Abstract
Mice lacking the RelA (p65) subunit of NF-κB die between days 14 and 15 of embryogenesis because of massive liver destruction. Fibroblasts and macrophages isolated from relA−/− embryos were found to be highly sensitive to tumor necrosis factor (TNF) cytotoxicity, raising the possibility that endogenous TNF is the cause of liver cell apoptosis. To test this idea, we generated mice lacking both TNF and RelA. Embryogenesis proceeds normally in such mice, and TNF/RelA double-deficient mice are viable and have normal livers. Thus, the RelA-mediated antiapoptotic signal that protects normal cells from TNF injury in vitro can be shown to be operative in vivo.
One of the defining characteristics of tumor necrosis factor (TNF) is its cytostatic/cytotoxic activity in vitro for certain mouse and human transformed cells (1, 2). Early studies showed that cytotoxicity could be enhanced by inhibitors of protein or RNA synthesis (3) and that cell killing had the features of an apoptotic process (4, 5). Although suspicions were raised that TNF cytotoxicity might be an epiphenomenon not relevant to the biological functions of TNF, the discovery of a growing family of ligands and receptors related to TNF (6)—most notably Fas/Fas ligand with potent apoptosis inducing activity—provided support for the idea that TNF-mediated cytotoxicity has physiological significance. Further support comes from recent findings implicating NF-κB in TNF cytotoxicity. Although activation of NF-κB was known to mediate several activities of TNF (7), recent studies indicate that NF-κB activation by TNF protects cells from TNF cytotoxicity (8–11). Thus, fibroblasts from mice with a deletion of the RelA component of NF-κB are highly sensitive to TNF killing, in contrast to the resistance of normal fibroblasts (8). In addition, inhibition of NF-κB activity by overexpression of IκB greatly increases the sensitivity of cells to TNF cytotoxicity (9–11). Mice lacking the RelA component of NF-κB die in utero at days 14–15, and the major morphological abnormality in these mice at the time of death is massive liver degeneration associated with apoptosis of hepatocytes (12, 13). Because of the sensitivity of fibroblasts and macrophages from RelA-deficient mice to TNF-mediated cytotoxicity, Beg and Baltimore (8) suggested that endogenous production of TNF was responsible for the killing of hepatocytes in these mice. We tested this hypothesis by mating TNF-deficient and RelA-deficient mice to determine what effect absence of TNF has on the RelA-deficient phenotype.
MATERIALS AND METHODS
Derivation and Maintenance of Knockout Mice.
TNF-deficient mice and RelA-deficient mice have been described in detail elsewhere (13, 14). relA+/− mice backcrossed to C57 BL/6 mice for 10 successive generations and TNF−/− mice backcrossed to C57BL/6 mice for 5 generations were used in this study. Mice were maintained in a specific pathogen-free mouse facility at Aichi Cancer Center Research Institute.
DNA and RNA Blot Analyses.
DNA and total RNA were isolated by using proteinase K/SDS and guanidium thiocyanate/CsCl procedures, respectively (15, 16). For genotyping, DNA from developing embryos and limbs and tails of postnatal mice was digested with restriction enzymes and separated by agarose gel electrophoresis and then transferred to nitrocellulose filters (Schleicher & Schuell). To confirm disruption of the TNF gene, DNA digested with BamHI was examined with a 1,218-bp SmaI fragment from the lymphotoxin-β gene (nucleotides 3,181–4,399, GenBank accession no. U06950), a kind gift of Sergei Nedospasov. For analyzing relA disruption, DNA digested with PstI was analyzed with an 850-bp PstI–SmaI fragment of the 5′-untranslated region of the relA gene (13). Total RNA was separated by 2.2 M formaldehyde agarose gel electrophoresis and transferred to nitrocellulose filters. RNA blots were analyzed by using cDNA probes for relA (codons 185–277, ref. 17), c-rel (codons 144–277, ref. 18), and relB (codons 458–580, ref. 19).
Immunohistochemistry and Histology.
