Abstract
Myosin heavy chains (MyHCs) are highly conserved ubiquitous actin-based motor proteins that drive a wide range of motile processes in eukaryotic cells. MyHC isoforms expressed in skeletal muscles are encoded by a multigene family that is clustered on syntenic regions of human and mouse chromosomes 17 and 11, respectively. In an effort to gain a better understanding of the genomic organization of the skeletal MyHC genes and its effects on the regulation, function, and molecular genetics of this multigene family, we have constructed high-resolution physical maps of both human and mouse loci using PCR-based marker content mapping of P1-artificial chromosome clones. Genes encoding six MyHC isoforms have been mapped with respect to their linear order and transcriptional orientations within a 350-kb region in both human and mouse. These maps reveal that the order, transcriptional orientation, and relative intergenic distances of these genes are remarkably conserved between these species. Unlike many clustered gene families, this order does not reflect the known temporal expression patterns of these genes. However, the conservation of gene organization since the estimated divergence of these species (≈75–110 million years ago) suggests that the physical organization of these genes may be significant for their regulation and function.
Myosin heavy chains (MyHCs) are ubiquitous actin-based motor proteins that convert the chemical energy derived from hydrolysis of ATP into mechanical force that drives diverse motile processes including cytokinesis, vesicular transport, and cellular locomotion in eukaryotic cells. The MyHCs have been divided into 9 to 11 classes (see refs. 1–3). Class II or “conventional” MyHCs include the extensively studied group of sarcomeric MyHCs that self associate to form filaments and function enzymatically to promote contraction in striated (cardiac and skeletal) muscles. Muscle myosin is a heterohexamer consisting of two MyHCs and two associated nonidentical pairs of myosin light chains. The seven MyHC isoforms that predominate in mammalian skeletal muscles include two developmental isoforms, MyHC-embryonic and MyHC-perinatal; three adult skeletal muscle isoforms, MyHC-IIa, MyHC-IIb, MyHC-IIx/d; and MyHC-β/slow, which is also expressed in cardiac muscle (reviewed in ref. 4). MyHC-extraocular is expressed primarily in extrinsic eye muscles and has been detected in pharyngeal muscle as well (5, 6). MyHC isoforms are differentially regulated in response to diverse stimuli, including physiological, hormonal, mechanical, neurological and other signals (4). The ATPase activity conferred by the MyHCs in a given muscle correlates with speed of contraction; thus MyHCs are major determinants of the functional properties of the muscles in which they are expressed. Recently, genetic inactivation of the adult fast IIa and IIb genes has demonstrated that these genes are functionally distinct and required for normal muscle function (7, 8).
In contrast to smooth muscle MyHC, nonmuscle MyHC, and most other contractile protein families, sarcomeric MyHC genes are clustered in the genomes of those mammals examined. Two “cardiac” genes, MyHC-α and MyHC-β, are arranged tandemly with 4.5-kb intergenic separation on chromosomes 14 of humans and mice (9–13) and are similarly linked in rat and rabbit (14, 15). Genes encoding isoforms expressed in skeletal muscle have been located in clusters on chromosomes 17 and 11 in humans and mice, respectively (13, 16, 17). Sequence comparisons of human, mouse, and rat MyHC cDNA sequences reveal that MyHCs are highly conserved and that orthologous isoforms encoded by genes in the cardiac and skeletal clusters have been maintained between mammalian species (see ref. 18). However, sequence data for the coding, noncoding, and regulatory regions of these gene loci are incomplete and the significance of the clustered nature of the sarcomeric MyHCs remains to be determined.
A yeast artificial chromosome (YAC)-based contiguous set of overlapping chromosomes (contig) representing the human skeletal locus had previously placed six MyHC genes within a 500-kb segment on chromosome 17, a region encompassing a polymorphic genetic marker mapped to 17p13.1 (19). This contig ordered sequence-tagged site (STS) markers for four of six human skeletal genes (MyHC-embryonic, MyHC-perinatal, MyHC-IIa, and MyHC-IIx/d) but the identities of the remaining two genes were unknown and their relative locations unresolved. Likewise, a tentative YAC-based map had ordered six mouse skeletal MyHC genes on a segment of chromosome 11 (20). However, accurate high-resolution physical maps describing the order and orientation of these genes have been lacking.
Toward obtaining an improved understanding of the genomic organization, regulation, and diversity of the mammalian MyHC gene family, we have used PCR-based marker content mapping of P1-artificial chromosomes (PACs) with MyHC gene-specific and other STS primers to generate high-resolution physical maps of both human and mouse skeletal MyHC loci. The PAC-based maps in this report revise the gene order derived from previous YAC-based maps and reveal that human and mouse MyHC loci are extremely conserved in terms of linear order, transcriptional polarity, and spacing of genes, suggesting that maintenance of these characteristics may be important for the function of the mammalian skeletal MyHC gene family. We have found that, unlike other clustered genes, the MyHC genes are not arranged according to their temporal expression patterns, but their organization does reflect the extent of sequence similarity among family members.
MATERIALS AND METHODS
Nomenclature.
In this report, the six skeletal MyHC genes are designated: MyHC-embryonic (MyHC-emb), MyHC-IIa, MyHC-perinatal (MyHC-peri or MyHC-pn), or exoc MyHC-IIb, MyHC-IIx/d, and MyHC-extraocular (MyHC-eo). Human genes MyHC-IIa, MyHC-IIx/d, MyHC-IIb, and MyHC-extraocular correspond to genes MYHas8, MYHa, MYH?, and MYH2 from the previously published YAC contig by Yoon et al. (19). MyHCs IIa, IIb, and IIx/d may elsewhere be referred to as MyHC-2A, MyHC-2B, and MyHC-2X/D, respectively.
Hybridization Screening of PACs.
