Abstract
The human PTEN tumor suppressor gene is mutated in a wide variety of sporadic tumors. To determine the function of PTEN in vivo we have studied a PTEN homolog in Caenorhabditis elegans. We have generated a strong loss-of-function allele of the PTEN homolog and shown that the deficient strain is unable to enter dauer diapause. An insulin-like phosphatidylinositol 3-OH kinase (PI3′K) signaling pathway regulates dauer-stage entry. Mutations in either the daf-2 insulin receptor-like (IRL) gene or the age-1 encoded PI3′K catalytic subunit homolog cause constitutive dauer formation and also affect the life span, brood size, and metabolism of nondauer animals. Strikingly, loss-of-function mutations in the age-1 PI3′K and daf-2 IRL genes are suppressed by loss-of-function mutations in the PTEN homolog. We establish that the PTEN homolog is encoded by daf-18, a previously uncloned gene that has been shown to interact genetically with the DAF-2 IRL AGE-1 PI3′K signaling pathway. This interaction provides clear genetic evidence that PTEN acts to antagonize PI3′K function in vivo. Given the conservation of the PI3′K signaling pathway between C. elegans and mammals, the analysis of daf-18 PTEN mutant nematodes should shed light on the role of human PTEN in the etiology of metabolic disease, aging, and cancer.
The human PTEN (also known as Mutated in Multiple Advanced Cancers 1) tumor suppressor gene was cloned because of its frequent mutation in late stage glioblastomas and prostate tumors (1, 2). Subsequent studies have identified PTEN mutations in a wide variety of sporadic human cancers, including endometrial tumors (3), malignant melanomas (4), and bladder carcinomas (5). Germ-line human PTEN mutations also have been identified as the underlying genetic lesions of both Cowden disease (6) and Bannayan–Zonana syndrome (7). These autosomal dominant syndromes result in a high predisposition to hamartomas, benign tumors that contain differentiated but disorganized cells. Cowden disease also is characterized by an increased incidence of breast and thyroid tumors. This predisposition to malignancies is recapitulated in mice carrying targeted mutations within the murine PTEN gene (8). PTEN heterozygous mutant mice develop dysplasias in a wide spectrum of tissues and have a high incidence of prostate and colon tumors (8, 9). PTEN homozygous mutant mouse embryos die between embryonic day 7.5 and 9.5 with patterning defects (8, 9), an overgrowth of cells in the caudal and cephalic regions of the embryo, and defects in placentation (9). Thus, PTEN is required for a number of developmental processes, but the mechanism by which it exerts its effects is unknown.
The predicted human PTEN protein sequence contains a 200-aa region called a TAG domain (10) that shares homology with tensin, auxilin, and G cyclin-associated kinase (1, 2). Nested within the TAG domain is a dual-specificity protein phosphatase signature motif (refs. 1 and 2; Fig. 1). In vitro studies have shown that human PTEN displays phosphatase activity against a number of substrates, including focal adhesion kinase (11). PTEN preferentially dephosphorylates phosphoamino acids surrounded by acidic residues (12). Consistent with this preference, PTEN can dephosphorylate the D3 position of the acidic substrate phosphatidylinositol (3,4,5) triphosphate (PtdIns-3,4,5-P3; ref. 13), a product of phosphatidylinositol 3-OH kinase (PI3′K; ref. 14).
Figure 1.
Sequence analysis of the Caenorhabditis elegans PTEN homolog and structure of the nr2037 deletion allele. The C. elegans PTEN homolog from cosmid T07A9 is aligned with human PTEN. Identical amino acids are indicated in bold; identities and conservative changes are shaded. The phosphatase signature motif and probable catalytic aspartic acid are indicated by a box. Conserved tyrosine phosphorylation sites are indicated by asterisks. Amino acids encoded by sequences deleted in the nr2037 allele are overlined. The deletion removes bp 1,991–2,980 according to GenBank’s numbering of cosmid T07A9.
In C. elegans, a PI3′K signal transduction pathway modulates the ability of nematodes to enter the developmental dauer-larval stage (15). The insulin/IGF-I receptor-like (IRL; IGF-I is insulin-like growth factor I) molecule DAF-2 (16) transduces a signal via a PI3′K, AGE-1 (15), and two Akt homologues, AKT-1 and AKT-2 (17), to prevent dauer formation induced by the DAF-16 transcription factor (17–19). Loss-of-function mutations in daf-2 IRL or age-1 PI3′K lead animals to form dauers constitutively and arrest development (20, 21). In adult animals, disruption of daf-2 IRL or age-1 PI3′K increases fat storage as well as longevity (16, 22). The modulation of nematode metabolism by an insulin-like pathway is similar to the control of mammalian glucose homeostasis by insulin (16). This similarity suggests that evolutionarily conserved factors that impinge on the DAF-2 IRL AGE-1 PI3′K signaling cascade in C. elegans may play a causal role in human metabolic diseases, such as type II diabetes (16), in which insulin signaling is impaired.
To study the in vivo function of PTEN, we have identified a C. elegans PTEN homolog. We find that animals that carry a deletion in the PTEN gene are unable to form dauers and that this mutation suppresses mutations in daf-2 IRL and age-1 PI3′K. The PTEN homolog maps close to a gene known to be required for dauer formation, daf-18 (20), and both sequence and genetic analyses indicate that daf-18 encodes the PTEN homolog. Thus, the C. elegans PTEN homolog acts to antagonize signaling through the PI3′K pathway. Intriguingly, daf-18 PTEN mutant animals exhibit a number of secondary phenotypes that may be independent of the DAF-2 IRL AGE-1 PI3′K signaling cascade. This pleiotropy suggests that PTEN may play a role in a second signal transduction pathway in vivo. We discuss the implications of C. elegans daf-18 PTEN biology for cancer and type II diabetes.
