Abstract
Leishmania are evolutionarily ancient protozoans (Kinetoplastidae) and important human pathogens that cause a spectrum of diseases ranging from the asymptomatic to the lethal. The Leishmania genome is relatively small [≈34 megabases (Mb)], lacks substantial repetitive DNA, and is distributed among 36 chromosomes pairs ranging in size from 0.3 Mb to 2.5 Mb, making it a useful candidate for complete genome sequence determination. We report here the nucleotide sequence of the smallest chromosome, chr1. The sequence of chr1 has a 257-kilobase region that is densely packed with 79 protein-coding genes. This region is flanked by telomeric and subtelomeric repetitive elements that vary in number and content among the chr1 homologs, resulting in an ≈27.5-kilobase size difference. Strikingly, the first 29 genes are all encoded on one DNA strand, whereas the remaining 50 genes are encoded on the opposite strand. Based on the gene density of chr1, we predict a total of ≈9,800 genes in Leishmania, of which 40% may encode unknown proteins.
The Kinetoplastidae are flagellated protozoans found in terrestrial and aquatic environments that cause diseases in organisms ranging from plants to vertebrates. These diseases result in widespread human suffering and death, as well as considerable economic loss from infection of livestock, wildlife, and crops. In addition, kinetoplastids have been particularly valuable for the study of fundamental molecular and cellular phenomena, such as RNA editing (1), mRNA transsplicing (2), glycosylphosphatidylinositol-anchoring of proteins (3), antigenic variation (4), and telomere organization (5). The early evolutionary divergence of these organisms makes comparison of their sequences with those of other eukaryotes, as well as prokaryotes, useful for the identification of ancient conserved motifs, and their protein sequences may be a useful source of diversity for protein engineering.
The numerous human-infective Leishmania spp. cause a spectrum of diseases with pathologies ranging from the asymptomatic to the lethal, and there are correlations between species and disease type and severity (6). The Leishmania haploid genome content is ≈34 megabases (Mb; ref. 7), consisting of 36 chromosomes ranging in size from 0.3 Mb to 2.5 Mb (8). It contains ≈30% repeated sequence (9), half of which is a series of telomeric hexamer repeats, whereas the remainder comprises other simple sequence repeats, transposons, as well as tandem and dispersed gene families such as rRNA, spliced-leader, tubulin, and gp63. The Leishmania molecular karyotype is conserved between Leishmania strains and species (10) with most genes syntenic among species (8). There are modest chromosome size polymorphisms between strains and larger size polymorphisms between species. Thus, this organism is an ideal candidate for a genome-sequencing project to elucidate its full genetic complement. The Leishmania Genome Network, established with the support of the World Health Organization, initiated a coordinated effort to map and sequence the Leishmania genome (see www.ebi.ac.uk/parasites/leish.html). Leishmania major MHOM/IL/81/Friedlin (LmjF) was selected as the reference strain to be sequenced and a first-generation contig map of the LmjF genome was constructed by cosmid fingerprinting (7). We report here the complete sequencing of chromosome 1 (chr1), the smallest chromosome.
MATERIALS AND METHODS
Cosmid Mapping.
A genomic library was constructed in the shuttle cosmid cLHYG (11) by using partial Sau3AI-digested genomic DNA from LmjF, and 9,216 clones were picked from this library, arrayed, and transferred to nylon membranes (7). These filters were hybridized with clamped homogenous electric field gel electrophoresis-isolated chr1b DNA, chr1-specific sequence-tagged site probes (ST113, ST124, ST126, ST129, ST132, and ST143) from Leishmania infantum (12), and probes prepared from cosmids by PCR amplification with primers derived from cosmid end sequencing (L549-T7 PCR, L915-T3 PCR, L1439-T7, L2602-T7 PCR, L2759-T7, L4171T7, L549-T3 PCR, L5754-T3 PCR, L5842-T3, and L8856-T7) or cosmid subclones (1018C6N/St, 1018F7, 1020A10, 1020A10, 1020A11, 1020A12, 1020B1, 1025D6, 1025F1, L1439AB, L5701P25, and L7563AB). Hybridizations were done in 1 M NaCl, 1% SDS, and 100 μg/ml denatured salmon sperm DNA for 18 h at 65°C. Filters were washed twice for 20 min in 2× standard saline citrate (0.15 M sodium chloride/0.015 M sodium citrate, pH 7) and 1% SDS at 65°C and then twice for 20 min in 0.1× standard saline citrate (0.15 M sodium chloride/0.015 M sodium citrate, pH 7) and 0.2% SDS at 65°C. Hybridization data were compiled with segmap (13), and maps were compared manually to those obtained by cosmid fingerprinting (7). Precise map locations of cosmid clones were determined by comparing their end sequences with the consensus sequence obtained from selected cosmids.