High-density filters containing human (RPCI/1 and 3) and mouse (RPCI21/Segment 2) PAC libraries (purchased from the Rosewell Park Cancer Institute, Buffalo, NY) were screened by hybridization with random-primed sarcomeric MyHC-specific probes. Human filters were screened with MyHC consensus probe LL1000 (19), yielding 28 positive PACs. The same probe was then hybridized to a slot blot of these 28 clones, confirming 18 positive PAC clones. Mouse filters were probed with a mixture of eight MyHC gene-specific PCR products (see gene-specific STSs listed in Table 1b), which enabled identification of 30 MyHC-positive mouse PAC clones.
Table 1.
STS sequences and PCR conditions for MyHC contigs
| STS/Gene name | Primer Sequence 5′ → 3′ | Anneal temp.,°C | Product size, bp |
|---|---|---|---|
| A. Human | |||
| MyHC-emb5′ | F-GATGTGGTTCCAGCACTGG | ‡58 | 174 |
| R-CTGGCCTACAGTGAACGTG | |||
| MyHC-emb3′ | F-GCCAGCCCTTCTGGAGCA | 55 | 93 |
| R-GAGTCACATGGACATTAAGT | |||
| †MyHC-IIa5′ | F-TGTCGGGACTCTCAGGTAG | 58 | 200 |
| R-CAGCATCTATTAGCGTGTCC | |||
| MyHC-IIa3′ | F-TCATGTCCTGATGCCATGGAATG | 63 | 119 |
| R-CTACCCTATGCTTTATTTCCTTTG | |||
| *A288E1L | F-TAGCCATTGCCTATATCAAG | 55 | 108 |
| R-GCATCGAGCTCATCGAGAAG | |||
| MyHC-IIx/d3′ | F-ATCTAACTGCTGAAAGGTGACC | 56 | 71 |
| R-TAAGTACAAAATGGAGTGACAAAG | |||
| MyHC-IIb5′ | F-CCTAAGTACTGTGTTAGTTCTCAT | 65 | 1,000 |
| R-TCTGAACATGCTTGGAGCATCA | |||
| MyHC-IIb3′ | F-GTTATCTCAAATGCATCTCTGTTC | 66 | 150 |
| R-GATATACAGGACAGTGACAAAGAACT | |||
| MyHC-pn5′ | F-CAGGGAGAAAATACTCCC | 58 | 171 |
| R-GTTGGGAAGGGAAGACATG | |||
| MyHC-pn3′ | F-TATCAAGAGGCTGAAGAAAG | 47 | 112 |
| R-AATGTATACATTTACTGTAG | |||
| MyHC-eo5′ | F-GAGCATGCCCATATTATCAAAGAC | 67 | 300 |
| R-TGCAGTCATGAGCTCTGAGCGA | |||
| †401014T7 | F-CCCTTCTGGTTCTGCAGACT | 57 | 73 |
| R-CGGTCATAGAGAAGACCAGAGA | |||
| MyHC-eo3′ | F-CATGAGCTAGAGGAGGCCGC | 63 | 885 |
| R-GTATTTAGGAGAATTTATTTCTCA | |||
| B. Mouse | |||
| †475-D-1T7 | F-ATGAACTGGAAACCCTCCTC | 56 | 158 |
| R-CTTGAGAAGCTGAAGTAGTAG | |||
| †538-C-3T7 | F-GTATCTCCAGCACCCTAC | 50 | 276 |
| R-TGATGGAGTCACAAAGATCAC | |||
| MyHC-emb3′ | F-TGAGCAAGACCTCCTGGTG | 56 | ∼100 |
| R-TGCATGTGGAAAAGTGATACG | |||
| MyHC-IIa5′ | F-CTGTACAAAGATCTGCTGT | 50 | ∼300 |
| R-CAAGTCAGCGAGTGACCA | |||
| MyHC-IIa3′ | F-GGCGAGGCACAAAATGTGAA | 56 | ∼100 |
| R-GCAAAGGAACTTGGGCTCTT | |||
| MyHC-IIx/d5′ | F-ATTTCTCTCTCTGCCAGGCA | 56 | ∼200 |
| R-TCAAACTAGCCCCGCAGTG | |||
| †380-N-16T7 | F-TGGACAAGGGTATGAACTTCG | 56 | 120 |
| R-GAGACATTGATGGGTCCTTG | |||
| MyHC-IIx/d3′ | F-ATTGATCCAAGTGCAGGAAAG | 56 | ∼200 |
| R-TCCTGATCTACAAATGATCGG | |||
| †630-D-9T7 | F-CTTAGCAGGTCTCCACGTC | 56 | 328 |
| R-GTGATGCTTGCAAAGGAGTC | |||
| MyHC-IIb5′ | F-TCTGTCACTCGGTGCTTCC | 56 | ∼200 |
| R-AGGGTTTTTGGAGGCTGTTT | |||
| †433-B-7T7 | F-GTCGTGCCTAAGTCCTAGC | 56 | 109 |
| R-AGGCCCAAGAGAGAATTTGC | |||
| †561-O-1T7 | F-CTGTCCACCTACCTTGAGG | 56 | 366 |
| R-GAGTTCTCTTCTGCTCAGTG | |||
| MyHC-pn3′ | F-ACACATCTTGCAGAGGAAGG | 56 | ∼100 |
| R-TAAACCCAGAGAGGCAAGTG | |||
| †386-N-23T7 | F-GCACTGTTAACCGTGAGCC | 56 | 185 |
| R-GGTAAATCAAACCAGGCTCC | |||
| †645-M-6T7 | F-AGAACGAGGAAAAGCAAGGC | 56 | 218 |
| R-CAGATATTCCTTCTGGGCAC | |||
| †410-O-9T7 | F-AGTTCACTGCCCTAAGCATG | 56 | 177 |
| R-AAGGCAGCTGTGGATATGAC | |||
| MyHC-eo3′ | F-CAGGTAAAAGCAGACACCTG | 56 | ∼300 |
| R-AGCTTTAAGGAATAGGCTTCG | |||
| †524-K-7T7 | F-AACATTGATGACTGCCCTGC | 56 | 299 |
| R-TTGGAGGTAGAGTTGTCAGG |
*, STS used in YAC contig (19), (†) STS from PAC End Rescue. All PCR used 1.5mM MgCl2 (detailed in Materials and Methods) except (‡) 2.0mM MgCl2.