Five-micrometer frozen sections of whole embryos were air-dried, fixed with cold acetone, and reacted with hamster anti-murine TNF mAb (ref. 20; kindly provided by Dr. Robert Schreiber) or a polyclonal rabbit anti-mouse TNF antibody (Genzyme). Binding was visualized by an avidin–biotin enzyme complex method (Vectastain Elite ABC kit; Vector Laboratories), with subsequent counterstaining with hematoxylin. For histological examination, tissues were fixed in 4% paraformaldehyde and embedded in paraffin. The sections (5 μm) were stained with hematoxylin/eosin.
Reverse Transcription–PCR (RT-PCR) Analysis of the Expression of TNF, TNF Receptor (TNFR)1, and TNFR2.
The expression of TNF, TNFR1, and TNFR2 was determined by using RT-PCR analysis. Total RNA (10 μg) isolated from tissues or cultured cells of 14.5-day postcoitum (dpc) embryos or from cultured cells of 3-day postnatal mice was used for cDNA synthesis using oligo(dT) primers and 200 units of SuperScript II (Life Technologies, Gaithersburg, MD). PCR was performed in a final volume of 50 μl containing all four dNTPs, 2 mM MgCl2, 2.5 units of Taq polymerase (Takara Shuzo, Otsu, Japan), and each primer at 0.2 μM by using a Program Control System PC-700 (Astec, Fukuoka, Japan). Each amplification cycle consisted of 93°C for 1 min, 60°C for 1 min, and 72°C for 1 min. After 30 cycles, PCR products were separated by electrophoresis on a 1.5% agarose gel and visualized by ethidium bromide staining. The primers used in these experiments were designed to distinguish authentic RT-PCR products from PCR products generated from contaminating genomic DNA as described (21).
TNF Cytotoxicity.
Cells from liver and brain and fibroblasts of 14.5-dpc embryos and cells from spleen, kidney and heart of 3-day old mice were prepared according to methods described previously (22). Briefly, liver, kidney, and heart cells were plated on collagen type I-coated plastic dishes (Becton Dickinson Labware) and cultured in DMEM (Life Technologies, Gaithersburg, MD) with 10% fetal calf serum. Fibroblasts and cells from brain were plated on plastic dishes in DMEM with 10% fetal calf serum. Spleen cells were cultured in RPMI medium 1640 (Life Technologies, Gaithersburg, MD) with 10% fetal calf serum. Cultured cells from embryonic liver expressed α-fetoprotein, and cells from heart continued to show pulsation in vitro. After 24 hours in culture, recombinant murine TNF (Sigma) was added at a final concentration of 1, 10, or 100 ng/ml. Viability was examined by using trypan blue exclusion.
Injection of TNF.
Newborn mice from the mating of TNF−/−relA+/− mice were injected intravenously with 1 μg or 0.2 μg of recombinant murine TNF (Sigma). Mice were monitored for 24 hours after injection and then genotyped by Southern blot analysis.
RESULTS
TNF/RelA Double-Deficient Mice.
Mice lacking relA die in utero around 14.5 days postcoitum (dpc), as shown in Table 1, confirming previous results (12, 13). To determine the role of TNF in the embryonic death of RelA-deficient mice, mice heterozygous for the relA gene deletion were crossed to TNF-deficient mice for two successive generations. From the second generation, TNF−/−relA+/− mice were selected and intercrossed. As shown in Table 1, matings of TNF−/−relA+/− gave rise to viable TNF−/−relA−/− progeny in the predicted Mendelian ratio. This is in contrast to the embryonic lethality of mice with the TNF+/+relA−/− genotype (Table 1). In a broad survey of tissues from double-deficient mice, no morphological abnormalities were found, either in liver or other tissues, in contrast to the liver degeneration in mice with TNF+/+ relA−/− genotype (Fig. 1). Thus, elimination of TNF rescues RelA-deficient mice from embryonic lethality by abolishing TNF-mediated liver degeneration. Furthermore, in the absence of TNF, RelA is dispensable for embryonic and postnatal development.