Cosmid Sequencing.
Intact cosmid DNA (L549, L2602, L2759, L4171, L5701, L1439, L3162, L9003, and L9259) or restriction fragments thereof (L2602, L1231, and L3169) were sheared by sonication, repaired with the Klenow fragment of Escherichia coli DNA polymerase and T4 polynucleotide kinase, and fractionated by agarose gel electrophoresis. Fragments of 0.8–2.0 kilobase (kb) were purified and ligated to SmaI-digested, phosphatase-treated pBluescriptII SK(−) DNA (Stratagene; L549, L2602, L2759, L1231, L4171, L5701, L3169, and L1439) or M13mp18 replicative form DNA (L549, L2602, L3162, L9003, and L9259). Clones obtained from each ligation were screened for the presence of inserts (indicated by the color white in the presence of isopropyl β-d-thiogalactoside), and 400–1,000 clones were selected for automated DNA sequencing. Plasmid clones were sequenced in both directions by using dyeTerminator chemistry with KS (5′-CGAGGTCGACGGTATCG-3′), ZR30 (5′-CGGTGGCGGCCGCTCTA-3′), and SMR29 (5′-GCTCCACCGCGGTGGC-3′) primers; M13 clones were sequenced by using dyeTerminator and dyePrimer chemistry with −21M13 (5′-GTAAAACGACGGCCAGT-3′) primer. Cosmid end sequences were obtained from cosmid DNA by using dyeTerminator chemistry with HygT3 (CCGTTGCGCCGTAGAAG) or HygT7a (CGATGATAAGCTGTCAAACATG) primers. Generally, 600–1,000 sequencing reactions were performed during the shotgun phase before assembly by using the phred/phrap/consed software suite (14). Gaps and regions of poor sequence quality were finished by sequencing existing clones with different chemistries and/or different primers or by sequencing PCR products amplified from the cosmid DNA. The clones L1231 and L3162 contained tandem repeats that failed to assemble correctly with phrap; these regions were assembled manually by using the program seqman (DNASTAR, Madison, WI). The clones L9003 and L9259 contained large (>10-kb) regions that consisted of tandem and interspersed repeats that failed to assemble correctly. Only the first two copies of the 272/282-bp repeat from L9003 are included in the chr1 consensus sequence. The sequence of L9259 has not yet been determined definitively and was not included. Sequence from a clone containing telomeric and subtelomeric sequences, obtained by using a recently reported PCR-based method (15, 16), overlapped with the “left” end of L549 and was included in the final chr1 consensus. This sequence has been deposited in the GenBank sequence database (accession no. AE001274).
Sequence Analysis.
The chr1 consensus sequence was examined for putative protein-coding ORFs by using the GCG programs testcode and codonpreference. The predicted amino acid sequences from each putative gene were used for blastp searches of the nonredundant protein databases and tblastn searches of all the nucleotide databases (see www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-newblast?Jform=0). Generally, hits with blast scores of >50 and e values of <1 × e−6 were considered potentially significant, although some exceptions were made on visual inspection of the alignments. blast searches of the Clustered Orthologous Group database (www.ncbi.nlm.nih.gov/COG/cognitor.html) and hmm searches of the Pfam 2.1 database (genome.wustl.edu/Pfam/cgi-bin/hmm_page.cgi) were also performed. Each protein sequence was scanned for motifs in the prosite database by using generunner 3.0 and membrane-spanning domains were predicted by using tmpred (ulrec3.unil.ch/software/TMPRED_form.html).
