Abstract
We have cloned a mutS homolog from Drosophila melanogaster called spellchecker1 (spel1) and have constructed spel1 mutant flies. MutS proteins promote the correction of DNA mismatches and serve important roles in DNA replication, recombination, and repair. The spel1 gene belongs to a subfamily of mutS first characterized by the MSH2 gene of yeast and which also includes hMSH2, one of the two major hereditary nonpolyposis colon cancer loci of humans. Like msh2 mutants in other species, we find that flies lacking the spel1 gene suffer a highly increased rate of instability in long runs of dinucleotide repeats when analyzed after 10–12 fly generations. Using a new assay, we have also discovered that mutations in spel1 decrease the stability of a dinucleotide repeat when it is copied into the site of a double-strand break during gene conversion. Contrary to the case in mammalian cells, spel1 deficiency does not affect tolerance of flies to a methylating agent nor does it affect resistance to γ-irradiation.
Keywords: mismatch repair, double-strand break repair, homologous recombination, gene conversion, hereditary nonpolyopsis colon cancer
The MutS protein of Escherichia coli recognizes base-pair mismatches in DNA as well as mismatches formed by small insertions or deletions. It binds to these structures and recruits other proteins, including MutL, needed to resolve the contradiction. This protein complex can identify which strand to repair by sensing the methylation state of the two strands or by the presence of single-strand breaks. A region of up to a few kilobases is removed from the “mutant” strand by exonuclease activity; DNA polymerase then replaces the sequence, thereby resolving the mismatch (reviewed in ref. 1). The fidelity of replication is severely compromised in mutS and mutL mutants, leading to an increased frequency of point mutations and to instability of simple DNA repeats (2, 3).
In addition to promoting faithful replication, MutS, the MutS homologs (MSHs) and associated mismatch repair proteins also are involved in processing intermediates of homologous recombination, promoting meiotic crossing over and double-strand break repair, and repairing oxidative and UV-induced DNA lesions (4–17). For example, heteroduplex DNA formed during gene conversion at the MAT locus in yeast is processed by mismatch repair at an early stage (10). Msh2 of yeast appears to regulate the forward movement of Holliday junctions (4). In vitro, MutS can regulate the extent of heteroduplex formation by blocking RecA-catalyzed branch migration through regions with mismatches (18, 19). This may be the basis for the inhibition of recombination between diverged sequences that is absent in mismatch repair-deficient mutants (6, 20–23). This activity may safeguard genome stability by preventing recombination between related genes within an organism and homologous genes between species.
This mismatch repair system has been conserved throughout evolution. At present, MSHs have been found in 14 species of bacteria and 10 species of eukaryotes ranging from fungi to humans. In some species, e.g., Saccharomyces cerevisiae, as many as six MSH genes have been found. In general, when multiple MSH genes are found in a species, they each have a distinct function. Of the six genes in yeast, MSH1 caries out the repair of mitochondrial DNA (24), while MSH3 and MSH6 act as specificity subunits for nuclear mismatch repair. They both form heteromers with MSH2, and as alternative partners they determine the affinity of the complex toward base–base mismatches (MSH2+MSH3) (25) or insertion/deletion mismatches (MSH2+MSH6) (26). MSH4 and MSH5 are expressed exclusively during meiosis and are required for normal levels of meiotic exchange and chromosome disjunction (7). MSH4 and MSH5 form a heteromeric complex with each other, and both are required for function.
The MSH2 gene limits the rate of spontaneous mutation (24). MSH2 mutants also have an increased rate of postmeiotic segregation, indicating the presence of unrepaired mismatches in heteroduplex DNA (24). Mutations in MSH2 as well as the mutL homologs, MLH1 and PMS1, lead to a dramatic increase in the instability of simple DNA repeats (3, 27).
In humans, similar repeat instability is known to occur in tumors from patients with hereditary nonpolyposis colon cancer (28). This form of cancer, as well as some sporadic cancers that also show repeat instability, were found to be induced by mutations in the human mutS and mutL homologs: hMSH2, hMLH1, hPMS1, and hPMS2 (29–31). These mutations are inherited as if dominant, resulting in a predisposition to hereditary nonpolyposis colon cancer in heterozygous individuals, but they act recessively at the level of the cell because expression of the functional copy is absent in the cancers.