MATERIALS AND METHODS
Isolation of a PTEN Deletion Mutant and Sequence Analysis of the daf-18(e1375) Allele.
A library of C. elegans mutants was generated by trimethylpsoralen treatment and UV irradiation (L.X.L., J. Spoerke, E. Mulligan, J. Chen, B. Reardon, M. Basson, R. Clover, and C.D.J., unpublished work). Worm populations were arrayed in microtiter plates with 20 F1 animals per well, and genomic DNA was sampled from pools of nematodes. This DNA was subjected to PCR analysis by using nested primers designed to assay for deletions in the phosphatase and TAG homology domain of the PTEN homolog. External primers were ATGCAGTGAGAAGAGAGGCGTTCG and ACCGACTCCTCGAATATCTCCAC, and internal primers were ACCCGTGCCGTTTGAATTAGC and CCTGATTCCGTGTATGATTTGTCTC. A deletion amplicon of 2.2 kb (as opposed to a wild-type PCR product of 3.2 kb) was identified from a library of 192,000 genomes that had been exposed to mutagens. Sibling selection and single-worm PCR were used to isolate individual worms bearing the deletion mutation, which was given the allele name nr2037. The sequence of the PTEN homolog in daf-18(e1375) was determined by single-worm PCR amplification of exons and intron–exon junctions, followed by direct sequencing.
Strains and Maternal Rescue.
The mutations used in this study were, for linkage group II, age-1(m333, mg44), sqt-1(sc13), and mnC1[dpy10(e128) and unc-52(e444)]; for linkage group III, daf-2(e1370ts); and for linkage group IV, daf-18(e1375 and nr2037) and dpy9(e12). Mutations are described in ref. 23, except for daf-18(nr2037), whose generation is described above. Strains were provided by the Horvitz lab (Massachusetts Institute of Technology, Cambridge, MA) or by the Caenorhabditis Genetics Center (St. Paul, MN). Single-worm PCR was performed for the nr2037 deletion allele with the primers GATTGGTGTCTACGTGGAACGG,GCCAACGAAGTGCTAAATCGAC, and AATTCAATCGTCGAGCGG. The combination of these three primers yielded wild-type amplicons of 718 bp and 2,077 bp and/or a deletion amplicon of 1,087 bp. Single-worm PCR was performed for the daf-18(e1375) allele with the primers AAGTGTCGACATTTGACGGCTCCTCTACTG and CTTCGAGATGTACTGCGTTGC. The mutant amplicon was 28 bp larger than the wild-type amplicon and could be separated on a 3% agarose gel. Details concerning the construction of daf-2;daf-18 or age-1;daf-18 double-mutant animals are available on request. The presence of daf-2 or age-1 in the double-mutant animals was confirmed by complementation. Maternal rescue effects were delineated by inspecting the self-progeny of age-1(m333)/age-1(m333);daf-18/+ hermaphrodites.
Germ-Line Transformation.
Germ-line transformation was performed as described in ref. 24, with DNA concentrations of 100 μg/ml or 20 μg/ml T078KBSP or 100 μg/ml pRF4 (see Table 3). T078KBSP contains an 8-kb SmaI–SacII subclone of C. elegans cosmid T07A9. The pRF4 plasmid contains the rol-6(su1006) allele as a dominant marker (24). Rol progeny from injected adults were picked singly to plates, and lines that segregated the extrachromosomal arrays were used in the phenotypic tests.
Table 3.
Rescue of daf-18(e1375) by an 8-kb subclone of cosmid T07A9
| Genotype | Rol animals, no.*
|
Non-rol, no.†
|
||
|---|---|---|---|---|
| Dauers | Adults/L4 | Dauers | Adult/L4 | |
| daf-2 | — | — | 134 | 0 |
| daf-2; daf-18 | — | — | 3‡ | 392 |
| daf-2; daf-18; array 1 | 21 | 0 | 0 | 16 |
| daf-2; daf-18; array 2 | 37 | 0 | 5 | 21 |
| daf-2; daf-18; array 3 | 33 | 0 | 1§ | 9 |
| daf-2; daf-18; array 4 | 7 | 0 | 3 | 15 |
Adults hermphrodites (n = 4–6) were allowed to lay eggs for 5–6 h. Progeny were placed at 25°C and scored visually 78 h after eggs were laid. These data are from one representative experiment. Experiments were performed at least twice.
Transgenic animals were marked with the rol-6 transgene.
Non-roll animals are largely not transgenic, although a small proportion may be mosaic for the extrachromosomal array.
These were “partial dauers.” They were lighter and moved more than true dauers.
This animal looked like an L2d larva, because it was darker than a normal L2 larva.
Analysis of the Dauer-Defective Phenotype and Complementation Tests.
For assay of dauer formation under starved, crowded conditions, animals were picked singly to a nematode growth media-agar plate. Animals were allowed to deplete the bacteria on the plate, and 5 days later, they were treated with 3 ml of 1% SDS. Plates were inspected visually 30–60 min later for live dauers. For complementation tests, nr2037/+ males were mated to e1375 homozygote hermaphrodites. Individual hermaphrodite progeny were assayed for dauer formation as above. Plates were scored for the presence of the nr2037 deletion by PCR analysis of a “scoop” of progeny by using deletion-specific primers. Initial complementation tests used the DR235 strain, which has been designated daf-18(e1375) dpy-9(e12). However, the DR235 and daf-18(e1375) (strain CB1375 from the Caenorhabditis Genetics Center) complemented each other in a dauer-formation test. Furthermore, DR235 animals did not burst. These results indicate that DR235 does not contain the e1375 mutation.
