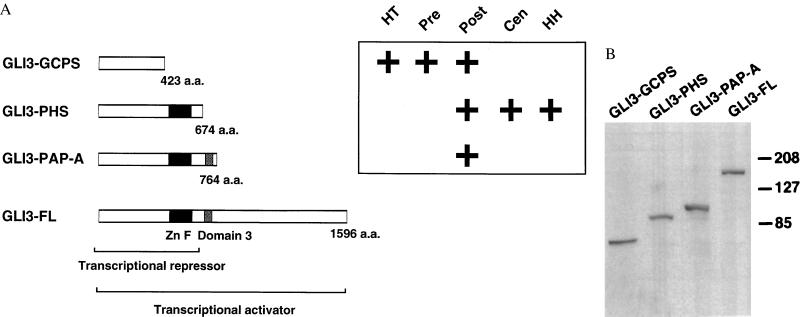
The GLI3 gene originally was cloned by homology to GLI and subsequently was shown to be interrupted by translocations in patients with the Greig cephalopolysyndactyly syndrome (GCPS) (1). Subsequently, it was demonstrated that mutations in GLI3 can cause the Pallister–Hall syndrome (PHS) (2) and postaxial polydactyly type A (PAP-A) (3). These findings raised the question of how distinct human developmental disorders (GCPS and PHS) and isolated anomalies (PAP-A) can be caused by mutations in a single gene. The GCPS mutations that have been described (truncations, deletions, point mutations, and translocations) support the previous assertions that haploinsufficiency of GLI3 causes GCPS whereas frameshift mutations alone are known to cause PHS and PAP-A. Because all three disorders can be caused by frameshift mutations, we chose to investigate the potential correlation of the position of truncation mutations and the effect of those mutations on GLI3 localization and function as a possible explanation for the distinct phenotypes (4). The model is based on the homology of GLI3 to the Drosophila cubitus interruptus gene product, Ci (5). In Drosophila, Ci exists as a 155-kDa full-length form (Ci155) that has been shown to be anchored indirectly to the microtubular apparatus in the cytoplasm in a complex with the costal2, fused, and suppressor of fused [su(fu)] gene products (6–8). The Ci protein can undergo proteolytic processing and be released as a 75-kDa truncated product (Ci75) that represses downstream genes (ptc, gsb, and dpp), or it may be released in the Ci155 form and activate transcription of these genes (6, 9). The balance of activator and repressor forms is regulated by hedgehog signaling, mediated through patched/smoothened transduction (7). We hypothesized that the human truncation mutations in GLI3 cause distinct disorders of human development because of the presence or absence of key functional domains and that these proteins activate or repress downstream genes in a manner predicted by the dual forms of Drosophila Ci. Phenotypic analyses show that although PHS and GCPS share some features (e.g., postaxial polydactyly), they have nonoverlapping manifestations and are distinct syndromes (10, 11). PAP-A is a nonsyndromic form of autosomal-dominant postaxial polydactyly (3) and could be considered a mild form of either GCPS or PHS. GCPS and PHS cannot be placed on a single phenotypic continuum of severity, implying that there are distinct pathogenetic mechanisms producing the different anomalies in these two disorders. On the basis of these data, we hypothesized that the truncation mutations caused distinct developmental anomalies because of the inclusion or loss of these functional domains (4). We chose to model these mutations by comparing the effects of truncated GLI3 proteins: GCPS is modeled after the truncation mutation that deletes the zinc fingers and domains 3–7 (12), PHS deletes domains 3–7 (2), and PAP-A deletes domains 4–7 (3) (see Fig. 1A).
Figure 1.