Cell extracts from Drosophila have very high levels of mismatch-repair activity (32), so studying mismatch-repair genes in flies with the powerful genetic approaches that they offer should illuminate the complex interactions that have been hinted at in other organisms. To begin this study, we cloned a mutS homolog from Drosophila called spellchecker1 (spel1) and found that it is adjacent to a gene known as lethal(2)35Aa. We have constructed lines of Drosophila that have deletions of spel1. Although spel1-null mutants are viable and relatively fertile, they suffer a highly increased rate of instability in dinucleotide repeats when these repeats are transmitted normally through the germ line or when copied into the site of a double-strand break during gene conversion. However, these mutants are not significantly altered in their sensitivity to the methylating agent methyl methanesulfonate (MMS) or to γ-irradiation.
MATERIALS AND METHODS
Nucleic Acid Techniques.
DNA fragment isolation, ligation, subcloning, and blotting were performed by using standard methods (33). The sequences of the degenerate primers used for the initial PCR are ATCACNGGNCCNAAYATGGG and GTCATYTCNACCATRAANGT. The enhanced chemiluminescence (ECL) direct-labeling system (Amersham Pharmacia) was used to label gel-purified DNAs for screening the cDNA library. The cDNA library was constructed in the Thomas Schwarz laboratory (Stanford University) from a mixture of random primed and oligo(dT)-primed mRNA from Drosophila heads. Other cDNAs were obtained from the Berkeley Drosophila Genome Project/HHMI EST Project (unpublished data) and from Jeff Sekelsky in the Scott Hawley laboratory (University of California, Davis).
DNA Sequence Determination and Analysis.
The sequence of cloned genomic DNA and cDNA was determined by the dideoxy chain-termination method (34) as modified for Sequenase 2.0 (United States Biochemical) and for fluorescent DyeDeoxy terminators (Applied Biosystems). DNA sequence was compiled using the uwgcg software (35). The DNA sequence was assigned GenBank accession no. U17893.
Drosophila Stocks.
Fly stocks were maintained on cornmeal-molasses agar at 23°C. Mutations and abbreviations not explained here are described in Lindsley and Zimm (36) and in FlyBase (37). The chromosomes used are as follows: two ethyl methylsulfonate-induced mutants of l(2)35Aa; b l(2)35AaSF12 Adhn4, and b l(2)35AaSF32 Adhn2 pr cn (38) and four deficiencies, Df(2L)TE146(Z)GW7 al dp b l(2)pwn cn [abbreviated GW7 (39)], Df(2L)b84h1 [abbreviated b84h1 (40)], Df(2L)b80e3, pr bw (abbreviated b80e3), and Df(2L)A400, b cn bw; T(2;3;4)CA4 [abbreviated A400 (41)].
In Situ Hybridization, Drosophila transformation, and tests for rescue of l(2)35Aa.
Hybridization of a biotin-labeled DNA probe to polytene chromosomes from Drosophila salivary glands and its detection was carried out as described (42). Both the spel1 genomic region (from the SalI site at −247.5 to the EagI site at −242.3) and the GalNAc-T genomic region (from the BglII site at −250.9 to the EcoRI site at −245.7) were cloned into the P element vector, CaSpeR4, to create P{w+mC GalNAc-T = CaGal} and P{w+mC spel1 = CaSpel}. The standard method of P element transformation was used (43). Insertions of P{CaGal} and P{CaSpel} on the third chromosome were then crossed into stocks containing the ethyl methylsulfonate-induced alleles of l(2)35Aa and the GW7 and b84h1 deficiencies. Test crosses were performed by mating two stocks (each heterozygous for a different mutant allele). One of the stocks also carried the transgene to be tested. These crosses produced compound heterozygotes, deficient for l(2)35Aa, which die as pupae unless rescue occurs. To facilitate construction of spel1 null lines, an insertion of GalNAc-T on the second chromosome, P{CaGal}10.6, was recombined onto the GW7-deficiency chromosome.
Tests for Sensitivity to MMS or γ-Rays.
Crosses were performed as above with GW7 P{CaGal}10.6/CyO and b84h1/CyO, or with mei9AT1 or mie41D5, (mutants known to be sensitive to both MMS and γ-rays). The progeny were treated as early larvae, and phenotypes of the surviving adults were scored to determine the ratio of surviving homozygous mutants to heterozygotes. Treatments were as described in ref. 44, and the following doses were used: 0.0%, 0.05%, and 0.1% MMS and 0, 1,000, 1,500 and 2,000 rad.
Microsatellite Analysis.
Lines of spel1−/− flies were established by crossing the two deficiency stocks, Df(2L)GW7 P{CaGal}10.6/CyO and Df(2L)b84h1/CyO. From the offspring of this cross, spel1−/− sons and daughters were selected and mated individually to begin 23 lines. Single male and female offspring from these crosses were in turn mated to establish over 200 sublines. Control lines were set up in the same manner using an alternative deficiency [Df(2L)A400 in place of Df(2L)GW7] to produce spel1−/+ heterozygotes. All of these stocks were then maintained by transferring adults to fresh vials every 21 days. This resulted in populations averaging ≈5–50 flies per line.