STS-Content Mapping of PACs.
Human and mouse PACs identified by hybridization were screened by PCR with STS primers listed in Table 1. Human gene-specific primers for the following STSs: MyHC-embryonic 5′ and 3′(X13988), MyHC-perinatal 3′(M36769, M35250, Y00821), MyHC-IIa 3′ (S73840, AF111784), and MyHC-IIx/d 3′ (X03740, AF111785), were designed from their corresponding published MyHC sequences (GenBank accession numbers indicated in parentheses). Human MyHC-IIa 5′ primers and MyHC-IIb and MyHC-extraocular gene-specific primers were designed from cDNA and genomic sequences corresponding to these genes (AF111782, AF111783). Mouse MyHC gene-specific STSs designed from published sequences include MyHC-emb 3′ (21), MyHC-pn 3′ (13), MyHC-IIa 5′, MyHC-IIx/d 5′, MyHC-eo (20), MyHC-IIa 3′ and MyHC-IIx/d 3′ (22). Primers for mouse STS MyHC-IIb 5′ were designed from the published IIb promoter sequence (23).
PCR was carried out in 20–50 μl by using 100 ng each primer, 0.2 mM dNTPs, 1.5 mM MgCl2, Perkin-Elmer AmpliTaq Gold, 10XPCR buffer II, and 10 ng PAC DNA prepared by alkaline lysis (protocol available online from http://bacpac.med.buffalo.edu/hdhf_prot.html). Cycling conditions were as follows (see Table 1 for annealing temperatures). Human PCR: 94°C/3 min (1 ×), 94°C/1 min, anneal temp/1 min (35 ×), 72°C 1 min (1 ×). Mouse PCR: 94°C/45 sec (1 ×), 94°C/20 sec, annealing temperature/20 sec, 72°C/20 sec (30 ×), 72°C/3 min. STS markers and PAC clones were ordered by analyzing the marker content of each PAC clone, and transcriptional orientations were established by resolution of 5′ and 3′ markers from the same gene to different PACs or by resolution of PAC end sequences containing MyHC sequences from gene-specific markers (as described for each gene in the Results section).
PAC End Rescue.
PAC end sequences were used to generate additional STSs (indicated by † in Table 1 and by squares in Fig. 1) for both mouse and human contigs. Likewise, PAC end sequences were used to establish that groups of human PACs that contain identical marker content (left end group: 429-F-6, 410-C-3, 280-D-10; right end group: 517-H-15, 484-B-19, 182-F-4, 401-O-14) are comprised of unique PAC clones. End-rescue STSs are named for the PAC end from which they were derived. Ligation-mediated vectorette/bubble PCR (24) was performed on all mouse PACs and human PACs 269-H-13 and 401-O-14. The vectorette was comprised of primers RK 331 (5′-CTCTCCCTTCTCCGCAAATCGATCTCGAG TCTAGAGTC GACGTCCTCTC CTT-3′) and RK532 (5′-AAGGAGAGGACGCTGTCTGTCGA AGGTAAGGAACGGA CGAGAG AAGGGAGAG-3′). Vectorette PCR was carried out by using a primer internal to the vectorette, RK332 (5′-CCGCAAATCGATCTCGAGTCTA GAGTCGAC-3′) and either Sp6 or T7 primers from the PAC vector. PCR products containing PAC ends were purified by using the Qiaquick PCR Purification kit (Qiagen, Chatsworth, CA) and sequenced directly by using an ABI-Prism 377 automated sequencer. The Primer program [Whitehead Institute for Biomedical Research/Massachusetts Institute of Technology (MIT) Center for Genome Research, Cambridge, MA] was used to design PCR primers, which were then used to rescreen PAC clones.
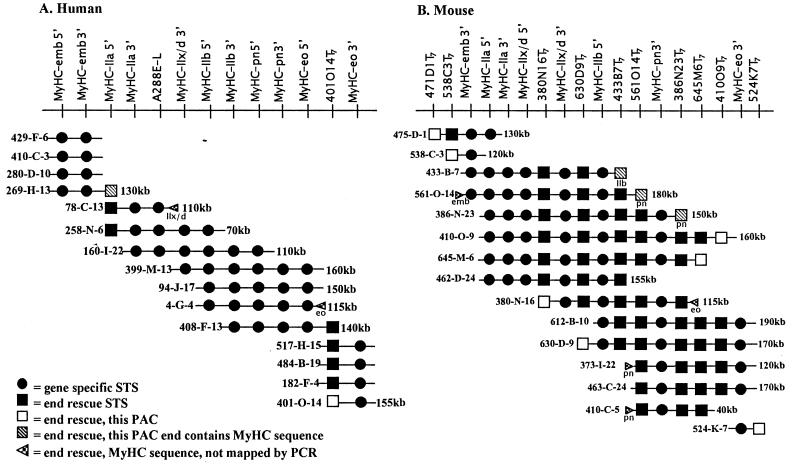
Figure 1.
Physical maps of skeletal MyHC gene clusters. The top line in each map shows the order of markers (listed in Table 1) established by STS content mapping of PACs aligned below. PAC names and sizes (if determined) are indicated at the left and right ends of PACs, respectively. MyHC genes identified in end rescue sequences (hatched symbols) are indicated below symbol. Spacing of markers does not reflect actual scale.