Construction and expression of mutant and full-length GLI3. (A) GLI3 constructs including full-length (GLI3-FL) and three truncations that model mutations in human developmental malformations including GLI3-GCPS, GLI3-PHS, and GLI3-PAP-A. All constructs include an amino-terminal enterokinase recognition site (anti-Xpress epitope) tag. The zinc finger domain (Zn F) and domain 3 are shown as their presence or absence varies among the human truncation mutations. It has been hypothesized previously that full-length GLI3 is a transcriptional activator and truncated GLI3 (GLI3-PHS) is a transcriptional repressor. The major phenotypic features are shown to the left of the figure. HT, hypertelorism; Pre, preaxial polydactyly; Post, postaxial polydactyly; Cen, central polydactyly; HH, hypothalamic hamartoma. (B) Western blot of transfected cell lines with anti-Xpress antibody showing the production of proteins of the predicted sizes from GLI3-GCPS, GLI3-PHS, GLI3-PAP-A, and GLI3-FL constructs. The size of full-length GLI3 was reported previously as 190 kDa (5), similar to the result shown here.
MATERIALS AND METHODS
Construction of GLI3 Truncation Mutants.
A series of mutants that generate C-terminal truncated proteins of GLI3 was constructed by PCR amplification and subcloning of full-length human GLI3 cDNA pGli3bs-2 (5). Primers GFP5′ (5′-TAGCTGACGAGCTCAGAAGACATCATGGAGG-3′) and GFPtrunc3′ (5′-GATCGCTAGAGCTCAACCAAGGGCTCCCTGAGT-3′) were used to amplify GLI3 cDNA from the initiator codon to codon 675. This fragment was cloned into the SacI site of pEGFP-C2 (CLONTECH) to generate plasmid EGLI3-PHS. GLI3 cDNA from the initiator codon to codon 423 was cloned by excising a SacI and PstI fragment from EGLI3-PHS and ligating it into the SacI and PstI sites of pEGFP-C2 to generate plasmid EGLI3-GCPS. Primers GFP5′ and R764ter (5′-GCGCGCGTCGACCTATTGCAAAGCAAGGGCTGTGGT-3′) were used to generate a SacI and SalI fragment of GLI3 cDNA from the initiator codon to codon 764, which was cloned into pEGFP-C2 to generate plasmid EGLI3-PAP-A. A 4.8-kb EcoRI fragment of pGli3bs-2 was cloned into the EcoRI site of EGLI3-PHS construct to generate EGLI3-FL, full-length GLI3. Correct insert orientation was determined by digestion with SacI and SalI to generate a 4.9-kb GLI3 fragment and a 4.6-kb pEGFP backbone vector. To generate amino-terminal enterokinase-tagged constructs, EGLI3-GCPS, EGLI3-PAP-A, and EGLI3-FL were digested by SacI, blunted with T4 DNA polymerase, digested with SalI, and ligated into EcoRV and XhoI-digested pcDNA3.1HisC vector (Invitrogen) to generate GLI3-GCPS, GLI3-PAP-A, and GLI3-FL, respectively. EGLI3-PHS was digested with SacI, blunted with T4 DNA polymerase, and ligated into pcDNA3.1HisC, which was digested with ApaI, blunted with T4 DNA polymerase, and digested again with EcoRV to generate GLI3-PHS. All PCR-derived products were confirmed by sequencing.
Western Blotting.
Lysates were prepared from cells 24 hr after transfection. Aliquots of lysates were separated on 4–15% SDS-polyacrylamide gel, transferred to poly(vinylidene difluoride) membrane (Millipore), and probed with a 1:5,000 dilution of Anti-Xpress antibody (Invitrogen). Immunodetection with sheep anti-mouse horseradish peroxidase conjugate and enhanced chemiluminescence (Amersham) was performed according to the manufacturer’s instructions.
Immunofluorescence.