Determination of Brood Size.
L4 larvae were picked singly to a nematode growth media-agar plate and grown at 25°C. Adults were moved once a day for 3 days. Progeny were counted 40–48 h after eggs were laid. Numbers of progeny were counted on at least 4 days for each hermaphrodite parent in each experiment.
Analysis of the Dauer-Constitutive and Bursting Phenotypes.
For each genotype, 1–10 adults were allowed to lay eggs for 5–6 h. Progeny were grown at 25°C and inspected visually for dauers, L4 larva, or adults 54–56 h later. Where applicable, progeny were inspected again at 78–80 h and 102–104 h later for dauer morphology or the bursting phenotype.
RESULTS
Mutation of the C. elegans PTEN Homolog Prevents Entry into the Dauer Stage.
A search of A Caenorhabditis elegans Database (ACeDB) identified a homolog of the human PTEN gene on cosmid T07A9 (GenBank accession no. AF036706). The sequence of the genomic locus and that of a full-length cDNA (yk400b8, kindly provided by Y. Kohara, National Institude of Genetics, Mishima, Japan) predict a 962-aa protein (Fig. 1). The C. elegans protein is 43% identical to human PTEN over the 200-aa TAG domain and differs in only 1 of 11 residues in the phosphatase signature motif. There is significantly less homology in the C-terminal portion of these proteins, but a potential tyrosine phosphorylation site is conserved. This site is mutated specifically in a tumor-derived form of human PTEN, suggesting that it plays an important role in the regulation of this protein (2).
To isolate a defective PTEN allele, we used a PCR-based approach (ref. 25; L.X.L., J. Spoerke, E. Mulligan, J. Chen, B. Reardon, M. Basson, R. Clover, and C.D.J., unpublished work) to screen a library of C. elegans mutants for animals carrying a deletion in the TAG/phosphatase domain. We identified a 990-bp deletion, designated nr2037, that spans sequences from intron 2 to exon 3 (Fig. 1). This deletion removes much of the TAG domain, including the entire phosphatase signature motif. We believe nr2037 to be a null mutation of the C. elegans PTEN homolog. Animals carrying the nr2037 deletion were outcrossed seven times and subjected to phenotypic analysis. These mutants were found to have a number of partially penetrant phenotypes. A small proportion of the nr2037 animals was sterile, was defective in laying eggs, or had a tendency to form “bags-of-worms” (data not shown). In addition, 17% of these mutants burst from the vulva as adults (Table 1). None of the nr2037 animals were able to form dauers, as judged by SDS resistance (Table 1). We therefore conclude that the C. elegans PTEN homolog is necessary for normal entry into dauer diapause.
Table 1.
Vulval bursting and complementation tests
| Genotype | Animals that burst from vulva, % | n | Number of plates with indicated number of SDS-resistant dauers
|
|||
|---|---|---|---|---|---|---|
| 0 | 1–2 | 3–10 | >100 | |||
| +/+ | 0 | 552 | 0 | 0 | 0 | 3 |
| nr2037/nr2037 | 17.8 | 337 | 5 | 0 | 0 | 0 |
| e1375/e1375 | 14.4 | 660 | 0 | 2 | 2 | 0 |
| nr2037/e1375 | ND | — | 4 | 1 | 0 | 0 |
Adult hermaphrodites of a given phenotype were allowed to lay eggs for 4–6 h. Progeny were placed at 25°C and scored 2–4 days later. ND, not determined.
The C. elegans PTEN Homolog Is a Component of the age-1/P13 Kinase Pathway.
Entry into the C. elegans dauer stage is regulated by an IRL protein DAF-2 that transduces its signal through a PI3′K homolog, AGE-1 (15, 16). Mutation of either of these genes leads to constitutive dauer formation (20, 21). PI3′K is known to phosphorylate phosphatidylinositol (3, 4)bisphosphate (16, 26) to generate PtdIns-3,4,5-P3 (14), and human PTEN can catalyze the reverse reaction in vitro (13). We hypothesized that the dauer-formation defect of nr2037 mutant animals results from changes in the regulation of the age-1 PI3′K pathway. To test this hypothesis, we examined the phenotype of animals with both the nr2037 deletion mutation and a loss-of-function mutation in either daf-2 IRL or age-1 PI3′K. The nr2037 deletion allele completely suppressed the dauer constitutive phenotype of these daf-2 IRL and age-1 PI3′K mutations (Table 2). nr2037 also rescued the brood-size reduction that has been described for the daf-2(e1370) mutants (refs. 27 and 28; Table 2). We therefore conclude that the PTEN homolog functions either downstream of or parallel to daf-2 IRL and age-1 PI3′K in vivo. Because PtdIns-3,4,5-P3 is both the product of PI3′K (14) and a target for dephosphorylation by PTEN (13), it is likely that the PTEN homolog contributes to the regulation of dauer formation by directly affecting the levels of this phosphatidylinositol.
Table 2.