Determination of PAC Sizes.
Human and mouse PAC inserts were excised from the vector by digestion with NotI and resolved by pulsed-field gel electrophoresis to determine sizes. Human insert sizes were confirmed by Southern hybridization by using random-primed total human DNA.
Determination of Intergenic Distances.
PCR using primers MyHC-IIa5′-F [3′ untranslated region (UTR) sense primer; Table 1a] and LL773 (MyHC-IIx/d exon 3 antisense primer; 5′-TCCACCCAAA GACTGATGT-3′) were used with PAC template 258-N-6 to determine the intergenic distance between human MyHC-IIa and MyHC-IIx/d. Primers MyHC-IIx/d 3′-F (3′UTR sense primer; Table 1a) and LL778 (MyHC-IIb intron 2 reverse primer; 5′-CCAGTCTTCTAAA TACCAGCCCA-3′) were used with the same template for intergenic PCR between human MyHC-IIx/d and MyHC-IIb. Mouse PCR primers MyHC-IIa3′-F and MyHC-IIx/d5′-R, and MyHC-IIx/d3′-F and MyHC-IIb5′-F (see Table 1b) were used with PAC template 561-O-14 to determine the intergenic distances between mouse MyHC-IIa and MyHC-IIx/d and MyHC-IIx/d and MyHC-IIb, respectively. PCR was carried out as described for STS-content mapping with the following exceptions. BIO-X-ACT polymerase and 10× Optibuffer (Bioline, London) were used, as were modified annealing temperatures and extension times of 57°C and 6 min and 56°C and 7 min for IIa–IIx/d and IIx/d–IIb reactions, respectively.
Radiation Hybrid Mapping.
The GeneBridge 4 human radiation hybrid panel (Whitehead Institute/MIT Center for Genome Research) was screened in duplicate by PCR with human STS MyHC-embryonic 5′ (Table1a, PCR cycling carried out as described for STS content mapping) and scores were submitted electronically for analysis by the Whitehead Institute.
RESULTS
To compare the number, order, and transcriptional orientations of the skeletal MyHC multigene families in humans and mice, marker content mapping was used to assemble PAC contigs for both human and mouse loci. PAC clones, which are less likely to recombine (undergo deletions and translocations), have smaller insert sizes, and in general provide more reliable mapping information than YAC clones, were used to ensure the resolution of these closely linked genes.
Human Map.
The human PAC map (Fig. 1a) refines several aspects of the previously described YAC-based physical map. A total of 13 markers were ordered by STS content mapping of 18 PACs isolated by hybridization screening of a human PAC library (see Materials and Methods). One published STS and 10 new gene-specific STSs were used for the primary PCR screen, which generated three contigs comprised of 15 PACs. The secondary PCR screen with two STS markers derived from PAC end-rescue sequences 269-H-13Sp6 (marker MyHC-IIa 5′) and 401-O-14T7 was used to establish the final contig with an average depth of 4 PACs per marker. Examination of end sequences (see Materials and Methods) of the two groups of human PACs that contain identical marker content (left end group: 429-F-6, 410-C-3, 280-D-10; right end group: 517-H-15, 484-B-19, 182-F-4, 401-O-14) establishes that each PAC clone is unique.
The resulting linear order of the genes as shown in Fig. 1 is embryonic–IIa–IIx/d–IIb–perinatal–extraocular. This gene order differs from the YAC-based order [embryonic–IIa–IIx/d–perinatal–MYH?– MYH2, with MYH? (MyHC-IIb) and MYH2 (MyHC-eo) unresolved and unidentified; ref. 19] by placement of the MyHC-IIb gene (MYH?) between MyHC-IIx/d and MyHC-perinatal genes. The use of two new gene-specific STSs for MyHC-IIb, as well as the improved resolution of PACs, facilitated this reordering. Likewise, two new gene-specific STSs and the end-rescue sequence 4-G-4Sp6 enabled the placement of MyHC-extraocular (MYH2 in ref. 19) on the right side of MyHC-perinatal.
The transcriptional directions (5′ to 3′) of the MyHC-IIa, MyHC-IIb, MyHC-perinatal, and MyHC-extraocular genes as left to right with respect to the contig were discerned by resolution of their distinct 5′ and 3′ gene-specific STSs. PAC end-rescue sequences containing MyHC sequences (Fig. 1a, hatched arrowheads) were also used to establish transcriptional polarity. PAC end 78-C-13Sp6 contains MyHC-IIx/d sequence that is 5′ (upstream) of the sequence in STS MyHC-IIx/d 3′, establishing the orientation of this gene as left to right with respect to the contig. The 4-G-4Sp6 PAC end contains MyHC-extraocular sequence that is 3′ (downstream) of the sequence in STS marker MyHC-eo 5′, which further supports the left-to-right polarity of MyHC-extraocular. In addition, intergenic PCR between the 3′ and 5′ ends of MyHC-IIa and MyHC-IIx/d, and MyHC-IIx/d and MyHC-IIb (see Materials and Methods) further confirms the orientations of these genes. As the MyHC-embryonic 5′ and 3′ STSs cannot be resolved on our map, the orientation of human MyHC-embryonic remains undetermined.
Mouse Map.
The PAC map of the mouse cluster (Fig. 1b) orders and orients all six MyHC skeletal genes from mouse chromosome 11. Although a tentative YAC-based map was previously constructed (20), this is the first high-resolution map available for this mouse locus. Thirty PACs were initially selected by hybridization screening of a mouse PAC library (see Materials and Methods). These PACs were rescreened by PCR with gene-specific primers (Fig. 1b, circles) from 5′ UTR, 3′ UTR, or intron sequences, and a contig containing 15 clones was constructed by marker content mapping. Eleven STSs derived from PAC end sequences (Fig. 1b, squares) were added to increase resolution and marker density, resulting in a final contig with an average depth of six PACs per marker. The eight gene-specific STSs used in the primary screen were sufficient to determine the linear order of the six mouse genes as embryonic–IIa–IIx/d–IIb–perinatal–extraocular, which differs from the previously constructed YAC map that had placed the MyHC-perinatal gene between MyHC-embryonic and MyHC-IIa genes.