HeLa cells (ATCC CCL2) were cultured according to the distributor and seeded onto glass coverslips in six-well plates 12 hr before the calcium phosphate transfection (Stratagene) with plasmid DNAs, using the manufacturer’s instructions. After 24 hr, cells were fixed with 1% paraformaldehyde for 20 min, washed with PBS twice, and permeated with 0.05% Triton X-100 for 10 min. The cells were blocked with 5% BSA in PBS for 30 min, rinsed in 1% BSA in PBS, immunostained with 1:400 dilution of Anti-Xpress antibody (Invitrogen), washed with 1% BSA five times, incubated with 1:400 secondary antibody coupled to tetramethylrhodamine B isothiocyanate (Sigma), and washed with 1% BSA once. Fluorescence microscopy was performed by using a Zeiss Axiophot microscope, and images were captured with a Photometrics CH250 liquid-cooled charge-coupled device camera. Images were processed with the ip lab (Scanalytics, Fairfax, VA) software package on a Macintosh 8100/110 computer. At least 100 transfected cells were counted for each construct. Cells were scored as nuclear, cytoplasmic, or mixed (nuclear and cytoplasmic) signal pattern. These experiments were repeated in 293 cells lines and gave similar results (data not shown).
GLI3 Transcriptional Activity.
The PTCH1 genomic region was cloned as described previously (13). A 4.3-kb fragment of the 5′ regulatory region was subcloned into the pGL3-Basic vector (Promega) upstream of the firefly luciferase reporter gene. It was demonstrated that this construct can mediate activation of the reporter gene in different cell lines in response to expression of GLI1 and SHH cDNA (P.K. and R.T., unpublished observations).
Human 293 cells (kidney epithelial cell line) were obtained from American Type Culture Collection and grown in DMEM supplemented with penicillin and streptomycin and 10% fetal bovine serum (GIBCO/BRL). Cells were passaged to 24-well plates the day before experiments, and transfections were carried out with DNA complexed to the Superfect transfection reagent (Qiagen) according to the manufacturer’s instructions. Twenty-four hours after transfection, the medium was removed and the cells were lysed in Somalyze lysis buffer (BioOrbit, Turku, Finland). Luciferase activity was determined on a BioOrbit 1250 Luminometer by using BioOrbit Luciferin Substrate and ATP reagents according to the manufacturer’s instructions. The results from at least six experiments from two separate transfections were compiled. The results were normalized for transfection efficiency and protein expression by measuring β-galactosidase activity generated from an Rous sarcoma virus–lacZ expression construct that was cotransfected into each well. This activity was measured by using the luminometric GalactoLight Plus assay (Tropix, Bedford, MA) according to the manufacturer’s instructions.
Statistical analyses were performed by using Student’s t test unless data showed evidence of unequal SDs (F test, p < 0.05). In these cases, the Mann–Whitney nonparametric test was used. All calculations were performed by using the instat program (GraphPad, San Diego).
RESULTS
Subcellular Localization of GLI3 Proteins.
The model predicted that GLI3 proteins would be localized into subcellular compartments similarly to processed and unprocessed Ci. We tested this model first by determining the subcellular localization of various truncated mutants of the GLI3 protein in HeLa cells. A series of GLI3 expression constructs was made to mimic the frameshift mutations found in GCPS (423 aa), PHS (674 aa), and PAP-A (764 aa) in addition to full-length GLI3 (1596 aa) (Fig. 1A). The hypothesis predicted that GLI3-FL would be localized to the cytoplasm as seen in intact Ci155, that a GLI3-PHS protein would localize to the nucleus, as does Ci75, and that GLI3-GCPS protein would show cytoplasmic or nonspecific localization because it is lacking the putative nuclear localization signal in the C-terminal end of the zinc finger domain.