Epistasis analysis with daf-2 IRL, age-1 PI3′K, and daf-18 PTEN
| Daf-c genotype | Daf-d genotype | L4 larvae and adults, % | Dauers, % | Young larvae, % | n | Average brood size |
|---|---|---|---|---|---|---|
| + | + | 99.3 | 0.0 | 0.7 | 876 | 233.6 ± 6.0 |
| + | daf-18(e1375) | 99.8* | 0.0 | 0.2 | 1030 | 184.5 ± 17.3 |
| + | daf-18(nr2037) | 99.5* | 0.0 | 0.5 | 943 | 211.7 ± 12.8 |
| daf-2(e1370) | + | 0.0 | 97.6 | 2.4 | 449 | 32.7 ± 10.6 |
| daf-2(e1370) | daf-18(e1375) | 1.2 | 98.8† | 0.0 | 571 | 164.7 ± 22.6 |
| daf-2(e1370) | daf-18(nr2037) | 97.3* | 0.0 | 2.7 | 754 | 222.2 ± 16.9 |
| age-1(m333)‡ | + | 0.0 | 100.0 | 0.0 | 85 | ND |
| age-1(m333) | daf-18(e1375) | 99.6§ | 0.0 | 0.4 | 280 | ND |
| age-1(m333) | daf-18(nr2037) | 99.4* | 0.0 | 0.6 | 360 | ND |
| age-1(mg44)‡ | + | 0.0 | 97.4 | 0.6 | 79 | ND |
| age-1(mg44)¶ | daf-18(nr2037) | 100.0 | 0.0 | 0.0 | 44 | ND |
For dauer counts, 1–10 adults per plate were allowed to lay eggs for 5 to 6 h. Progeny were scored 54–56 h after the eggs were laid. These data are the totals of at least two assays (generally two plates per assay) conducted on different days. n is the total number of progeny scored. See Materials and Methods for details regarding brood-size assays. Mean brood sizes are shown ±SEM. Experiments were performed at least twice with one representative data set shown. There were 4–10 parental hermaphrodites scored. ND, not determined.
A few adults (0.6–1.5) had burst.
These were “partial dauers.” They were lighter and moved more spontaneously than true dauers; 99.2% grew into adults or L4 larva by 78 h after eggs were laid.
The adults that laid these eggs were the progeny of age-1/mnC1 hermaphrodites (mnC1 is a balancer for age-1). Plates were scored only if mnCl did not segregate; therefore, these are the progeny of age-1 homozygotes.
Of these, 46.9% were L4 larva. In contrast, for other genotypes that did not form dauers, the worms were overwhelmingly adults by this time.
These animals were only scored once.
daf-18 Encodes the C. elegans PTEN Homolog.
A search of A Caenorhabditis elegans Database (28) found that daf-18 (20), a known suppressor of daf-2 IRL and age-1 PI3′K mutations (21, 29, 30), maps close to the PTEN homolog. In addition to being dauer-formation defective, the only previously identified allele of daf-18, e1375, has several secondary phenotypes similar to those observed in the nr2037 animals (21, 29, 30). We showed that daf-18 corresponds to the C. elegans PTEN homolog by using four criteria.
First, we directly compared the vulval-bursting phenotype of daf-18(e1375) (30) and nr2037 animals, because it is a phenotype that has not been reported for other dauer-formation mutants (26). The daf-18(e1375) and nr2037 mutant animals were found to have a similar incidence of vulval bursting (Table 1). Animals from both strains also had a predisposition to form bags of worms, and a small proportion of mutants from each strain was sterile or defective in laying eggs (ref. 21 and data not shown).
Second, we tested whether a genomic DNA fragment encoding the C. elegans PTEN homolog could rescue the defects of the daf-18(e1375) mutant allele. For ease of scoring, this experiment was conducted with a daf-2(e1370);daf-18(e1375) double mutant. As shown in Table 3, an 8-kb subclone of cosmid T07A9, in which the PTEN homolog is the only complete ORF, was sufficient to suppress the daf-18(e1375) phenotype and restore the temperature-sensitive dauer-constitutive phenotype of the daf-2(e1370) allele in four independent transgenic lines. The rescued animals maintained the dauer state at 25°C for at least 2 weeks, then recovered, and resumed development with kinetics similar to the daf-2(e1370) control when shifted to the permissive temperature (data not shown). We therefore conclude that the C. elegans PTEN homolog is sufficient to rescue the dauer-formation defects of the daf-18 mutation.
Third, we showed that daf-18(e1375) and nr2037 fail to complement one another in a dauer-formation test (Table 1). daf-18(e1375)/nr2037 heterozygote animals formed very small numbers of dauers at a frequency similar to that of the e1375 homozygotes (Table 1). Furthermore, both daf-18(e1375) and nr2037 mutant animals show maternal rescue of the dauer-formation defects (see below).
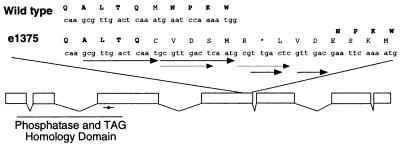
Finally, sequence analysis of the PTEN locus in daf-18(e1375) animals showed that e1375 consists of a complex series of tandem duplications in exon 4 of the C. elegans PTEN ORF (Fig. 2). The resulting alteration causes a frameshift that leads to a 6-aa insertion followed by a premature stop codon (Fig. 2). This change does not alter the sequences encoding the TAG/phosphatase domain but instead maps to the C-terminal portion of the PTEN ORF. No function has been ascribed to the C-terminal half of the PTEN protein but it is a frequent site of mutations in human tumors (1, 2, 5). These data prove that daf-18(e1375) and nr2037 are allelic and that daf-18 encodes the C. elegans homolog of the PTEN tumor suppressor.