After 10–12 generations, DNA was prepared from one male from each line and analyzed by PCR. New alleles were detected as novel-sized PCR fragments not present in either ancestor from the progenitor pair.
The primers used to amplify microsatellite regions are as follows: acagcaacaacggagcaac (elf1-f); tctgcaacctgggagtctgg (elf1-r); cgtcgatctcaagcgtctgc (mam-Dc); ggaagttggccgccgcattg (mam-Gc); aagatacatccgtgcgcgtat (sev-f); cccaactgaaaagcaactcc (sev-r); ccaccttagggcgtggctgt (35F-f); gacatatccaaacaccaatgcac (35F-r); cttcctgtgacaatggctgg (white-f); acacacacttttatactctctccgc (white-r); gggtctttctgcttcagttacc (U1a-f); ggaatacacgaatccccctt (U1a-r); ctcttagtgcgcagggattc (tenA-f); gagtcgctcaatggcaggc (tenA-r); ttccaagtcacacggacggg (AbdB-f); gcacaccgacaacacaagac (AbdB-r). Amplification was performed with one primer of each pair fluorescently labeled with either 6-carboxyfluorescein (6-FAM) or 5-hexachlorofluorescein (HEX), and the products were analyzed along with DNA size standards on a GeneScan gel (Applied Biosystems).
RESULTS
Isolation of spel1 cDNA and Genomic Clones.
We initially amplified a 191-bp fragment of the spel1 gene from Drosophila genomic DNA by using degenerate PCR. The primers were designed to hybridize to highly conserved regions of the MutS coding sequence based on an alignment of MutS members from bacteria, yeast, mouse, and human. This fragment was cloned, and its DNA sequence confirmed that it was from a gene of the MutS family. We used a PstI restriction site present in the cloned fragment (Fig. 1) to test whether the degenerate PCR amplified multiple homologs of mutS. No PstI-resistant fragments could be detected, suggesting that only a single mutS family member had amplified.
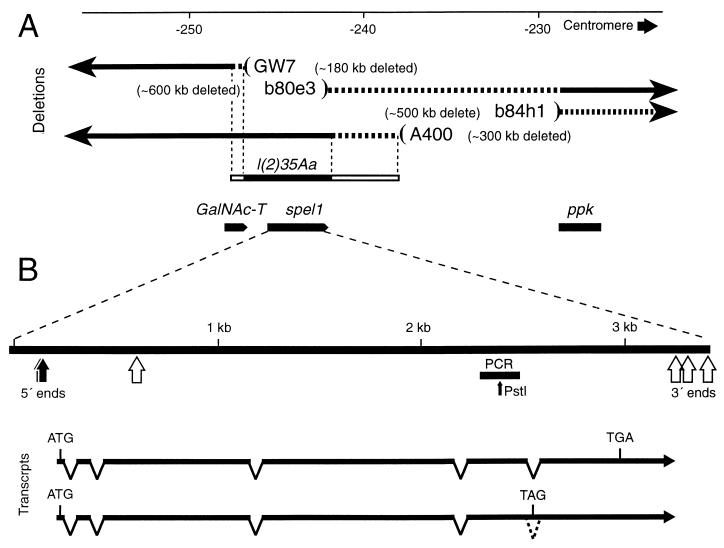
Figure 1.
Gene and transcript map of spel1. (A) The genomic region around spel1 is shown. Deletion-containing chromosomes used are displayed as black arrows representing the retained regions; on the opposite side, beyond the parentheses, blank areas indicate sequences that are absent. The regions of dashed line indicate the extent of uncertainty in the deletion end-points. The l(2)35Aa is a genetically defined locus known from ethyl methylsulfonate-induced mutations and was mapped by the GW7 and A400 deficiencies as shown. The solid and open portions represent the minimal and maximal extent of this locus. The neighboring genes GalNAc-T and ppk are localized from the DNA sequence. The numbering system is the one presented in Davis et al. (45). (B) The spel1 gene is enlarged with 5′ ends and sites of 3′ poly(A) in cDNAs indicated. Directly below the gene, the black bar represents the PCR fragment that was used as the initial probe. The PstI restriction site was used to test the homogeneity of the product of degenerate PCR. Examples of full-length or nearly full-length transcripts are shown below (one with the fifth intron retained and one completely spliced).