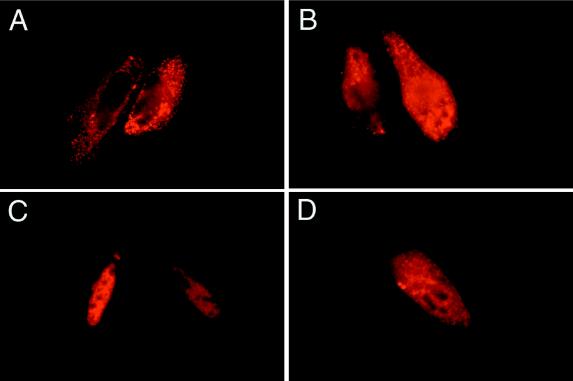
Subcellular localization was performed by using GLI3 cDNA constructs fused with an amino-terminal enterokinase (EK) epitope. The constructs produced epitope-GLI3 chimeric proteins of the predicted size (Fig. 1B). Amino-terminal EGFP-tagged GLI3 proteins yielded similar results (data not shown). Transfection of the GLI3-FL construct showed punctate cytoplasmic fluorescence in 97% of transfected cells (n = 211) (Fig. 2A). In contrast, the GLI3-PHS protein showed nuclear fluorescence in 86% of transfected cells (n = 273) (Fig. 2C). The GLI3-GCPS protein showed a mixed pattern with about 1/3 of transfected cells (n = 139), each showing nuclear, cytoplasmic, and mixed nuclear and cytoplasmic signals (Fig. 2B). The GLI3-PAP-A construct produced a protein that showed inconsistent staining, with most cells showing either a cytoplasmic pattern (49%, n = 270) or a nuclear pattern (41%) and fewer (10%) showing a mixed nuclear and cytoplasmic pattern (Fig. 2D). The dramatic difference between the GLI3-FL and GLI3-PHS localization data shows that the targeting of normal and truncated GLI3 proteins mimics that of Ci155 and Ci75, respectively. These observations are consistent with the proposed model.
Figure 2.
GLI3 subcellular localization. (A) The full-length, wild-type GLI3-FL transfection showed cytoplasmic staining consistent with the known subcellular localization of the Drosophila Ci155 protein. (B) Fluorescence microscopy image of GLI3-GCPS protein showing nonspecific and variable cytoplasmic and nuclear immunofluorescence signal. This staining pattern is consistent with a model that predicts aberrant targeting of GLI3-GCPS protein resulting from absence of both the cytoplasmic anchor and the nuclear localization signal. (C) The GLI3-PHS protein gave a strong and consistent nuclear staining that mimics the effect of the Drosophila Ci75 processed protein, which is targeted to the nucleus. (D) The GLI3-PAP-A construct gave a result similar to the GLI3-GCPS construct, with a mix of cytoplasmic and nuclear staining. The GLI3-PAP-A pattern is attributed to inefficient cytoplasmic anchoring because of partial function of the cytoplasmic anchor. The Results section includes data on cell counts for each subcellular localization pattern. Each transfection was scored in at least 139 cells.
GLI3 Transcriptional Regulation of PTCH1.