RESULTS AND DISCUSSION
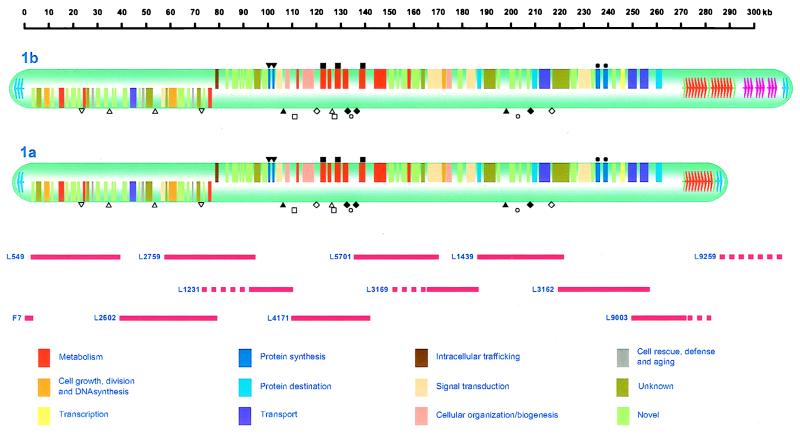
A cosmid contig map of LmjF chr1 was constructed by a combination of cosmid fingerprinting, hybridization with chr1-specific probes, and cosmid end sequencing, and 11 cosmids representing a tile-path for most of chr1 were selected for shotgun sequencing (see Fig. 1). As the cosmid library was underrepresented for telomeric sequences, clones containing telomeric and subtelomeric sequences were obtained by using a recently reported PCR-based method (15, 16). The sequences obtained from 10 cosmid clones and 1 telomere clone were assembled into a final consensus sequence of 268,984 bp, which represents 94% of the sequence of the smaller homologue, chr1a. The balance comprises telomeric and subtelomeric repeats (see below). The assembled consensus sequence was validated by Southern blot analysis of LmjF genomic DNA by using the cosmid DNAs as probes. In all cases, the restriction patterns matched those predicted from the sequence (data not shown). Within the 45,095 bp of chr1 sequence represented in overlaps between individual cosmid sequences, only 22 nucleotide differences were observed. With only one exception, these were confined to two overlaps (L4171/L5701 and L3162/L9003) and occurred only in intergenic regions. The data suggest that the overlapping cosmids with sequence differences may be derived from the different chr1 homologs. Thus, with the exception of the subtelomeric regions (see below), the sequences of the chr1 homologs seem to be almost identical. This apparently low level of allelic variation could perhaps be explained by frequent recombination between homologous chromosomes and the absence of detectable sexual reproduction in Leishmania.
Figure 1.
Gene organization of LmjF chr1. The location and coding strand of the 79 protein-coding ORFs are indicated by the boxes, which are color-coded according to functional categories that were adapted from the Saccharomyces cerevisiae functional catalogue (muntjac.mips.biochem.mpg.de/ycd/funcat/index.html). Genes encoding predicted proteins with homology to proteins of unknown function or containing uninformative motifs are classified in the “Unknown” category, and those with no homology or other identifying features are classified as “Novel”. The locations of the sequenced cosmids and PCR clones are shown below the map. Dotted lines represent the portions of cosmids L1231, L3169, L9003, and L9259 that were not sequenced fully. Repeats within the “informational” region are indicated by the following symbols: I (▿); II (▵); III (▾); IV (▴); V (▫); VI (◊); VII (▪); VIII (⧫); IX (○); and X (●). Telomeric hexamer/octamer repeat regions are indicated by blue arrows, and regions containing subtelomeric repeats are indicated by red (272-bp), green (282-bp), and magenta (LiSTIR1-related and other) arrows.
Analysis of the sequence revealed 908 ORFs greater than 225 nt (75 aa) in length. The GCG programs testcode and codonpreference (which calculate period-three constraint and third-codon-position GC bias, respectively) accurately predict protein-coding ORFs in Leishmania (17, 18). When these analyses were applied to the chr1 sequence, 79 putative protein-coding genes were identified (Fig. 1). These genes are contained within a 257,163-bp informational region, flanked by 3,703-bp and 8,118-bp noninformational regions at the left and right ends, respectively, which are in turn flanked by telomeric and subtelomeric repetitive sequences. No genes encoding structural RNAs were identified. As seen for other protein-coding regions in Leishmania and other Kinetoplastidae, no evidence for introns was found. Much of the informational region (55%) is protein-coding, and the ORFs are flanked by pyrimidine-rich tracts. All of the intergenic regions contained several tracts of 10 or more pyrimidine nucleotides (Y10), and all but three (TKRP1-L549.8, L3169.1-DDI1, and L3169.3-L3169.4) contained one or more tracts of ≥15 consecutive pyrimidine nucleotides (Y15). Pyrimidine tracts of this size were rare within the protein-coding ORFs (only one contained a Y15 tract). Polypyrimidine tracts provide at least part of the processing signals for the addition of the 39-nt spliced-leader (or mini-exon) sequence that is transspliced to the 5′ end of all mRNAs (2) and for 3′ polyadenylation (19–21).