Figure 2.
daf-18(e1375) consists of a complex series of duplications in the C. elegans PTEN homolog. Standard one-letter codes for both DNA and amino acid sequences are shown, with the DNA sequences in lower case. e1375 consists of a complex repeat structure in exon 4 of the daf-18 PTEN gene. bp 4995 is deleted, followed by a 13-bp duplication of bp 4,983–4,995 (corresponding to the new mutant sequence; based on GenBank’s numbering of T07A9). The first 10 bp of the repeat are duplicated again. This repeat is followed by a 6-bp subrepeat and then the insertion of an adenine. The complex repeat leads to a 6-aa insertion in exon 4, followed by a stop codon. The remainder of the exon is frameshifted with respect to the insertion. The active site of DAF-18 PTEN is indicated by an asterisk.
Comparison of the Dauer-Epistasis and Maternal-Rescue Effects of the Two daf-18 Alleles.
We consistently found that the daf-18(e1375) animals were able to form dauers at low frequency under conditions in which the daf-18(nr2037) animals did not enter dauer diapause (Table 1). This finding, combined with the fact that the daf-18(e1375) mutation leaves the C. elegans PTEN phosphatase domain intact, suggested that the e1375 allele may have residual PTEN activity. To address this issue, we compared the ability of the daf-18(e1375) and the daf-18(nr2037) alleles to suppress the phenotypes of daf-2 IRL or age-1 PI3′K loss-of-function mutations (Table 2). Consistent with previous studies (21, 29, 31), the e1375 mutation poorly suppressed the constitutive dauer formation of daf-2(e1370) but largely rescued the dauer-constitutive phenotype of age-1(m333) (Table 2). The suppression of age-1 PI3′K loss of function by the weak daf-18(e1375) allele was not complete, however, as these doubly mutant animals were somewhat developmentally delayed in comparison with daf-18 single-mutant or wild-type nematodes (Table 2). In contrast, the daf-18 PTEN nr2037 deletion allele completely suppressed the constitutive dauer-formation phenotypes of both daf-2 and age-1 mutants. This result suggests that the e1375 allele retains residual PTEN function. Both alleles of daf-18 PTEN can suppress the decreased brood-size phenotype of daf-2(e1370) mutant animals (Table 2; 27, 28). Thus, fertility may be less sensitive to PtdIns-3,4,5-P3 levels than dauer formation.
While constructing the double mutant animals used above, we noticed that both alleles of daf-18 PTEN showed a strong maternal rescue of the daf-18 mutant dauer-formation defect. Because daf-18 loss of function suppresses age-1(m333), 25% of the progeny of age-1(m333);daf-18/+ hermaphrodites are predicted to be homozygous for the daf-18 mutation and, therefore, to form normal adults rather than dauers. Instead, we found that nearly all the progeny of age-1(m333);daf-18/+ hermaphrodites arrested as dauers. After 5 days, only 2.4% of these progeny grew to adults, and these were sterile (data not shown). This maternal rescue of daf-18 function may explain why daf-18 mutant alleles have not been identified in genetic screens for suppressors of daf-2 IRL mutations, despite our finding that a strong loss-of-function daf-18 allele can suppress daf-2 IRL and age-1 PI3′K loss-of-function mutations. A genetic screen for suppressors of daf-2 IRL and age-1 PI3′K mutations that takes into account the possibility of maternal rescue should yield other alleles of daf-18 PTEN, as well as heretofore unidentified components of the daf-2 IRL age-1 PI3′K signaling pathway.
DISCUSSION
DAF-18/PTEN Acts in the C. elegans PI3′K Signaling Pathway.
We have identified a C. elegans homolog of the mammalian PTEN tumor suppressor and isolated animals that carry germ-line deletions within this gene. Based on four criteria, we conclude that the C. elegans PTEN homolog corresponds to a previously identified gene, daf-18 (20): (i) the PTEN deletion mutant, nr2037, and the daf-18(e1375) alleles give rise to a common, diverse set of phenotypes; (ii) transformation with genomic DNA containing the C. elegans PTEN homolog can rescue the phenotypic effects of the daf-18(e1375) mutant allele; (iii) the daf-18(e1375) and nr2037 alleles fail to complement one another for dauer formation; and (iv) daf-18(e1375) animals contain a mutant version of the C. elegans PTEN homolog that results in the premature termination of the ORF.
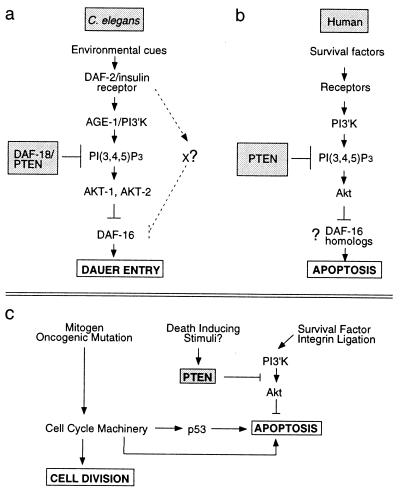
The daf-18(nr2037) deletion allele results in the loss of sequences encoding the PTEN phosphatase catalytic signature motif as well as much of the TAG homology domain. Accordingly, the daf-18(nr2037) deletion allele is likely a null mutation. Loss of the DAF-18 PTEN catalytic function abolishes dauer diapause. Epistasis analysis indicates that DAF-18 PTEN functions as a component of a PI3′K signaling pathway downstream of or parallel to DAF-2 IRL and AGE-1 PI3′K (Table 2; refs. 21 and 31). Because PtdIns-3,4,5-P3 is both a product of PI3′K (14) and a substrate of PTEN (13), we propose that DAF-18 PTEN acts downstream of DAF-2 IRL to antagonize the activity of AGE-1 PI3′K (Fig. 3a).