The primary STS screen also established the transcriptional orientations of MyHC-IIa and MyHC-IIx/d genes as left to right with respect to the contig. PAC end-rescue sequences (Fig. 1b, arrowheads) were used to orient the remaining genes. PAC end 561-O-14Sp6 contains MyHC-embryonic sequence that is 5′ (upstream) of the sequence from marker MyHC-emb 3′. Similarly, PAC end 380-N-16Sp6 contains MyHC-extraocular sequence that is 5′ (upstream) of that found in marker MyHC-eo 3′ (3′ UTR), establishing the transcriptional polarity of these genes from left to right. Four PAC ends contain sequences corresponding to MyHC-perinatal. 561-O-14T7 and 386-N-23T7 (with sequences 5′ and 3′ of each other, respectively) were mapped. Two additional PAC ends (373-I-22T7 and 410-C-5T7) contain perinatal sequences 5′ (upstream) of sequences from 561-O-14T7 and 386-N-23T7 and map to the left of these markers, suggesting that MyHC-perinatal is oriented from left to right. PAC end 433-B-7 contains MyHC-IIb 3′ sequence and maps to the right of marker MyHC-IIb 5′, indicating that the orientation of this gene is also left to right. Thus, the linear order of all six mouse skeletal MyHC genes is identical to that of human. Likewise, their transcriptional orientations are the same as that determined for five of the six human genes, from left to right with respect to the contig, with the human MyHC-embryonic orientation unresolved. However, the conservation of transcriptional polarity of the other five genes between human and mouse contigs makes it seem probable that the polarity of the embryonic gene is also conserved.
Gene Cluster Sizes and Spacing of Genes.
To define the size of each locus, individual PAC clone sizes were determined (described in Materials and Methods; see Fig. 1 right ends of PACs for size), and a set of contiguous PACs that spans each locus with the minimum amount of overlap (minimum tiling path) was identified as follows: human PACs 269-H-13/258-N-6/4-G-4/408-F-13/401-O-14 and mouse PACs 538-C-3/561-O-14/373-I-22. Based on the sizes of clones in the minimum tiling paths and assuming a minimal amount of overlap of clones, we conclude that the six genes in each locus are located within a region not larger than ≈350 kb.
The intergenic distances between the MyHC-IIa and MyHC-IIx/d (4.5 kb in mouse and human) and between MyHC-IIx/d and MyHC-IIb (5.0 kb in mouse, 9.0 kb in human) were determined by PCR for both human and mouse loci (described in Materials and Methods). In addition, the mouse MyHC-IIa–MyHC-IIx/d intergenic region was sequenced by using a genomic clone containing this region (Leinwand Laboratory, unpublished sequence data). These distances are consistent with those estimated based on PAC sizes and the locations of gene-specific markers in the contigs. For example, in the human contig, PAC 258-N-6, which is ≈70 kb, contains 5′ and 3′ markers for the MyHC-IIa gene, the entire MyHC-IIx/d gene, and the MyHC-IIb 5′ marker. These data and the assumption that each gene is ≈20–26 kb (see refs. 25–29 for complete genomic MyHC sequences) would suggest that the three “adult” skeletal genes may be more tightly linked than embryonic, perinatal, and extraocular genes. By the same criteria, we estimate the intergenic distances between the other genes (for which long-range intergenic PCR was unsuccessful) to range from ≈15–60 kb. The extent of conservation in their spacing between human and mouse remains to be determined.
Human MyHC Radiation Hybrid Mapping.
The human MyHC locus had been previously physically mapped to 17p13 (18, 30, 31) and genetically linked to a restriction fragment length polymorphism marker, D17S1 (32). However, the locus had not been placed on the high-resolution integrated maps of human chromosome 17. Therefore, we have used a gene-specific STS (MyHC-embryonic 5′, Table1a) to place the human MyHC locus on the current radiation hybrid map at the Whitehead Institute at a position 5.66cR from the marker W1–4633 with a lod score of over 3.0. This location is consistent with previous genetic mapping data and has allowed the MyHC genes to be positioned for the current chromosome 17 sequencing effort as part of the Human Genome Project (Bruce Birren, Whitehead Institute/MIT Center for Genome Research, personal communication).
DISCUSSION
The conservation of order, orientation, and overall spacing of the human and mouse skeletal MyHC genes presented here (schematized in Fig. 2) supports the idea that these loci were formed by gene duplications that occurred before the divergence of these species between ≈75–110 million years ago (33, 34) and suggests that the gene order and spacing in this locus may be important for the function of this gene family. Other well known examples of clustered gene families include the globin (35) and homeobox genes (36, 37), in which linear order can be related to regulation of temporal expression patterns. Likewise, the organization of T cell receptor genes, which are arranged in clusters that are highly conserved in human and mouse, is important for the regulation of splicing and recombination essential for T cell maturation (38, 39). Genes encoding individual family members of many other multigene family proteins and other contractile proteins (summarized in ref. 4), such as the myosin light chains, actin, and troponin I, are not clustered. In light of this, the organization of the highly conserved tightly linked skeletal MyHC cluster is intriguing.
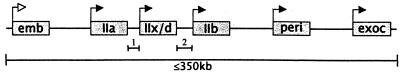
Figure 2.