This fragment was used to probe a Drosophila head-specific cDNA library at high stringency, and five clones were obtained, all of which were from the same gene as the PCR fragment based on their DNA sequence. However, none of these appeared to represent the entire mRNA as judged by the absence of a poly(A) tail and by comparison to the other mutS family members. We performed 5′ and 3′ rapid ampplification of cDNA ends and obtained further cDNA clones from other libraries and found that the spel1 transcript appears to have multiple initiation and polyadenylation sites (Fig. 1B). It is also interesting that of seven cDNAs that extend nearly to the 3′ end, two of these retain the fifth (final) intron (Fig. 1B), which would lead to premature termination of translation, eliminating the final 182 amino acids. The first half of this region is highly conserved in the MSH2 family, but we do not yet know whether these transcripts are translated or what affect the truncated protein may have.
The amino acid sequence of spel1 is highly similar to the human Msh2 protein (65% similar and 43% identical residues) and all Msh2s from other species. The MSH2s and spel1 are more closely related to each other than to other members of the MutS family. They therefore compose a distinct subfamily, which may indicate that they are true functional homologs.
Mapping the spel1 Gene and Constructing spel1-Null Flies.
A spel1 cDNA was labeled and used to probe Drosophila polytene chromosomes from larval salivary gland squashes. The probe hybridized to a single site on the left arm of chromosome 2 at cytological position 35A4-B1. This region of the Drosophila genome has been the focus of much attention including extensive screens for recessive lethal or sterile mutants (45, 46). We obtained a λ clone from the Ashburner lab to determine the sequence of the spel1 genomic DNA and the surrounding area and discovered an ORF abutting the 5′ end of spel1. The amino acid sequence of this ORF is very similar to the human and bovine polypeptidyl N-acetylglucosamine transferase (GalNAc-T). Furthermore, a gene known as lethal(2)35Aa had been mapped to the same 10-kb region as the spel1 and the GalNAc-T genes. To test whether either of these genes could rescue the recessive lethality caused by l(2)35Aa mutations, transgenic flies were made that carried an ectopic copy of either the spel1 or GalNAc-T genomic sequence. We found that genomic DNA containing GalNAc-T could rescue the lethality, whereas the spel1 gene could not.
We were able to create spel1-null mutants by combining two existing deficiencies (Fig. 1A). These chromosomes have large deletions extending about 500 kb distally (b84h1) or about 180 kb proximally (GW7), but they only overlap in a small region that includes spel1 and the adjacent GalNAc-T gene. The GalNAc-T gene is supplied as a transgene to allow survival of the spel1 mutant created by this deficiency pair (Fig. 1A).
In some experiments, control flies were constructed by combining deletion b80e3 with deletion A400. This creates a spel1−/+ heterozygote that has a similar configuration in the surrounding region as exists in the null mutant (Fig. 1A).
Mutants Are Not Affected in Sensitivity to MMS or γ-Rays.
We tested spe1−/− mutants for survival after treatment with agents that lead to DNA double-strand breaks. Crosses were set up that produced both spel1−/− and heterozygous spel1−/+ progeny. The relative survival of these two genotypes was scored after treatment with the methylating agent MMS or exposure to γ-rays. We found the ratio of spel1−/− mutant offspring to heterozygotes was not significantly affected by MMS doses of 0.0%, 0.05%, and 0.1% or by γ-ray doses of 0, 1,000, 1,500, and 2,000 rad (with sample sizes of 4,307, 3,084, 2,779, and 236, 233, 188, and 10 surviving flies). As positive controls, mei9AT1 and mei41D5 mutants were tested in the same way, and both showed very dramatic sensitivity to both agents.
Microsatellites Are Unstable in spel1 Mutants.
MSH2 deficiency in yeast and humans leads to an increased rate of mutation. The most extensively characterized type of mutation occurs by the gain or loss of repeat units in simple repetitive sequences (microsatellites). To test whether spel1-null mutations also affect microsatellite stability, we chose eight loci with naturally occurring microsatellites and measured mutations arising over the course of 10–12 fly generations. More than 200 independent spel1−/− and spel1−/+ lines were established and later tested for new mutations by identifying novel-sized PCR fragments caused by the gain or loss of repeats. At one locus, no mutations were found in either spel1−/− or the control, but at each of the remaining seven loci, more mutations arose in the spel1-null background, and at some loci the rate was much higher in spel1−/− than in spel1−/+ (Table 1). For the seven informative loci combined, the microsatellite instability is significantly greater in spel1-null mutants as judged by the conservative sign test (P = (1/2)7 = 0.0078).
Table 1.