Figure 3.
PI3′K signaling modules in mammals and nematodes have implications for cancer. (a) The C. elegans AGE-1 PI3′K homolog is recruited by the DAF-2 IRL protein. PI3′K generates PtdIns-3,4,5-P3, whereas the DAF-18 PTEN homolog degrades this second messenger. PtdIns-3,4,5-P3 and its derivative PtdIns-3,4-P2 lead to the activation of Akt-family kinases. AKT-1 and AKT-2 may directly phosphorylate the DAF-16 forkhead transcription factor, thereby antagonizing its function (17). DAF-16 forkhead activity leads to induction of the dauer state (18, 19). Loss of DAF-16 activity prevents dauer formation. (b) In mammals, PTEN and PI3′K similarly antagonize one another to regulate local PtdIns-3,4,5-P3 levels and thus Akt activation. Although C. elegans Akt homologs have not been shown to play a role in nematode programmed cell death, mammalian Akt can potently protect cells against apoptosis. DAF-16 homologs (18) of the FKHR family (38) may play a role in the regulation of apoptosis by Akt. (c) PTEN and PI3′K regulate the integration of extracellular survival signals into the normal cell division cycle. Loss of PTEN results in increased Akt activity and increased cell survival even in the presence of the proapoptotic cues induced by aberrant cell division.
The recruitment of the AGE-1 PI3′K by DAF-2 IRL should increase the levels of PtdIns-3,4,5-P3. It is believed that this phosphatidylinositol and its derivatives positively regulate AKT-1 and AKT-2 (17), the C. elegans homologs of the mammalian Akt/PKB protooncogene (32), in a manner analogous to the activation of mammalian Akt (32). Once activated, AKT-1 and AKT-2 antagonize the function of the DAF-16 forkhead transcription factor, probably by direct phosphorylation (17). DAF-16 induces dauer-stage entry, and the inhibition of its activity by the AGE-1 PI3′K AKT signaling cascade is sufficient to prevent dauer formation (17). Based on our genetic studies and the known biochemical properties of PTEN, we propose that DAF-18 PTEN acts in opposition to PI3′K to reduce the levels of PtdIns-3,4,5-P3, thereby down-regulating AKT-1 and AKT-2 and restoring the activity of the DAF-16 forkhead transcription factor (Fig. 3a; refs. 17 and 18). In this manner, the relative levels and activities of DAF-18 PTEN and AGE-1 PI3′K will play a critical role in determining whether animals enter dauer diapause. Our data indicate that normal dauer formation depends absolutely on daf-18 PTEN (Tables 1 and 2) and, therefore, presumably the down-modulation of PtdIns-3,4,5-P3 levels.
The daf-2 Signaling Pathway.
Recent genetic studies have raised questions about the complexity of the DAF-2 IRL AGE-1 PI3′K pathway (17). Early work suggested that a single linear pathway exists downstream of DAF-2 IRL in which AGE-1 PI3′K acts to antagonize DAF-16 forkhead function (refs. 18 and 19; Fig. 3a). However, two published observations seem to be at odds with this model. First, both a gain-of-function akt-1 mutation, akt-1(mg144), and the weak daf-18(e1375) allele suppress age-1/PI3′K but not daf-2/IRL mutations (refs. 17, 21, 27, and 31; Table 2). Second, screens for daf-2 IRL suppressors have yielded mutations within the daf-16-encoded transcription factor but not other components of the PI3′K pathway, such as daf-18 PTEN (26). To explain the first observation, Paradis and Ruvkun (17) have proposed that DAF-2 IRL signaling may be transduced to the DAF-16 transcription factor by a second pathway that acts in parallel to PI3′K. Our findings suggest alternative explanations for these two observations that are consistent with the existence of a single DAF-2-dependent pathway.
The ability of akt-1(mg144) and daf-18(e1375) to suppress age-1 PI3′K but not daf-2 IRL mutations probably is due to the fact that many of these alleles result in only partial loss or gain of function. We have shown that daf-18(e1375) contains residual activity and that a strong loss-of-function daf-18 PTEN allele can suppress both age-1 PI3′K and daf-2 IRL. The akt(mg144) mutant also may have intermediate activity in a manner analogous to daf-18(e1375); however, because akt-1(mg144) is a gain-of-function allele, its strength is difficult to quantitate. Furthermore, we suggest that the age-1(m333) and age-1(mg44) animals are not null with respect to the AGE-1 PI3′K product PtdIns-3,4,5-P3. This conclusion is based on the understanding that DAF-18 PTEN degrades PtdIns-3,4,5-P3 and that, therefore, daf-18 PTEN loss must rescue the age-1 PI3′K mutant phenotype by the stabilization of some preexisting pool of PtdIns-3,4,5-P3.
The failure of daf-2 IRL genetic suppressor screens to identify daf-18 PTEN mutant alleles may be explained by the strong maternal rescue that we have observed for the daf-18 PTEN alleles. This effect would cause both the F1 and F2 progeny of daf-2 IRL mutant animals exposed to mutagens to adopt the dauer state irrespective of the status of their daf-18 PTEN loci. A daf-2 IRL genetic suppressor screen that accounts for possible maternal rescue should yield daf-18 mutant alleles and also may lead to the identification of proteins involved in the recruitment or regulation of DAF-18 PTEN function.