Perhaps the most remarkable feature of the chr1 sequence is the gene organization. From the left end of the chromosome, the first 29 genes are all located on the same DNA strand (between 6 and 79 kb from the left telomere), whereas the remaining 50 genes are all on the other strand (between 81 and 263 kb from the left telomere). As far as we are aware, this sequence is the only reported case of two “head-to-head” units of protein-coding genes that span an entire chromosome with absolute strand polarity. This organization is consistent with the polycistronic gene organization of trypanosomatid protein-coding genes seen previously in smaller regions (22), and the organization of LmjF chr3 seems similar, except that the units are organized in a “tail-to-tail” manner (data not shown). Very little is known about the transcription of protein-coding genes by RNA polymerase II (pol II) in trypanosomatids, other than that it is polycistronic. Although a few candidates for pol II promoters have been reported (23–25), their identification remains controversial. Regulation of gene expression in these parasites is primarily posttranscriptional, occurring at the levels of transsplicing, polyadenylation, mRNA stability, translation, or protein stability (26). Transcriptional regulation has been observed only in cases of specialized pol I promoters (e.g., variable surface glycoproteins and procyclic acidic repetitive proteins; ref. 26) and then only at the level of transcript elongation. There are two general possibilities that exist for the transcription of chr1. There may be a single pol II promoter region upstream of each unit of colinear genes, where transcription initiates and proceeds toward each telomere. Alternatively, transcription may initiate at multiple sites along the chromosome. Indeed in the extreme, initiation may occur somewhat randomly on both strands, resulting in the transcription of most, or all, of their length. At this time, it is uncertain whether the coding-strand polarity reflects transcriptional processes, the cotranscriptional nature of mRNA processing in trypanosomatids, or other processes. The signals for polyadenylation and transsplicing of adjacent genes are colocalized, and the two processes seem to be linked (19–21). Thus, processing of transcripts from protein-coding genes that are clustered on the same DNA strand may be more efficient than if they were dispersed. Experiments to investigate transcription along chr1 are underway. The region where the coding-strand changes (between XPP and PAXP) is of particular interest, because it is an obvious candidate for a transcription initiation site for both (divergent) sets of genes. However, another interesting, and not necessarily unrelated, possibility is that the region between XPP and PAXP may contain a replication origin and/or centromere. The size of this region (1.6 kb) is within the range of other intergenic regions (0.8–4.6 kb). Interestingly, however, it was found (along with the neighboring region between XPP and L2759.8) to have a higher sequence complexity than other intergenic regions (David Landsman, National Center for Biotechnology Information, personal communication).
The chr1 informational region contains two to three copies of 10 different repeats of 117–522 bp, which are separated by up to 200 kb (see Fig. 1), but no large (>50 bp) inverted repeats were found; three sets of these repeats (III, VII, and X) represent tandem duplication of protein-coding genes (RPS7, LCFACAS, and EIF-4A, respectively). One (VIII) represents a duplication of part of a gene (LCFACAS3) into the adjacent intergenic region, whereas the others (I, II, IV, V, VI, and IX) occur in intergenic regions. The significance of these latter groups of repeats is not known; however, they may reflect genomic rearrangement associated with chromosome evolution, or they may represent common regulatory elements.