Table 1.
Influence of TNF genotype on survival of RelA-deficient mice. Results of matings of TNF+/+ or TNF−/− mice with relA+/− mice
| Mouse age |
TNF+/+relA+/− (♀) × TNF+/+relA+/− (♂)
|
TNF−/−relA+/− (♀) × TNF−/−relA+/− (♂)
|
||||
|---|---|---|---|---|---|---|
| TNF+/+relA−/− | TNF+/+relA+/− | TNF+/+relA+/+ | TNF−/−relA−/− | TNF−/−relA+/− | TNF−/−relA+/+ | |
| 14.5 dpc | 11 | 31 | 12 | 7 | 13 | 6 |
| 15.5 dpc | 3 | 24 | 6 | — | — | — |
| 16.5 dpc | 0 | 25 | 11 | 10 | 22 | 12 |
| Newborn | 0 | 18 | 9 | 25 | 62 | 29 |
| 21 Days old | 0 | 146 | 77 | 9 | 22 | 10 |
Data are given as number of mice generated.
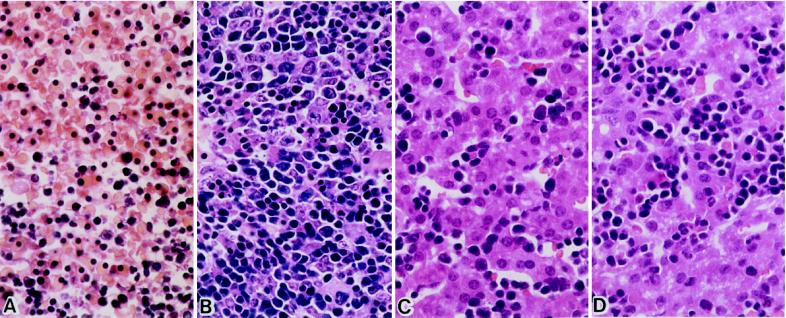
Figure 1.
Normal development of the liver in TNF−/−relA−/− mice. (A) TNF+/+relA−/− 14.5-dpc embryo. No viable hepatocytes can be seen. (B) TNF−/−relA−/− 14.5-dpc embryo. (C) TNF−/−relA−/− newborn mouse. (D) TNF−/−relA+/+ newborn mouse. Sections are hematoxylin/eosin-stained. (×400.)
TNF−/−relA−/− mice are healthy and active during the first 4–5 weeks of life. After approximately 40 days of age, however, the mice become sick and die. At 40 days of age, infiltration of lymphocytes was found in both portal areas and lobules of the liver (Fig. 2 A–C). In the lung, thickening of alveolar walls with inflammatory cell infiltration was observed (Fig. 2E). Inflammatory infiltrates also were seen around the bronchi and the pulmonary blood vessels. In the kidney, glomerular abnormalities with expansion of mesangial areas and thickening of capillary walls were seen (Fig. 2G). No such abnormalities were observed in TNF−/−relA+/+ mice (Fig. 2 D, F, and H). The inflammatory changes are most likely related to the compromised immune system of TNF/RelA double-deficient mice. As mice deficient in TNF (14, 23) or mice reconstituted with relA−/− lymphocytes (13, 24) have impaired immune systems, mice lacking both TNF and RelA would be expected to be highly susceptible to microbial and viral infections.
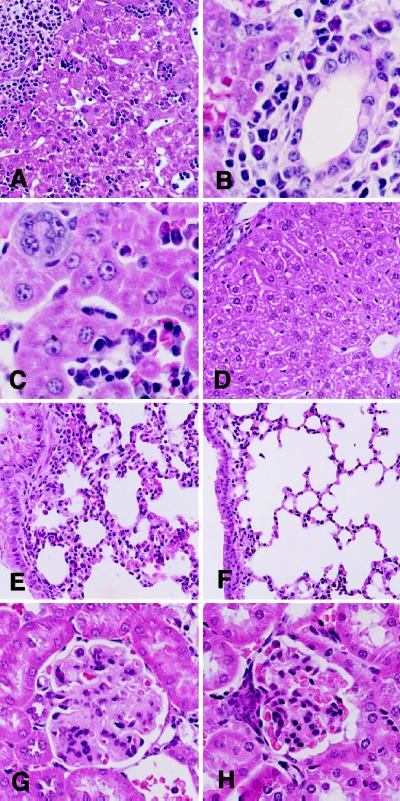
Figure 2.