Based on our findings, we favor a model in which DAF-2 IRL acts via a single linear pathway in which AGE-1 PI3′K and DAF-18 PTEN regulate AKT-1 and AKT-2 activity and, thus, the induction of dauer-stage entry by DAF-16 (Fig. 3a; refs. 16 and 17). However, it is also possible to modify the dual-pathway model to incorporate our data. Given the ability of the daf-18 null allele to suppress daf-2 IRL, we propose that the activity of the second (non-PI3′K) pathway is unmasked only when there is an intermediate level of AGE-1 PI3′K pathway activity (17). Alternatively, DAF-18 could be acting as a component of both the PI3′K and the non-PI3′K pathways. Further exploration of this pathway is required to distinguish between these models.
daf-18 PTEN May Function in Another Signaling Pathway Independent of the DAF 2 IRL Cascade.
Previously, it has been noted that many daf-18(e1375) animals burst from the vulva as adults (ref. 30; Table 1). We have shown that the independently isolated daf-18(nr2037) deletion animals share the same phenotype, indicating that it is a bona fide consequence of daf-18 PTEN loss of function. This secondary phenotype is not modified by the presence of daf-2 IRL or age-1 PI3′K mutant alleles (ref. 30 and data not shown) and has not been reported for animals carrying mutations in daf-16, the only other known dauer-formation-defective gene in the PI3′K pathway (26), suggesting that daf-18 PTEN may be involved in a second signal transduction pathway that is independent of the daf-2 IRL age-1 PI3′K dauer-entry signaling cascade.
Implications for Cancer.
The components involved in the PI3′K signaling module are conserved remarkably between humans and C. elegans with regard to order of action and structure (16, 17). Despite the conservation of this pathway, there are clear differences in the phenotypic output of Akt function. Although there is no evidence for the involvement of nematode AKT homologs in the regulation of programmed cell death, mammalian Akt can protect cells against a variety of apoptosis-inducing stimuli (32).
Many oncogenic mutations induce programmed cell death, and the outgrowth of malignancies depends on the inactivation of this apoptotic program (Fig. 3c; ref. 33) This proapoptotic program can be circumvented by the up-regulation of survival cues, such as growth factor or integrin ligation (Fig. 3c). These survival cues are mediated frequently through the PI3′K/Akt pathway (32). Because Akt is regulated directly by PtdIns-3,4,5-P3 levels (32), our results suggest that mammalian PTEN will function upstream of Akt to inhibit its death-protective activity (Fig. 3 b and c). Consistent with this idea, recent in vitro studies have shown that PTEN −/− mouse embryonic fibroblasts have elevated PtdIns-3,4,5-P3, a concomitant increase in activated murine Akt, and heightened resistance to a variety of death-inducing stimuli (33).
It has been shown that a transforming growth factor β (TGF-β)-related signal transduction pathway (26) interacts with the daf-2 IRL age-1 PI3′K pathway to regulate entry into dauer diapause (17, 18, 26). It has been suggested that the convergence of the TGF-β and PI3′K signaling pathways may play a role in type II diabetes (17, 18, 34). We suggest that the TGF-β and PI3′K signaling pathways may also play a collaborative role in the suppression of human tumors. Intriguingly, loss of the TGF-β signaling component SMAD4 leads to juvenile polyposis coli (35), a disease phenocopied by loss-of-function mutations in PTEN (36, 37).
Implications for Type II Diabetes.
The regulation of metabolism and longevity are linked in both C. elegans and mammals (22). Loss-of-function mutations in daf-2 IRL or age-1 PI3′K significantly increase the life span of mutant animals (22), and this increased longevity can be suppressed by daf-18 PTEN mutation (29, 30). Both the C. elegans DAF-2 IRL molecule and the mammalian insulin receptor play roles in metabolic control. DAF-2 IRL signaling seems to be transduced largely by AGE-1 PI3′K (15, 16), and it has been proposed that factors in the corresponding human PI3′K pathway may be misregulated in type II diabetes (16, 18). Given the role of DAF-18 PTEN in the C. elegans DAF-2 insulin-like pathway, human PTEN may play a role in mammalian glucose homeostasis. Therefore, deregulation of human PTEN may predispose individuals to type II diabetes, and thus PTEN may be a rational pharmacological target for this disease.
Acknowledgments
We thank H. R. Horvitz and the members of the Horvitz lab for extensive intellectual and technical support, Y. Kohara for cDNA clones, J. H. Thomas for providing temporary laboratory space and supplies, R. Wisotzkey for generating the alignments used in Fig. 1, and H. R. Horvitz and S. P. Bell for critical reading of this manuscript. We also thank the Sanger Center for C. elegans cosmids. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Center for Research Resources of the National Institutes of Health.
ABBREVIATIONS
- IRL
insulin receptor-like
- PI3′K
phosphatidylinositol 3-OH kinase
- PtdIns-3
4,5-P3, phosphatidylinositol (3,4,5) triphosphate
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF126286).