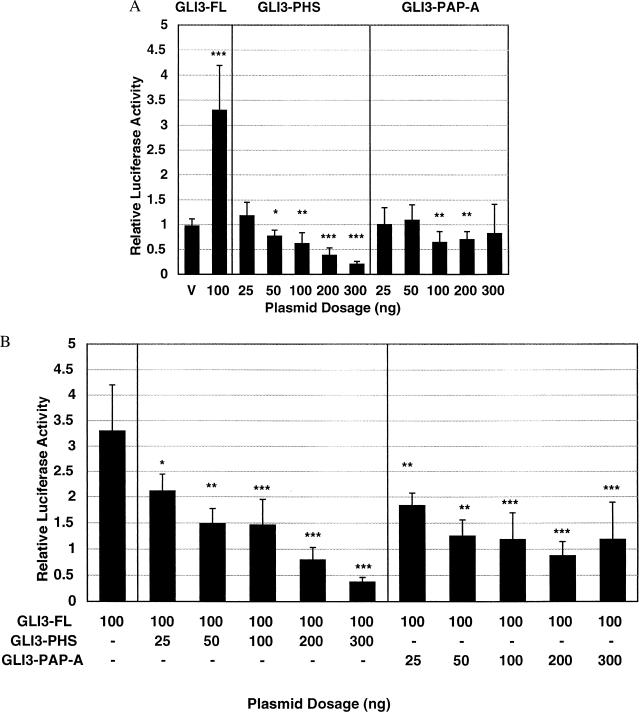
To further address the issue of the distinct developmental consequences of various GLI3 mutations, we next determined the transcriptional activation properties of full-length and truncated forms of GLI3. We hypothesized that full-length and truncated GLI3 proteins can have activator and repressor activities similar to full-length and truncated forms of Ci, respectively. In this model, full-length GLI3 was proposed to activate transcription, similar to Ci155 and full-length GLI1 (14). The GLI3-PHS construct was predicted to repress transcriptional activity constitutively, based on the transcriptional repressor effect of processed Drosophila Ci75. In addition, the GLI3-GCPS and GLI3-PAP-A proteins have effects that can be predicted from what is known about Ci, although they do not reflect normal functions of Ci. Specifically, we hypothesized that the GLI3-GCPS construct would show no repression or activation, being effectively a null allele, whereas the GLI3-PAP-A construct was predicted to cause weak repression activity because of the lack of a transcription activation domain (4). To test these hypotheses, a 4-kb 5′ promoter region of the human PTCH1 gene containing several putative GLI-binding sites was coupled to a luciferase reporter and transfected into 293 cells along with GLI3 mutation constructs. The PTCH1 promoter was used for these experiments because PTCH1 is known to be regulated by GLI3 (15). The GLI3-FL transfection caused a statistically significant activation of PTCH1 reporter gene activity whereas the GLI3-PHS construct caused a dose-dependent repression of basal PTCH1 expression as well as inhibition of the transcriptional activation by GLI3-FL (Fig. 3). As predicted, GLI3-GCPS showed no effect on PTCH1 expression, attributable to its absence of DNA-binding domains and defective subcellular targeting (data not shown). The GLI3-PAP-A construct showed no repression of basal transcriptional, but it did repress PTCH1 transcription activation induced by GLI3-FL (Fig. 3). Similar data were obtained using full-length GLI1 as the activator (data not shown).
Figure 3.
Effects of mutant and full-length GLI3 constructs on the PTCH1 reporter expression. (A) GLI3 expression constructs were used to determine relative transcriptional activation or repression of PTCH1 gene expression. GLI3 constructs were cotransfected with a PTCH1 reporter construct, allowing determination of PTCH1 expression using the luciferase system. The GLI3-FL construct showed activation of PTCH1 expression that was similar to that of unprocessed Ci155. The GLI3-PHS construct showed dose-dependent repression of basal PTCH1 activity, which was consistent with the lack of a transactivation domain and the presence of a zinc finger DNA-binding domain reminiscent of the effect of Ci75. The GLI3-PAP-A construct did not consistently lower the basal level of PTCH1 expression. Asterisks denote statistical significance in level compared with vector alone; ∗, P < 0.02; ∗∗, P < 0.005; ∗∗∗, P < 0.0001 by Student’s t test or Mann–Whitney U test (see Materials and Methods). V, vector alone at 100 ng. (B) The GLI3-PHS construct caused a dose-dependent repression of GLI3-FL-induced PTCH1 promoter activity consistent with its lack of a transactivation domain and constitutive nuclear localization. In comparison, the GLI3-PAP-A construct (which was associated with a mixed cytoplasmic and nuclear localization; see Fig. 2D) did not cause a dose-dependent reduction in GLI3-FL-induced PTCH1 expression levels, although the levels are all significantly lower than the GLI3-FL-induced PTCH1 expression. Asterisks denote statistical significance in level compared with transfection of GLI3-FL (100 ng) alone; ∗, P < 0.01; ∗∗, P < 0.001; ∗∗∗, P < 0.0001 by Student’s t test or Mann–Whitney U test (see Materials and Methods).