Conservation of human and mouse skeletal MyHC gene organization. Closed arrows indicate transcriptional orientation determined for human and mouse genes. Open arrow indicates transcriptional orientation determined for mouse only. Intergenic distances as determined by long range PCR are 1, 4.5 kb (for human and mouse) between MyHC-IIa and MyHC-IIx/d, and 2, 9.0 kb (human) and 5.0 kb (mouse) between MyHC-IIx/d and MyHC-IIb. Unlabeled intergenic distances are estimated to be ≈15–60 kb. Each gene cluster spans at least 350 kb.
It is attractive to hypothesize that the organization of MyHC genes has been maintained for so long because it is essential for the function, regulation, and/or molecular evolution of this expanded gene family. The spatial arrangement of the MyHC genes does not reflect their temporal expression patterns, as with the globin and homeobox genes described above. Indeed, the two earliest expressed genes, MyHC-embryonic and MyHC-perinatal, are separated by the three adult genes (IIa, IIx/d, IIb). This organization does, however, reflect the extent of sequence conservation among the human coding sequences. The MyHC-embryonic and MyHC-extraocular genes (located at the extreme ends of the contig) are also the most divergent in their sequences (ranging in identity from ≈80–84% with any of the other isoforms). In contrast, the three adult isoforms, which are more closely linked, are also more similar to each other (ranging in identity from ≈92–95% to each other; see A. Weiss, S. Schiaffino, and L. Leinwand, unpublished work, for identities and GenBank accession numbers AF111784, AF111785, AF111783, and AF111782 for human IIa, IIx/d, IIb, exoc full-length cDNAs, respectively). The three tightly linked adult genes are also similar in that, despite their differential regulation, they have overlapping expression patterns in mammalian adult skeletal muscles (see 40, 41). Thus, the clustering of these MyHC genes may reflect their similar functional roles, rather than the timing of their developmental expression. The more divergent MyHC-embryonic and MyHC-extraocular genes are more peripherally located and have highly specialized expression patterns. Expression of MyHC-embryonic is restricted temporally to early embryonic stages, and the expression of MyHC-extraocular is primarily restricted to a specialized group of muscles that function in eye movement (5, 42). Because comparisons of available orthologous skeletal MyHCs thus far reveal that there has been an apparent maintenance of sequence conservation among orthologous MyHC isoforms of mammalian species (see ref. 18), we would predict the sequence relationships among the mouse coding regions to be similar to those described above for the human.
Whereas many trans-acting factors involved in muscle-specific transcription and some of their binding motifs have been identified (for reviews, see refs. 43, 44), the mechanisms dictating the complex expression patterns of the individual skeletal MyHC genes, which appear to be regulated independently, have not yet been deciphered. The most extensive progress has been made in identification and characterization of basal and muscle-specific regulatory elements in the mouse MyHC-IIb gene promoter (23, 45–47). However, other mammalian skeletal MyHC gene promoters have not been available for study, nor have any MyHC locus-specific elements been identified. The observation that mice homozygous null for MyHC-IIx/d or MyHC-IIb show compensation by MyHC-IIa and MyHC-IIx/d genes, respectively, illustrates that the regulation of these genes is not completely independent (8). Further study of the promoters of these genes reveals that they share common muscle-specific regulatory elements and contain elements that promote gene-specific expression patterns as well (C. Sartorius and L. Leinwand, unpublished work). Furthermore, the “nontemporal” linear order of the skeletal MyHC genes does not preclude their regulation by a locus control region (LCR). LCRs have been proposed to provide a chromatin structure-mediated role of ensuring a state that is permissive for transcription, with the possibility that gene-specific expression patterns are primarily determined by individual promoter regions (for review, see ref. 35). Investigation of the tightly linked mammalian cardiac MyHC genes suggests that multiple distinct regulatory elements are involved in the muscle-specific (cardiac vs. skeletal) expression of the MyHC-β cardiac gene (48, 49). Moreover, the 5.6-kb upstream region of the murine MyHC-β gene has been suggested to contain an LCR for this locus (50).
Finally, the clustering of the skeletal MyHC genes may also be important for maintenance of their conservation by concerted mechanisms such as gene conversion (i.e., nonreciprocal recombination events), which is considered a major mechanism for sequence homogenization in multigene families (see refs. 51–53 for reviews on mechanism), including rRNA (54) and IgV (55, 56) genes. Evidence also exists for the occurrence of gene conversion events among mammalian cardiac MyHC genes and among chicken skeletal MyHC genes (57, 58). The high levels of conservation observed among the three human adult genes suggests strongly that gene conversion events have played a role in the molecular evolution of these genes as well.
The MyHCs are major components of the contractile apparatus of muscle, and their individual biochemical properties likewise are major determinants of muscle physiology and function. Their diverse and complex expression patterns may reflect regulatory features that are distinct from genes of other contractile proteins and other linked genes as well. The finding that the physical organization of these genes is extremely conserved suggests that further comparative studies of these loci will broaden our understanding of the regulation and molecular genetics of clustered multigene families. The proximity of these genes would make it highly likely that their intergenic regions contain important regulatory elements, and comparative sequencing and the identification of conserved sequences in human and mouse loci are logical next steps toward identifying regulatory features that are important, and potentially unique for the function this locus (see ref. 59). Examination of the skeletal MyHC loci of other mammals should also shed light on the significance of the physical organization of these genes.
Acknowledgments
This work was supported by National Institutes of Health (N.I.H.) Grants P60 DA11015–02 to K.K., GM29090 to L.L., and N.I.H. training grant 5T32GM07135 to D. M.