The consensus sequence from the left end of the chr1 has 3.7 kb 5′ to the first protein-coding gene. This sequence terminates in five copies of the octamer sequence (ACCAGTAC) and eight copies of the hexamer sequence (ACCCTA), which are found at the telomeres of Leishmania chromosomes (15). Southern analyses indicate that this junction is located 2.3 kb from the end of chr1a and 3.3 kb from the end of chr1b (data not shown). The right end of the chr1 consensus sequence terminates in an undetermined number of tandem 272-bp repeats, which correspond to the 274-bp repeat described in another strain of L. major (15). The first copy of the repeat, which begins 7.6 kb from the last protein-coding gene, differs in size (282 bp) from the others and contains several regions of divergent sequence. Sequence analysis of several PCR clones (15, 16) showed that hexameric (TAGGGT) telomere repeats occur immediately adjacent to the 272/282-bp repeat array. Although we have not sequenced the entire 272-bp repeat array, Southern analysis of isolated chromosomal DNA indicates that the array covers 12.5 kb (≈46 copies) on chr1a, extending to the telomeric hexameric repeats (data not shown). The right end of chr1b contains two 272/282-bp repeat arrays (9.0 and 11.5 kb; ≈75 copies) and an ≈17-kb region that does not contain these repeats. Preliminary analysis of cosmid clone L9259, which seems to be derived from this region of chr1b, indicates that it consists of several different types of tandem and interspersed repetitive elements. One of these is similar to the 81-bp LiSTIR1 repeat described in L. infantum (27). Copies of the 272/282-bp repeat and LiSTIR1-related sequence are present on at least two other chromosomes, but the latter is not present on chr1a (data not shown). Thus, it seems that recombination between subtelomeric repeats accounts for most of the size difference between the chr1 homologs (S. Sunkin, P.K., P.J.M., and K.S., unpublished work). Whether similar recombinations provide a basis for all the commonly observed size differences between other chromosome homologs in Leishmania is uncertain. Nevertheless, the asymmetric location of the subtelomeric repeats is intriguing, as it highlights the possibility that they may have a specific function.
A combination of database searches and protein-sequence analyses was used to assign the 79 genes on chr1 to 12 different categories based on their putative biological function (Table 1); 38 (48%) were assigned specific functional roles, whereas 9 (11%) showed similarity to proteins with unknown function or contained uninformative motifs. The remaining 32 (41%) had no identifying features or similarities. The last category may either represent genes that have as yet unknown functions, perhaps specific to the trypanosomatids, or that are sufficiently diverged as to have no significant sequence similarity to their functional homologs in other species.
Table 1.
Classification of L. major chr1 genes
| Category | n |
|---|---|
| Metabolism | 11 |
| Propionyl-CoA carboxylase (α-chain) (PCACA) | |
| Pyrazinamidase/nicotinamidase (PXNC) | |
| Exopolyphosphatase (XPP) | |
| Long-chain fatty acyl CoA synthetase (LCFACAS1, 2, 3, 4, and 5) | |
| Mitochondrial tricarboxylate carrier (MTCC) | |
| Thymidylate kinase-related (TKRP1) | |
| Threonine aldolase-related (L4171.5) | |
| Cell growth, division, and DNA synthesis | 3 |
| Mitotic centromere associated kinesin (MCAK) | |
| DNA repair/recombination (DRP1) | |
| DNA repair (DDI1) | |
| Transcription | 1 |
| Zn finger transcription factor (L3162.8) | |
| Protein synthesis | 4 |
| Ribosomal protein S7 (RPS7A and B) | |
| Elongation initiation factor-4A (EIF-4A1 and 2) | |
| Protein destination | 3 |
| Mitochondrial-processing protease (MPP) | |
| Peptidyl dipeptidase DCP (DCPA) | |
| Ubiquitin-activating enzyme (UBAE) | |
| Transport | 4 |
| Chloride channel protein (CCP) | |
| Calcium-activated K+ channel (CAKC1 and 2) | |
| Unknown transporter (L1439.8) | |
| Intracellular trafficking | 1 |
| PolyA export protein (PAXP) | |
| Signal transduction | 5 |
| Peptidyl-prolyl cis-trans isomerase (cyclophilin)-related (PPCTIRP) | |
| CDC16-related (L3169.1) | |
| 150-kDa oxygen-regulated protein-related (ORP150RP) | |
| Serine-threonine kinases (STPK1, L1231.6) | |
| Cellular organization/biogenesis | 3 |
| Glycosomal membrane protein (L4171.1L) | |
| AAA family ATPase (L4171.3) | |
| Dynein light chain? (L3169.3) | |
| Cell rescue, defense, death and aging | 3 |
| Glutaredoxin-related (GRXRP1) | |
| Thioredoxin-related (TRXRP1) | |
| Arsenate reductase-related (L2602.5) | |
| Unknown | 9 |
| Mouse & human EST yeast hypothetical protein (L549.2) | |
| Heliobacter pylori hypothetical protein & Trypanosoma cruzi EST (L549.10) | |
| Yeast hypothetical protein (L2602.6) | |
| T4 bacteriophage orf1 & Trypanosoma brucei EST (L5701.8) | |
| Caenorhabditis elegans hypothetical protein (L3162.1) | |
| COG0564 gene family (L2759.7) | |
| ATP/GTP binding protein & T. cruzi EST (L1439.2) | |
| Leucine zipper-containing proteins (L2759.2, L1231.2) | |
| No Similarity | 32 |
| (L549.1, L549.4, L549.6, L549.7, L549.8, L549.12, L549.13, L549.14R, L2602.1, L2602.2, L2602.4, L2759.4, L2759.5, L2759.8, L2759.11, L2759.12, L2759.13, L2759.14, L1231.1, L1231.3, L5701.3, L5701.4, L5701.5, L5701.7, L3169.4, L1439.3, L1439.4, L1439.5, L1439.6, L3162.2, L3162.4, L3162.7) |
EST, expressed sequence tag.