References
- 1.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang S I, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 2.Steck P A, Perhouse M A, Jasser S A, Yung W K, Lin H, Ligon A H, Langford L A, Baumgard M L, Hattier T, Davis T, et al. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 3.Tashiro H, Blazes M S, Wu R, Cho K R, Bose S, Wang S I, Li J, Parsons R, Ellenson L H. Cancer Res. 1997;57:3935–3940. [PubMed] [Google Scholar]
- 4.Guldberg P, thor Straten P, Birck A, Ahrenkil V, Kirkin A F, Zeuthen J. Cancer Res. 1997;57:3660–3663. [PubMed] [Google Scholar]
- 5.Teng D F, Hu R, Lin H, Davis T, Iliev D, Frye C, Swedlund B, Hansen K L, Vinson V L, Gumpper K L, et al. Cancer Res. 1997;57:5221–5225. [PubMed] [Google Scholar]
- 6.Liaw D, Marsh D J, Li J, Dahia P L, Wang S I, Zheng Z, Bose S, Call K M, Tsou H C, Peacocke M, et al. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 7.Marsh D J, Dahia P L, Zheng Z, Liaw D, Parsons R, Gorlin R J, Eng C. Nat Genet. 1997;16:333–334. doi: 10.1038/ng0897-333. [DOI] [PubMed] [Google Scholar]
- 8.DiCristofano A, Pesce B, CordonCardo C, Pandolfi P P. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki A, de la Pompa J L, Stambolic V, Elia A J, Sasaki T, del Barco Barrantes I, Ho A, Wakeham A, Itie A, Khoo W, et al. Curr Biol. 1998;8:1169–1178. doi: 10.1016/s0960-9822(07)00488-5. [DOI] [PubMed] [Google Scholar]
- 10.Kanaoka Y, Kimura S H, Okazaki I, Ikeda M, Nojima H. FEBS Lett. 1997;402:73–80. doi: 10.1016/s0014-5793(96)01484-6. [DOI] [PubMed] [Google Scholar]
- 11.Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada K M. Science. 1998;280:1614–1617. doi: 10.1126/science.280.5369.1614. [DOI] [PubMed] [Google Scholar]
- 12.Myers M P, Stolarov J P, Eng C, Li J, Wang S I, Wigler M H, Parsons R, Tonks N K. Proc Natl Acad USA. 1997;94:9052–9057. doi: 10.1073/pnas.94.17.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maehama T, Dixon J E. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 14.Toker A, Cantley L C. Nature (London) 1997;387:673–676. doi: 10.1038/42648. [DOI] [PubMed] [Google Scholar]
- 15.Morris J Z, Tissenbaum H A, Ruvkun G. Nature (London) 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- 16.Kimura K D, Tissenbaum H A, Liu Y, Ruvkun G. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 17.Paradis S, Ruvkun G. Genes Dev. 1998;12:2488–2498. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogg S, Paradis S, Gottlieb S, Patterson G I, Lee L, Tiseenbaum H A, Ruvkun G. Nature (London) 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 19.Lin K, Dorman J B, Rodan A, Kenyon C J. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 20.Riddle D L, Swanson M M, Albert P S. Nature (London) 1981;290:668–671. doi: 10.1038/290668a0. [DOI] [PubMed] [Google Scholar]
- 21.Gottlieb S, Ruvkun G. Genetics. 1994;137:107–120. doi: 10.1093/genetics/137.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kenyon C. In: C. elegans II. Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 791–813. [Google Scholar]
- 23.Hodgkin J. In: C. elegans II. Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 881–1047. [Google Scholar]
- 24.Mello C, Fire A. In: Methods in Cell Biology. Epstein H F, Shakes D C, editors. San Diego: Academic; 1995. pp. 452–482. [Google Scholar]
- 25.Jansen G, Hazendonk E, Thijssen K L, Plasterk R H. Nat Genet. 1977;17:119–121. doi: 10.1038/ng0997-119. [DOI] [PubMed] [Google Scholar]
- 26.Riddle D L, Albert P S. In: C. elegans II. Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 739–768. [Google Scholar]
- 27.Tissenbaum H A, Ruvkun G. Genetics. 1998;148:703–717. doi: 10.1093/genetics/148.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eeckman F H, Durbin R. In: Caenorhabditis elegans: Modern Biological Analysis of an Organism. Epstein H F, Shakes D C, editors. San Diego: Academic; 1995. pp. 584–607. [Google Scholar]
- 29.Larsen P L, Albert P S, Riddle D L. Genetics. 1995;139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dorman J B, Albinder B, Shoryer T, Kenyon C. Genetics. 1995;141:1399–1406. doi: 10.1093/genetics/141.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vowels J J, Thomas J H. Genetics. 1992;130:105–123. doi: 10.1093/genetics/130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Downward J. Curr Opin Cell Biol. 1998;10:262–267. doi: 10.1016/s0955-0674(98)80149-x. [DOI] [PubMed] [Google Scholar]
- 33.Stambolic V, Suzuki A, de la Pompa J L, Brothers G M, Mirtsos C, Sasaki T, Ruland J, Penninger J M, Siderovski D P, Mak T W. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 34.Patterson G, Koweek A, Wong A, Liu Y, Ruvkun G. Genes Dev. 1997;11:2679–2690. doi: 10.1101/gad.11.20.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Howe J R, Roth S, Ringold J C, Summers R W, Jarvinen H J, Sistonen P, Tomlinson I P, Houlston R S, Bevan S, Mitros F A, et al. Science. 1998;280:1086–1088. doi: 10.1126/science.280.5366.1086. [DOI] [PubMed] [Google Scholar]
- 36.Olschwang S, Serova-Sinilnikova O M, Lenoir G M, Thomas G. Nat Genet. 1998;18:12–14. doi: 10.1038/ng0198-12. [DOI] [PubMed] [Google Scholar]
- 37.Eng C, Peacocke M. Nat Genet. 1998;19:223. doi: 10.1038/897. [DOI] [PubMed] [Google Scholar]
- 38.Anderson M J, Viars S V, Czekay S, Cavanne W C, Arden K C. Genomics. 1998;47:187–199. doi: 10.1006/geno.1997.5122. [DOI] [PubMed] [Google Scholar]