DISCUSSION
The activation of the PTCH1 promoter by GLI3-FL is the first demonstration of transcription activation by GLI3 and is consistent with our hypothesis of a dual function (activator and repressor) for this protein (4). Furthermore, this observation suggests that GLI3 could activate downstream genes in response to sonic hedgehog (Shh) signaling. Previous studies have failed to show activation of either HNF3β promoter (hepatocyte nuclear factor 3 beta) constructs (16) or multimerized consensus GLI-binding sites by GLI3 (17, 18), leading to suggestions that GLI3 is a monofunctional transcriptional repressor. A possible explanation for the different findings is that GLI3 may cooperate with some other factor(s) for a transactivation effect and thereby possesses a distinct target-gene specificity compared with GLI1 (19). The experiments in the present report used the natural PTCH1 promoter in contrast to synthetic multimers of GLI consensus-binding sites used in previous studies. Although the absolute values of the transcriptional repression and activation in our experiments are smaller than those obtained by using synthetic promoters, they are statistically significant and are more likely to reflect true biologic effects of GLI3 than are studies that rely on synthetic promoters.
The data obtained in the present study support a model in which GLI3 mimics the bifunctional nature of the Drosophila Ci protein. The ability of mutant GLI3 proteins to repress downstream target genes in contrast to the activating effect of full-length GLI3 is consistent with the distinct developmental anomalies that are caused by these mutant proteins. It has been demonstrated that in XtJ, a mouse model for GCPS, reduced GLI3 expression leads to ectopic activation of Shh in the anterior limb bud (20), which may be mediated through the relief of repression of HNF3β (16, 21), and results in preaxial digit duplication. According to our model, in PHS and PAP-A, the expression of mutant protein would be expected to result in repression of target genes. This, in turn, may have effects on postaxial digit formation, possibly by modulating Hoxd-12 expression, which promotes the formation of posterior chondrogenic condensations (22). It will be interesting to learn whether the observed subcellular localization and transcriptional effects of mutant GLI3 proteins are an indication that GLI3 is processed physiologically to a truncated repressor in the sonic hedgehog pathway as a mechanism of posttranslational regulation or whether these mutations merely mimic the mechanism of processed and unprocessed Ci proteins. Further elucidation of the mechanism of GLI3 subcellular translocation, interaction with potential mammalian homologues of the Drosophila fused, su(fu), and costal2 gene products, and the existence or absence of GLI3 proteolytic processing in vivo will shed light on these issues. In this way, clinical and molecular analyses of human malformations can be an effective tool to elucidate mechanisms of gene action in normal and aberrant embryonic development.
Acknowledgments
We thank Drs. M. Chamorro, K. Kinzler, R. Nussbaum, W. Pavan, M. Rosenberg, S. Suchy, P. Schwartzberg, B. Vogelstein, and T. Wynshaw-Boris for technical advice, critical review of previous versions of this manuscript, and provision of Gli3 cDNA clones. Financial support for P.K., E.L., and R.T. was obtained from the Swedish Cancer Fund, the Swedish Children’s Cancer Fund, and the National Swedish Board for Radiation Protection.