Relatively few genes encoding metabolic enzymes were identified. Of particular interest are five copies of long-chain fatty acyl CoA synthetase (LCFACAS). These occur as tandem repeats, with a gene showing similarity to threonine aldolase between the first two copies. The five copies have only limited DNA sequence similarity to each other (DNA repeat VII) and show varying degrees of amino acid similarity (29–54%). They are, however, more similar to each other than to the corresponding proteins in other organisms. The most diverged LmjF LCFACAS gene (L5701.2) contains an ≈400 amino acid N-terminal extension coupled with several other smaller insertions. Sequence analysis of chr3 has identified at least one additional LCFACAS gene. These observations suggest that LCFACAS is a diverse and dispersed multicopy gene family in the Leishmania genome. Proteins encoded by different family members may have distinct specificities for different fatty acid substrates.
A small number of genes with potential roles in DNA replication, RNA transcription, and protein synthesis/processing were identified. Of particular interest is L3162.8, whose predicted protein product has similarity to a Zn finger transcription factor, because little is known about pol II transcription in the Kinetoplastidae. Duplication of the RPS7 (L1231.4 and L1231.5) and EIF-4A (L3162.5 and L3162.6) genes is not surprising, because genes encoding ribosome-associated proteins are often duplicated in Leishmania (28, 29). There were six membrane proteins, mostly transporters, identified, whereas several other proteins were predicted to contain one to seven membrane-spanning domains and hence may be membrane-associated. The two tandem copies of the putative calcium-activated K+ channel protein (L3162.9 and L9003.1) showed very little homology at the DNA level and only limited (37%) similarity at the amino acid level, suggesting that the duplication is ancient and that they may have functional differences.
ORFs encoding proteins involved in a variety of cellular processes were also identified. Several of these seem to have roles as antioxidants (e.g., thioredoxin, glutaredoxin-related, and arsenate reductase). The last is of particular interest, because arsenical drugs are used in the treatment of leishmaniasis. End sequencing of cosmids from other chromosomes has identified several other potential genes encoding proteins with putative antioxidant roles, perhaps reflecting the parasite’s localization within macrophages during the vertebrate stage of the lifecycle. The mitotic centromere-associated kinesin (encoded by L549.3) is also of interest, because little is known about Leishmania centromeres and chromosome segregation. Of the genes with no similarity to those in the protein databases, two (L1439.5 and L1439.6) seem to represent an ancient tandem duplication, because they are related (35% similarity) at the protein-sequence level but show no significant DNA sequence similarity.