Instability of microsatellite repeats
| Locus |
spel1 −/−
|
spel1 +/−
|
||||
|---|---|---|---|---|---|---|
| Chromosomes changed | Total chromosomes | % changed | Chromosomes changed | Total chromosomes | % changed | |
| elf1 (CAG)7 | 0 | 378 | 0 | 0 | 192 | 0 |
| mam (CAG)7 | 1 | 188 | 0.5 | 0 | 190 | 0 |
| sev (AC)14 | 3 | 97 | 3.1 | 2 | 98 | 2.0 |
| 35F (AT)17 | 18 | 192 | 9.4 | 0 | 190 | 0 |
| w (AT)13 | 9 | 92 | 9.8 | 1 | 96 | 1.0 |
| U1a1 (AT)15 | 44 | 380 | 11.6 | 0 | 192 | 0 |
| tenA (AT)14 | 14 | 94 | 14.9 | 0 | 95 | 0 |
| AbdB (CA)26 | 100 | 378 | 26.5 | 0 | 192 | 0 |
One microsatellite-containing locus (yan, also known as aol) was omitted from the summary because we were unable to interpret the complex pattern of bands that it produced. It is possible that the mutation rate at this locus was so high that many of the flies tested were mosaics, i.e., with significant fractions of their cells having new mutations. Such flies could produce more than two sizes of alleles by PCR analysis. Total number of chromosomes tested is reported; not all lines were tested at each locus.
In a few cases, the progenitor of a new allele could not be inferred with certainty, because the starting population had more than one allele size. However, 90% of all new alleles were within a single repeat unit of a parental allele, suggesting that the change occurred by the addition or loss of a single unit. Overall, ≈58% were increases, but this proportion varied widely between loci. For example, the Ula1 locus had 36 apparent increases and only 7 decreases.
The spel1 Mutation Affects Repeat Stability During Double-Strand Break Repair.
Because DNA repair may be more error-prone than replication, we also tested the stability of a dinucleotide repeat when used as a template to repair a double-strand break (Fig. 2).
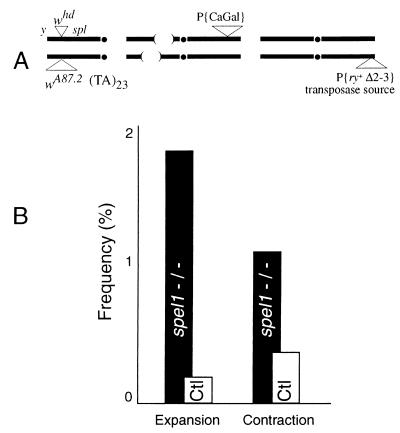
Figure 2.
Microsatellite instability during double-strand break repair. (A) Genotype of females in which a microsatellite template is copied into the site of a double-strand break. One of the X chromosomes carries whd, a P element insertion in the white locus that causes a double-strand break on excision (47). Its allele, wA87.2, was derived from whd as described (48). In this allele, the interior of the P element is replaced by a (TA)23 microsatellite. Only the outermost 17 bp of the P element remain at each end. Thus, double-strand breaks produced by excision at whd can be repaired by copying in the microsatellite sequence from the homolog. Chromosome 2 of these females is null for spel1 via the combination of overlapping deletions and has the rescuing GalNAc-T transgene in P{CaGal}, as described above. Finally, a source of P transposase is provided on chromosome 3 (49). (B) Sons from these females whose X chromosome was derived from the whd parent were selected using the yellow and split loci as markers (37). Among 4,377 such males tested individually by using PCR, we found that 40% had acquired the wA87.2 allele, as judged by agarose gel electrophoresis. This frequency is consistent with previous results (48). From these, we selected no more than one son from each female to analyze with the high-resolution genescan method as described above. That way, all gene conversions can be considered independent events. Of 276 such sons from spel1-null mothers, we found eight size changes: one 22-base expansion, four 2-base expansions, and three 2-base contractions. The controls (Ctl) were sons from females similar to those in A except that one or both of the spel1− deletions is replaced with a spel1+ chromosome. Among 541 sons in this group, there were three size changes, a 2-base expansion and contractions of 4 and 10 bp.
In this assay, P element excision creates a break at the white locus. The template for repair has an insertion of 23 copies of the dinucleotide AT. We looked for changes in the repeat number after events in which the entire dinucleotide repeat is copied into the homolog. We found an increase of >5-fold in the rate of repeat length changes (2.9% versus 0.5%) when this happened in a spel1-null background compared with heterozygotes with a wild-type spel1 allele. The difference was significant at P = 0.009 by Fisher’s exact test.