ABBREVIATIONS
- MyHCs
myosin heavy chains
- YAC
yeast artificial chromosome
- PAC
P1-artificial chromosome
- contig
contiguous set of overlapping clones
- UTR
untranslated region
- STS
sequence-tagged site
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Sellers J R, Goodson H V. Protein Profile. 1995;2:1323–1423. [PubMed] [Google Scholar]
- 2.Goodson H V. Soc Gen Physiol Ser. 1994;49:141–57. , 141–157. [PubMed] [Google Scholar]
- 3.Cheney R E, Riley M A, Mooseker M S. Cell Motil Cytoskeleton. 1993;24:215–223. doi: 10.1002/cm.970240402. [DOI] [PubMed] [Google Scholar]
- 4.Schiaffino S, Reggiani C. Physiol Rev. 1996;76:371–423. doi: 10.1152/physrev.1996.76.2.371. [DOI] [PubMed] [Google Scholar]
- 5.Wieczorek D F, Periasamy M, Butler-Browne G S, Whalen R G, Nadal-Ginard B. J Cell Biol. 1985;101:618–629. doi: 10.1083/jcb.101.2.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lucas C A, Rughani A, Hoh J F. J Muscle Res Cell Motil. 1995;16:368–378. doi: 10.1007/BF00114502. [DOI] [PubMed] [Google Scholar]
- 7.Sartorius C A, Lu B D, Acakpo-Satchivi L, Jacobsen R P, Byrnes W C, Leinwand L A. J Cell Biol. 1998;141:943–953. doi: 10.1083/jcb.141.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acakpo-Satchivi L J, Edelmann W, Sartorius C, Lu B D, Wahr P A, Watkins S C, Metzger J M, Leinwand L, Kucherlapati R. J Cell Biol. 1997;139:1219–1229. doi: 10.1083/jcb.139.5.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin H, Kemp J, Yip M Y, Lam P T P, Hoh J F, Morris B J. Cytogenet Cell Genet. 1990;54:74–76. doi: 10.1159/000132961. [DOI] [PubMed] [Google Scholar]
- 10.Matsuoka R, Yoshida M C, Kanda N, Kimura M, Ozasa H, Takao A. Am J Med Genet. 1989;32:279–284. doi: 10.1002/ajmg.1320320234. [DOI] [PubMed] [Google Scholar]
- 11.Saez L J, Gianola K M, McNally E M, Feghali R, Eddy R, Shows T B, Leinwand L A. Nucleic Acids Res. 1987;15:5443–5459. doi: 10.1093/nar/15.13.5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gulick J, Subramaniam A, Neumann J, Robbins J. J Biol Chem. 1991;266:9180–9185. [PubMed] [Google Scholar]
- 13.Weydert A, Daubas P, Lazaridis I, Barton P, Garner I, Leader D P, Bonhomme F, Catalan J, Simon D, Guenet J L. Proc Natl Acad Sci USA. 1985;82:7183–7187. doi: 10.1073/pnas.82.21.7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahdavi V, Chambers A P, Nadal-Ginard B. Proc Natl Acad Sci USA. 1984;81:2626–2630. doi: 10.1073/pnas.81.9.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman D J, Umeda P K, Sinha A M, Hsu H J, Jakovcic S, Rabinowitz M. Proc Natl Acad Sci USA. 1984;81:3044–3048. doi: 10.1073/pnas.81.10.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leinwand L A, Fournier R E, Nadal-Ginard B, Shows T B. Science. 1983;221:766–769. doi: 10.1126/science.6879174. [DOI] [PubMed] [Google Scholar]
- 17.Edwards Y H, Parkar M, Povey S, West L F, Parrington J M, Solomon E. Ann Hum Genet. 1985;49:101–109. doi: 10.1111/j.1469-1809.1985.tb01681.x. [DOI] [PubMed] [Google Scholar]
- 18.Weiss A, Leinwand L A. Annu Rev Cell Dev Biol. 1996;12:417–439. doi: 10.1146/annurev.cellbio.12.1.417. [DOI] [PubMed] [Google Scholar]
- 19.Yoon S J, Seiler S H, Kucherlapati R, Leinwand L. Proc Natl Acad Sci USA. 1992;89:12078–12082. doi: 10.1073/pnas.89.24.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Acakpo-Satchivi L. Ph.D. thesis. Bronx, NY: Albert Einstein College of Medicine; 1997. [Google Scholar]
- 21.Robbins J, Gulick J, Sanchez A, Howles P, Doetschman T. J Biol Chem. 1990;265:11905–11909. [PubMed] [Google Scholar]
- 22.Parker-Thornburg J, Bauer B, Palermo J, Robbins J. Dev Biol. 1992;150:99–107. doi: 10.1016/0012-1606(92)90010-e. [DOI] [PubMed] [Google Scholar]
- 23.Takeda S, North D L, Lakich M M, Russell SD, Kahng L S, Whalen R G. C R Acad Sci Ser III. 1992;315:467–472. [PubMed] [Google Scholar]
- 24.Riley J, Butler R, Ogilvie D, Finniear R, Jenner D, Powell S, Anand R, Smith J C, Markham A F. Nucleic Acids Res. 1990;18:2887–2890. doi: 10.1093/nar/18.10.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Epp T A, Dixon I M, Wang H Y, Sole M J, Liew C C. Genomics. 1993;18:505–509. doi: 10.1016/s0888-7543(11)80006-6. [DOI] [PubMed] [Google Scholar]
- 26.McCully J D, Wang R X, Kellam B, Sole M J, Liew C C. J Mol Biol. 1991;218:657–665. doi: 10.1016/0022-2836(91)90251-z. [DOI] [PubMed] [Google Scholar]