In addition to the chr1 sequence reported here, we have sequenced substantial (60- to 350-kb) regions on chr3 and chr35, and the gene distribution in all three regions indicates that in Leishmania genes do not tend to cluster into prokaryote-like operons of genes with similar function. Instead, the gene organization seems to resemble that of other eukaryotes, with the notable exception of the absolute coding-strand polarity discussed above. There do seem to be, however, regions with clusters of four to six genes with as yet unidentified functions; however, because this category represents >40% of all genes identified in Leishmania, they may not be statistically significant. The genes are packed tightly within the chr1 informational region, with a density of one gene per 3.26 kb. Similar gene densities are found for the regions of chr3 and chr35 that have been sequenced. This density is somewhat lower than that in S. cerevisiae (one per 2 kb; ref. 30), comparable to that in Plasmodium falciparum (one per 4 kb; ref. 31), and higher than that in C. elegans (one per 7 kb; ref. 32). The LmjF haploid genome size is ≈34 Mb (7). If one assumes that each of the 36 chromosomes contains ≈4 kb of telomeric sequence, ≈10 kb of subtelomeric repeats, and ≈10 kb of subtelomeric noninformational DNA (as seen with chr1a), then the informational size of the haploid genome would be reduced by ≈1 Mb. Allowing a further reduction by 0.75 Mb for RNA (≈70 copies of a 10-kb repeat) and spliced-leader (100 copies of 440-bp repeat) genes, the protein-coding portion of the genome would be ≈32 Mb. Thus, a gene density of one gene per 3.26 kb of informational DNA corresponds to a total of ≈9,800 genes in the entire Leishmania genome. The number of unique genes will be somewhat less than this, however, because a significant proportion of Leishmania genes are present in more than one copy (33). This conclusion is supported by the presence of three chr1 genes (RPS7, EIF-4A, and CAKC) that occur as tandem duplicates and of another (LCFACS) that occurs in five copies. It is interesting to note that some of these duplicated genes (LCFACS and CAKC) showed little homology at the DNA level and only limited similarity at the amino acid level, suggesting that the duplications are ancient and that individual gene products may have functional differences. Given that ≈50% of the protein-coding genes on chr1 have no currently identified function, it is reasonable to infer that completion of the Leishmania genome sequence will identify 4,000–5,000 genes with potentially parasite-specific function. Thus, the completion of the sequence of an entire chromosome is a step toward the elucidation of the entire complement of Leishmania genes. This step will provide the scientific community with a valuable resource and enable an entirely different set of approaches for the development of strategies for parasite control. For instance, a more complete understanding of the metabolic pathways present in the parasite should allow more rational drug design and targeting, and the antigens discovered may prove excellent candidates for DNA vaccines. In addition, this knowledge will be extremely valuable for future study of many aspects of basic molecular biology, such as transcription by pol II promoters, regulation of gene expression, and DNA replication and chromosome segregation.
Acknowledgments
This work received financial support from the United Nations Development Program/World Bank/World Health Organization Special Program for Research and Training in Tropical Diseases (T23/181/1 ID:940509), from the Burroughs–Wellcome Fund, and from National Institutes of Health Grant R01 AI40599.
ABBREVIATIONS
- chr1
chromosome 1
- kb
kilobase
- LmjF
Leishmania major MHOM/IL/81/Friedlin
- Mb
megabase
- pol II
RNA polymerase II
Footnotes
References
- 1.Stuart K. Annu Rev Microbiol. 1991;45:327–344. doi: 10.1146/annurev.mi.45.100191.001551. [DOI] [PubMed] [Google Scholar]
- 2.Perry K, Agabian N. Experientia. 1991;47:118–128. doi: 10.1007/BF01945412. [DOI] [PubMed] [Google Scholar]
- 3.Krakow J L, Hereld D, Bangs J D, Hart G W, Englund P T. J Biol Chem. 1986;261:12147–12153. [PubMed] [Google Scholar]
- 4.Borst P, Rudenko G. Science. 1994;264:1872–1873. doi: 10.1126/science.7516579. [DOI] [PubMed] [Google Scholar]