ABBREVIATIONS
- PHS
Pallister–Hall syndrome
- GCPS
Greig cephalopolysyndactyly syndrome
- PAP-A
postaxial polydactyly type A
References
- 1.Vortkamp A, Gessler M, Grzeschik K-H. Nature (London) 1991;352:539–540. doi: 10.1038/352539a0. [DOI] [PubMed] [Google Scholar]
- 2.Kang S, Graham J M, Jr, Olney A H, Biesecker L G. Nat Genet. 1997;15:266–268. doi: 10.1038/ng0397-266. [DOI] [PubMed] [Google Scholar]
- 3.Radhakrishna U, Wild A, Grzeschik K-H, Antonarakis S E. Nat Genet. 1997;17:269–270. doi: 10.1038/ng1197-269. [DOI] [PubMed] [Google Scholar]
- 4.Biesecker L G. Nat Genet. 1997;17:259–260. doi: 10.1038/ng1197-259. [DOI] [PubMed] [Google Scholar]
- 5.Ruppert J M, Vogelstein B, Arheden K, Kinzler K W. Mol Cell Biol. 1990;10:5408–5415. doi: 10.1128/mcb.10.10.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aza-Blanc P, Ramírez-Weber F-A, Laget M-P, Schwartz C, Kornberg T B. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- 7.Robbins D J, Nybakken K E, Kobayashi R, Sisson J C, Bishop J M, Thérond P P. Cell. 1997;90:225–234. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- 8.Monnier V, Dussillol F, Alves G, Lamour-Isnard C, Plessis A. Curr Biol. 1998;8:583–586. doi: 10.1016/s0960-9822(98)70227-1. [DOI] [PubMed] [Google Scholar]
- 9.Ruiz i Altaba A. Cell. 1997;90:193–197. doi: 10.1016/s0092-8674(00)80325-6. [DOI] [PubMed] [Google Scholar]
- 10.Biesecker L G, Graham J M., Jr J Med Genet. 1996;33:585–589. doi: 10.1136/jmg.33.7.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finnigan D P, Clarren S K, Haas J E. Am J Med Genet. 1991;40:395–400. doi: 10.1002/ajmg.1320400403. [DOI] [PubMed] [Google Scholar]
- 12.Wild A, Kalff-Suske M, Vortkamp A, Bornholdt D, König R, Grzeschik K-H. Hum Mol Genet. 1997;6:1979–1984. doi: 10.1093/hmg/6.11.1979. [DOI] [PubMed] [Google Scholar]
- 13.Hahn H, Wicking C, Zaphiropoulous P G, Gailani M R, Shanley S, Chidambaram A, Vorechovsky I, Holmberg E, Unden A B, Gillies S, et al. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 14.Lee J, Platt K A, Censullo P, Ruiz i Altaba A. Development. 1997;124:2537–2552. doi: 10.1242/dev.124.13.2537. [DOI] [PubMed] [Google Scholar]
- 15.Buscher D, Bosse B, Heymer J, Rüther U. Mech Dev. 1997;62:175–182. doi: 10.1016/s0925-4773(97)00656-4. [DOI] [PubMed] [Google Scholar]
- 16.Sasaki H, Hui C, Nakafuku M, Kondoh H. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- 17.Vortkamp A, Gessler M, Grzeschik K H. DNA Cell Biol. 1995;14:629–634. doi: 10.1089/dna.1995.14.629. [DOI] [PubMed] [Google Scholar]
- 18.Yoon J W, Liu C Z, Yang J T, Swart R, Iannaccone P, Walterhouse D. J Biol Chem. 1998;273:3496–3501. doi: 10.1074/jbc.273.6.3496. [DOI] [PubMed] [Google Scholar]
- 19.Liu F, Massagué J, Ruiz i Altaba A. Nat Genet. 1998;20:325–326. doi: 10.1038/3793. [DOI] [PubMed] [Google Scholar]
- 20.Masuya H, Sagai T, Moriwaki K, Shiroishi T. Dev Biol. 1997;182:42–51. doi: 10.1006/dbio.1996.8457. [DOI] [PubMed] [Google Scholar]
- 21.Chang B E, Blader P, Fischer N, Ingham P W, Strahle U. EMBO J. 1997;16:3955–3964. doi: 10.1093/emboj/16.13.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knezevic V, De Santo R, Schughart K, Huffstadt U, Chiang C, Mahon K, Mackem S. Development. 1997;124:4523–4536. doi: 10.1242/dev.124.22.4523. [DOI] [PubMed] [Google Scholar]