DISCUSSION
We have created flies that are mutant for the MSH2 homolog spel1 and established a phenotype by demonstrating that microsatellite sequences are less stable in this background under two circumstances. Instability was seen when repeats were simply replicated through several generations, similar to what has been shown for msh2 mutant yeast, mice, and human cells. We also discovered that spel1 is important for maintaining the integrity of repeats when they are copied into the site of a break during gene conversion. Recently, a similar study did not detect such a difference (T. Dray, C. Raynor, and G. Gloor, personal communication). One important distinction in that study may be that it examined a trinucleotide repeat, whereas ours was a dinucleotide. Trinucleotide repeats are intrinsically more stable than dinucleotides (27). In addition, the stability of different sequence repeat arrays varies widely (e.g., Table 1).
Humans with a mutant allele of hMSH2 are prone to certain cancers, most notably hereditary nonpolyposis colon cancer (29, 30). The cells in these cancers generally do not display gross rearrangements or chromosome loss typical of other cancers, but instead show heterogeneity in the number of copies of simple repeats (28). A similar fate has been found for mice with a Msh2 knockout except that the predominant form of cancer is lymphoma (50). Msh2-deficient rodent and human cell lines have been found to be resistant to killing by some methylating agents (51–54), but we see no effect of spel1 mutations on sensitivity to MMS or to γ-irradiation. This may indicate that flies process the damage differently, perhaps without the involvement of spel1.
Recent studies on mutation rates at microsatellite loci in wild-type Drosophila reported great variation between loci, with the longest dinucleotide repeats tending to be the least stable and average rates of about 6 × 10−6 changes per generation (55, 56). Although we cannot accurately determine a rate from our data because the lines were not independent in the first generations, the rates calculated by Schug et al. (56) and Schlotterer et al. (57) appear lower than we found in the spel1−/+ heterozygous controls. This suggests that a single copy of spel1 may not be sufficient to produce the wild-type level of stability. If this is true, mutations in spel1 have an even greater effect on repeat stability than we have calculated.
Preliminary evidence indicates spel1 also affects meiotic recombination. We measured recombination at four intervals on the X chromosome in spel1-null and in wild-type control flies. From 1,139 spel1− and 1,013 wild-type progeny, the rates of recombination were higher in spel1 mutants in the two distal intervals. The differences in rates obtained for interval 1 (7.3% spel1− vs. 4.2% wild-type) and interval 2 (9.7% spel1− vs. 5.3% wild-type) were highly significant according to Fisher’s exact test. We detected no differences in the rates over the two proximal intervals. Corresponding msh2 mutations in S. cerevisiae also cause defects in meiotic recombination, although a phenotype of altered recombination frequencies at different chromosome positions has not been reported. The yeast mutation increases postmeiotic segregation and increases recombination between diverged sequences—phenotypes we have not yet tested. Our experiments cannot explain why spel1 deficiency might cause the effect we see, but other mutations in Drosophila are known to affect recombination unequally in distal vs. proximal regions (e.g., mei-41).
Do Drosophila have multiple Msh genes, as seems to be the case in most eukaryotes? There are no reports yet of other Msh genes from flies, despite the fact that three other labs have independently isolated clones of spel1 while hunting for other homologs (G. Crouse, C. Osgood, and J. Sekelsky, personal communications). Still, it seems likely that there are more MsH genes yet to be found in flies, given the strong trend of eukaryotes to have multiple MsH members with diversified function.
MutS homologs play an important role in the correction of replication errors. Recent findings, some of which were quite surprising, show that they also function in recombination and in repair of DNA. They have at least three roles in recombination: preventing recombination between diverged sequences (6, 22, 23, 58), processing heteroduplex intermediates (4, 5), and trimming nonhomologous ends of certain recombination intermediates (11, 12, 59). The meiosis-specific MSHs may well have yet another function (7). There also is evidence that they can function in repair of oxidative damage (14–17) and “nucleotide excision repair” (13) and that Msh2 physically interacts with several nucleotide excision-repair proteins (60). Investigation of mismatch repair in bacteria and yeast has been very fruitful. By exploiting the powerful genetic tools available in Drosophila, we hope to complement studies in mammals and shed more light on the roles of mismatch repair proteins in higher eukaryotes.
Acknowledgments
We thank Christine Preston for probing polytene chromosomes and Saumil Mehta for help, Michael Ashburner and John Roote for supplying fly stocks and genomic clones, Tom Schwarz for the cDNA library, and Jeff Sekelsky and the Berkeley Drosophila Genome Project for cDNA clones. We also thank Christine Preston, Dena Johnson-Schlitz, Kohji Kusano, and James Crow for helpful suggestions on the manuscript. National Institutes of Health Grant GM30948 to W.E. supported this work. This paper is number 3,532 from the Laboratory of Genetics.