- 5.Blackburn E H. Nature (London) 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 6.Shaw J J, Lainson R. In: The Leishmaniases in Biology and Medicine. Peters W, Killick-Kendrick R, editors. Vol. 1. London: Academic; 1987. pp. 291–361. [Google Scholar]
- 7.Ivens A C, Lewis S M, Bagherzadeh A, Zhang L, Chang H M, Smith D F. Genome Res. 1998;8:135–145. doi: 10.1101/gr.8.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wincker P, Ravel C, Blaineau C, Pages M, Jauffret Y, Dedet J, Bastien P, Dedet J P. Nucleic Acids Res. 1996;24:1688–1694. doi: 10.1093/nar/24.9.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis J, Crampton J. In: Leishmaniasis: The Current Status and New Strategies for Control. Hart D T, editor. New York: Plenum; 1989. pp. 589–596. [Google Scholar]
- 10.Bastien P, Blaineau C, Pagès M. Subcell Biochem. 1992;18:131–187. doi: 10.1007/978-1-4899-1651-8_5. [DOI] [PubMed] [Google Scholar]
- 11.Ryan K A, Dasgupta S, Beverley S M. Gene. 1993;131:145–150. doi: 10.1016/0378-1119(93)90684-u. [DOI] [PubMed] [Google Scholar]
- 12.Ravel C, Macari F, Bastien P, Pages M, Blaineau C. Mol Biochem Parasitol. 1995;69:1–8. doi: 10.1016/0166-6851(94)00166-k. [DOI] [PubMed] [Google Scholar]
- 13.Bouffard G G, Idol J R, Braden V V, Iyer L M, Cunningham A F, Weintraub L A, Touchman J W, Mohr-Tidwell R M, Peluso D C, Fulton R S, et al. Genome Res. 1997;7:673–692. doi: 10.1101/gr.7.7.673. [DOI] [PubMed] [Google Scholar]
- 14.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, Bonfield J, Burton J, Connell M, Copsey T, Cooper J, et al. Nature (London) 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 15.Fu G, Barker D C. Nucleic Acids Res. 1998;26:2161–2167. doi: 10.1093/nar/26.9.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu G, Barker D C. BioTechniques. 1998;24:386–390. doi: 10.2144/98243bm10. [DOI] [PubMed] [Google Scholar]
- 17.Myler P J, Lodes M J, Merlin G, deVos T, Stuart K D. Mol Biochem Parasitol. 1994;66:11–20. doi: 10.1016/0166-6851(94)90031-0. [DOI] [PubMed] [Google Scholar]
- 18.Myler P J, Venkataraman G M, Lodes M J, Stuart K D. Gene. 1994;148:187–193. doi: 10.1016/0378-1119(94)90688-2. [DOI] [PubMed] [Google Scholar]
- 19.LeBowitz J H, Smith H Q, Rusche L, Beverley S M. Genes Dev. 1993;7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- 20.Ullu E, Matthews K R, Tschudi C. Mol Cell Biol. 1993;13:720–725. doi: 10.1128/mcb.13.1.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthews K R, Tschudi C, Ullu E. Genes Dev. 1994;8:491–501. doi: 10.1101/gad.8.4.491. [DOI] [PubMed] [Google Scholar]
- 22.Swindle J, Tait A. In: Molecular Biology of Parasitic Protozoa. Smith D F, Parsons M, editors. Oxford: Oxford Univ. Press; 1996. pp. 6–34. [Google Scholar]
- 23.Wong A K C, Curotto de Lafaille M A, Wirth D F. J Biol Chem. 1994;269:26497–26502. [PubMed] [Google Scholar]
- 24.Lee M G S. Mol Cell Biol. 1996;16:1220–1230. doi: 10.1128/mcb.16.3.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dresel A, Clos J. Exp Parasitol. 1997;86:206–212. doi: 10.1006/expr.1997.4161. [DOI] [PubMed] [Google Scholar]
- 26.Pays E, Vanhamme L. In: Molecular Biology of Parasitic Protozoa. Smith D F, Parson M, editors. Oxford: Oxford Univ. Press; 1996. pp. 88–114. [Google Scholar]
- 27.Ravel C, Wincker P, Bastien P, Blaineau C, Pagès M. Mol Biochem Parasitol. 1995;74:31–41. doi: 10.1016/0166-6851(95)02480-8. [DOI] [PubMed] [Google Scholar]
- 28.Myler P J, Tripp C A, Thomas L, Venkataraman G M, Merlin G, Stuart K D. Mol Biochem Parasitol. 1993;62:147–152. doi: 10.1016/0166-6851(93)90192-z. [DOI] [PubMed] [Google Scholar]
- 29.Soto M, Requena J M, Garcia M, Gómez L C, Navarrete I, Alonso C. J Biol Chem. 1993;268:21835–21843. [PubMed] [Google Scholar]
- 30.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq C, Johnston M, et al. Science. 1996;274:546. doi: 10.1126/science.274.5287.546. , 563–567. [DOI] [PubMed] [Google Scholar]
- 31.Gardner M J, Tettelin H, Carucci D J, Cummings L M, Aravind L, Koonin E V, Shallom S, Mason T, Yu K, Fujii C, et al. Science. 1998;282:1126–1132. doi: 10.1126/science.282.5391.1126. [DOI] [PubMed] [Google Scholar]
- 32.The C. elegans Sequencing Consortium. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 33.Tait A. Parasitol. 1983;86:29–57. doi: 10.1017/s0031182000050836. [DOI] [PubMed] [Google Scholar]