- 27.Liew C C, Sole M J, Yamauchi-Takihara K, Kellam B, Anderson D H, Lin L P, Liew J C. Nucleic Acids Res. 1990;18:3647–3651. doi: 10.1093/nar/18.12.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaenicke T, Diederich K W, Haas W, Schleich J, Lichter P, Pfordt M, Bach A, Vosberg H P. Genomics. 1990;8:194–206. doi: 10.1016/0888-7543(90)90272-v. [DOI] [PubMed] [Google Scholar]
- 29.Strehler E E, Strehler-Page M A, Perriard J C, Periasamy M, Nadal-Ginard B. J Mol Biol. 1986;190:291–317. doi: 10.1016/0022-2836(86)90003-3. [DOI] [PubMed] [Google Scholar]
- 30.Rappold G A, Vosberg H P. Hum Genet. 1983;65:195–197. doi: 10.1007/BF00286663. [DOI] [PubMed] [Google Scholar]
- 31.Karsch-Mizrachi I, Feghali R, Shows T B, Leinwand L A. Gene. 1990;89:289–294. doi: 10.1016/0378-1119(90)90020-r. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz C E, McNally E, Leinwand L, Skolnick M H. Cytogenet Cell Genet. 1986;43:117–120. doi: 10.1159/000132307. [DOI] [PubMed] [Google Scholar]
- 33.Britten R J. Science. 1986;231:1393–1398. doi: 10.1126/science.3082006. [DOI] [PubMed] [Google Scholar]
- 34.Li W H, Gouy M, Sharp P M, O’hUigin C, Yang Y W. Proc Natl Acad Sci USA. 1990;87:6703–6707. doi: 10.1073/pnas.87.17.6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin D I, Fiering S, Groudine M. Curr Opin Genet Dev. 1996;6:488–495. doi: 10.1016/s0959-437x(96)80072-4. [DOI] [PubMed] [Google Scholar]
- 36.Manak J R, Scott M P. Dev Suppl. 1994;61–77:61–77. [PubMed] [Google Scholar]
- 37.Krumlauf R. BioEssays. 1992;14:245–252. doi: 10.1002/bies.950140408. [DOI] [PubMed] [Google Scholar]
- 38.Hood L, Koop B F, Rowen L, Wang K. Cold Spring Harbor Symp Quant Biol. 1993;58:339–348. doi: 10.1101/sqb.1993.058.01.040. [DOI] [PubMed] [Google Scholar]
- 39.Koop B F, Rowen L, Wang K, Kuo C L, Seto D, Lenstra J A, Howard S, Shan W, Deshpande P, Hood L. Genomics. 1994;19:478–493. doi: 10.1006/geno.1994.1097. [DOI] [PubMed] [Google Scholar]
- 40.Schiaffino S, Reggiani C. J Appl Physiol. 1994;77:493–501. doi: 10.1152/jappl.1994.77.2.493. [DOI] [PubMed] [Google Scholar]
- 41.Pette D, Staron R S. Int Rev Cytol. 1997;170:143–223. doi: 10.1016/s0074-7696(08)61622-8. [DOI] [PubMed] [Google Scholar]
- 42.Brueckner J K, Itkis O, Porter J D. J Muscle Res Cell Motil. 1996;17:297–312. doi: 10.1007/BF00240928. [DOI] [PubMed] [Google Scholar]
- 43.Ludolph D C, Konieczny S F. FASEB J. 1995;9:1595–1604. doi: 10.1096/fasebj.9.15.8529839. [DOI] [PubMed] [Google Scholar]
- 44.Olson E N. Circ Res. 1993;72:1–6. doi: 10.1161/01.res.72.1.1. [DOI] [PubMed] [Google Scholar]
- 45.Lakich M M, Diagana T T, North D L, Whalen R G. J Biol Chem. 1998;273:15217–15226. doi: 10.1074/jbc.273.24.15217. [DOI] [PubMed] [Google Scholar]
- 46.Diagana T T, North D L, Jabet C, Fiszman M Y, Takeda S, Whalen R G. J Mol Biol. 1997;265:480–493. doi: 10.1006/jmbi.1996.0752. [DOI] [PubMed] [Google Scholar]
- 47.Takeda S, North D L, Diagana T, Miyagoe Y, Lakich M M, Whalen R G. J Biol Chem. 1995;270:15664–15670. doi: 10.1074/jbc.270.26.15664. [DOI] [PubMed] [Google Scholar]
- 48.Shimizu N, Prior G, Umeda P K, Zak R. Nucleic Acids Res. 1992;20:1793–1799. doi: 10.1093/nar/20.7.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rindt H, Knotts S, Robbins J. Proc Natl Acad Sci USA. 1995;92:1540–1544. doi: 10.1073/pnas.92.5.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knotts S, Rindt H, Robbins J. Nucleic Acids Res. 1995;23:3301–3309. doi: 10.1093/nar/23.16.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schimenti J C. Soc Gen Physiol Ser. 1994;49:85–91. , 85–91. [PubMed] [Google Scholar]
- 52.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 53.Dover G. Nature (London) 1982;299:111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- 54.Arnheim N, Krystal M, Schmickel R, Wilson G, Ryder O, Zimmer E. Proc Natl Acad Sci USA. 1980;77:7323–7327. doi: 10.1073/pnas.77.12.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andersson E, Matsunaga T. Immunogenetics. 1995;41:18–28. doi: 10.1007/BF00188428. [DOI] [PubMed] [Google Scholar]
- 56.Hood L, Campbell J H, Elgin S C. Annu Rev Genet. 1975;9:305–353. doi: 10.1146/annurev.ge.09.120175.001513. [DOI] [PubMed] [Google Scholar]
- 57.Epp T A, Wang R, Sole M J, Liew C C. J Mol Evol. 1995;41:284–292. doi: 10.1007/BF00186540. [DOI] [PubMed] [Google Scholar]
- 58.Moore L A, Tidyman W E, Arrizubieta M J, Bandman E. J Mol Biol. 1992;223:383–387. doi: 10.1016/0022-2836(92)90741-2. [DOI] [PubMed] [Google Scholar]
- 59.Wasserman W W, Fickett J W. J Mol Biol. 1998;278:167–181. doi: 10.1006/jmbi.1998.1700. [DOI] [PubMed] [Google Scholar]