ABBREVIATIONS
- MSH
MutS homolog
- MMS
methyl methanesulfonate
- GalNAc-T
N-acetylglucosamine transferase
Footnotes
Data Deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U17893).
References
- 1.Modrich P, Lahue R. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 2.Levinson G, Gutman G A. Nucleic Acids Res. 1987;15:5323–5338. doi: 10.1093/nar/15.13.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strand M, Prolla T A, Liskay R M, Petes T. Nature (London) 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 4.Alani E, Reenan R, Kolodner R. Genetics. 1994;137:19–39. doi: 10.1093/genetics/137.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alani E, Lee S, Kane M F, Griffith J, Kolodner R D. J Mol Biol. 1997;265:289–301. doi: 10.1006/jmbi.1996.0743. [DOI] [PubMed] [Google Scholar]
- 6.de Wind N, Dekker M, Berns A, Radman M, te Riele H. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 7.Ross-Macdonald P, Roeder G S. Cell. 1994;79:1069–1080. doi: 10.1016/0092-8674(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 8.Hunter N, Borts R H. Genes Dev. 1997;11:1573–1582. doi: 10.1101/gad.11.12.1573. [DOI] [PubMed] [Google Scholar]
- 9.Baker S M, Plug A W, Prolla T A, Bronner C E, Harris A C, Yao X, Christie D M, Monell C, Arnheim N, Bradley A, et al. Nat Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 10.Haber J E, Ray B L, Kolb J M, White C I. Proc Natl Acad Sci USA. 1993;90:3363–3367. doi: 10.1073/pnas.90.8.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saparbaev M, Prakash L, Prakash S. Genetics. 1996;142:727–736. doi: 10.1093/genetics/142.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugawara N, Paques F, Colaiacovo M, Haber J E. Proc Natl Acad Sci USA. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mellon I, Champe G N. Proc Natl Acad Sci USA. 1996;93:1292–1297. doi: 10.1073/pnas.93.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeWeese T L, Shipman J M, Larrier N A, Buckley N M, Kidd L R, Groopman J D, Cutler R G, te Riele H, Nelson W G. Proc Natl Acad Sci USA. 1998;95:11915–11920. doi: 10.1073/pnas.95.20.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson A L, Chen R, Loeb L A. Proc Natl Acad Sci USA. 1998;95:12468–12473. doi: 10.1073/pnas.95.21.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leadon S A, Avrutskaya A V. Mutat Res. 1998;407:177–187. doi: 10.1016/s0921-8777(98)00007-x. [DOI] [PubMed] [Google Scholar]
- 17.Leadon S A, Avrutskaya A V. Cancer Res. 1997;57:3784–3791. [PubMed] [Google Scholar]
- 18.Worth L J, Clark S, Radman M, Modrich P. Proc Natl Acad Sci USA. 1994;91:3238–3241. doi: 10.1073/pnas.91.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Worth L, Jr, Bader T, Yang J, Clark S. J Biol Chem. 1998;273:23176–23182. doi: 10.1074/jbc.273.36.23176. [DOI] [PubMed] [Google Scholar]
- 20.Feinstein S I, Low K B. Genetics. 1986;113:13–33. doi: 10.1093/genetics/113.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rayssiguier C, Thaler D S, Radman M. Nature (London) 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 22.Datta A, Adjiri A, New L, Crouse G F, Jinks Robertson S. Mol Cell Biol. 1996;16:1085–1093. doi: 10.1128/mcb.16.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selva E M, Maderazo A B, Lahue R S. Mol Gen Genet. 1997;257:71–82. doi: 10.1007/pl00008619. [DOI] [PubMed] [Google Scholar]
- 24.Reenan R A G, Kolodner R D. Genetics. 1992;132:975–985. doi: 10.1093/genetics/132.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. Curr Biol. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 26.Alani E. Mol Cell Biol. 1996;16:5604–5615. doi: 10.1128/mcb.16.10.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sia E A, Kokoska R J, Dominska M, Greenwell P, Petes T D. Mol Cell Biol. 1997;17:2851–2858. doi: 10.1128/mcb.17.5.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aaltonen L A, Peltomaki P, Leach F S, Sistonen P, Pylkkanen L, Mecklin J-P, Jarvinen H, Powell S M, Jen J, Hamilton S R, et al. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 29.Fishel R, Lescoe M, Rao M, Copeland N, Jenkins N, Garber J, Kane M, Kolodner M. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 30.Leach F, Nicolaides N, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen L, Nystrom-Lahti M, et al. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 31.Nicolades N C, Papadopoulos N, Liu B, Wei Y-F, Carter K C, Ruben S M, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, et al. Nature (London) 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 32.Bhui-Kaur A, Goodman M F, Tower J. Mol Cell Biol. 1998;18:1436–1443. doi: 10.1128/mcb.18.3.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 34.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindsley D L, Zimm G G. The Genome of Drosophila melanogaster. San Diego: Academic; 1992. [Google Scholar]
- 37.FlyBase. Nucleic Acids Res. 1996;24:53–56. doi: 10.1093/nar/24.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woodruff R C, Ashburner M. Genetics. 1979;93:133–149. doi: 10.1093/genetics/92.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gubb D, Roote J, Harrington G, McGill S, Durrant B, Shelton M, Ashburner M. Chromosoma. 1985;92:116–123. [Google Scholar]
- 40.Alexandrova M V. Drosophila Inf Serv. 1986;63:21–22. [Google Scholar]
- 41.Aaron C S. Mutat Res. 1979;63:127–137. doi: 10.1016/0027-5107(79)90109-x. [DOI] [PubMed] [Google Scholar]
- 42.Engels W R, Preston C R, Thompson P, Eggleston W B. Focus. 1986;8:6–8. [Google Scholar]
- 43.Rubin G M, Spradling A C. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 44.Boyd J B, Golino M D J, Nguyen T D, Green M M. Genetics. 1976;84:485–506. doi: 10.1093/genetics/84.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis T, Trenear J, Ashburner M. Genetics. 1990;126:105–119. doi: 10.1093/genetics/126.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chia W, Karp R, McGill S, Ashburner M. J Mol Biol. 1985;186:689–706. doi: 10.1016/0022-2836(85)90389-4. [DOI] [PubMed] [Google Scholar]
- 47.Gloor G B, Nassif N A, Johnson-Schlitz D M, Preston C R, Engels W R. Science. 1991;253:1110–1117. doi: 10.1126/science.1653452. [DOI] [PubMed] [Google Scholar]
- 48.Johnson-Schlitz D M, Engels W R. Mol Cell Biol. 1993;13:7006–7018. doi: 10.1128/mcb.13.11.7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robertson H M, Preston C R, Phillis R W, Johnson-Schlitz D, Benz W K, Engels W R. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Wind N, Dekker M, van Rossum A, van der Valk M, te Riele H. Cancer Res. 1998;58:248–255. [PubMed] [Google Scholar]
- 51.Branch P, Hampson R, Karran P. Cancer Res. 1995;55:2304–2309. [PubMed] [Google Scholar]
- 52.Aquilina G, Ceccotti S, Martinelli S, Hampson R, Bignami M. Cancer Res. 1998;58:135–141. [PubMed] [Google Scholar]
- 53.Andrew S E, McKinnon M, Cheng B S, Francis A, Penney J, Reitmair A H, Mak T W, Jirik F R. Proc Natl Acad Sci USA. 1998;95:1126–1130. doi: 10.1073/pnas.95.3.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaina B, Ziouta A, Ochs K, Coquerelle T. Mutat Res. 1997;381:227–241. doi: 10.1016/s0027-5107(97)00187-5. [DOI] [PubMed] [Google Scholar]
- 55.Schug M D, Mackay T F, Aquadro C F. Nat Genet. 1997;15:99–102. doi: 10.1038/ng0197-99. [DOI] [PubMed] [Google Scholar]
- 56.Schlotterer C, Ritter R, Harr B, Brem G. Mol Biol Evol. 1998;15:1269–1274. doi: 10.1093/oxfordjournals.molbev.a025855. [DOI] [PubMed] [Google Scholar]
- 57.Schug M D, Wetterstrand K A, Gaudette M S, Lim R H, Hutter C M, Aquadro C F. Mol Ecol. 1998;7:57–70. doi: 10.1046/j.1365-294x.1998.00304.x. [DOI] [PubMed] [Google Scholar]
- 58.Chambers S R, Hunter N, Louis E J, Borts R H. Mol Cell Biol. 1996;16:6110–6120. doi: 10.1128/mcb.16.11.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kirkpatrick D T, Petes T D. Nature (London) 1997;387:929–931. doi: 10.1038/43225. [DOI] [PubMed] [Google Scholar]
- 60.Bertrand P, Tishkoff D X, Filosi N, Dasgupta R, Kolodner R D. Proc Natl Acad Sci USA. 1998;95:14278–14283. doi: 10.1073/pnas.95.24.14278. [DOI] [PMC free article] [PubMed] [Google Scholar]