Morphological abnormalities in TNF−/−relA−/− 40-day-old mice. (A–C) Liver from TNF−/− relA−/− 40-day-old mouse. (A) Portal areas and parenchyma showing marked cellular infiltrates. (×150). (B) Predominantly lymphocytic infiltrate in portal areas with bile ducts showing regenerative changes. (×450). (C) Intrasinusoidal aggregates of inflammatory cells. Megakaryocytes were present, but other hematopoietic cells were not observed. (×450). (D) Liver from TNF−/−relA+/+ 40-day-old mouse. No abnormalities observed. (×150.) (E) Lung from TNF−/−relA−/− 40-day-old mouse. Thickened alveolar walls associated with lymphocytic infiltration, activated pneumocytes, and increased mucous cells in bronchi. (×150) (F) Lung from TNF−/−relA+/+ 40-day-old mouse. No abnormalities observed. (×150.) (G) Kidney from TNF−/−relA−/− 40-day-old mouse. Expanded mesangial areas in glomeruli with increased mesangial matrix and numbers of cells. Marked thickening of capillary walls with narrowing of the capillary lumen. (×300.) (H) Kidney from TNF−/−relA+/+ 40-day-old mouse. No abnormalities observed. (×300.)
Origin of TNF in Embryos.
To test whether TNF present in embryos is produced by the embryos themselves or produced by mothers and transmitted to the embryos through the placenta, two sets of crosses (female TNF−/−relA+/− mated with male TNF+/+relA+/− and female TNF+/+relA+/− with male TNF−/−relA+/−) were set up (Table 2). Maternal TNF was not the cause of death of TNF+/−relA−/− progeny, as the timing of embryonic lethality did not differ in the TNF+/−relA−/− progeny from both crosses. Thus, TNF is produced by the embryos themselves. During this study, it was noted that TNF+/−relA−/− mice survived longer than TNF+/+relA −/− mice, as some of the former were alive even up to birth whereas the latter died between 14.5 and 15.5 dpc (Tables 1 and 2). This gene dosage effect of TNF on the survival of relA−/− embryos is consistent with previous observations showing less TNF production in TNF+/− mice as compared with TNF+/+ mice (14).
Table 2.
Influence of parental TNF genotype and TNF gene dosage on embryonic lethality of relA−/− mice
| Mouse age |
TNF−/−relA+/− (♀) × TNF+/+relA+/− (♂)
|
TNF+/+relA+/− (♀) × TNF+/−relA+/+ (♂)
|
||||
|---|---|---|---|---|---|---|
| TNF+/−relA−/− | TNF+/−relA+/− | TNF+/−relA+/+ | TNF+/−relA−/− | TNF+/−relA+/− | −+/−relA+/+ | |
| 16.5 dpc | 5 | 10 | 7 | 7 | 19 | 8 |
| 18.5 dpc | 3 | 17 | 8 | 4 | 22 | 12 |
| Newborn | 2 | 24 | 16 | 1 | 39 | 19 |
| 21 Days old | 0 | 18 | 8 | 0 | 25 | 13 |
Data are given as number of mice generated.
To examine TNF production during development, embryos of 11.5- to 13.5-dpc mice were examined by immunohistochemistry using a hamster anti-TNF mAb (20) and a polyclonal rabbit anti-mouse TNF antibody. The specificity of these reagents for TNF was confirmed by their complete lack of reactivity with normal tissues from TNF−/− mice. Staining with these antibodies showed that immunoreactive TNF is present in most tissues of TNF+/+ embryos, including liver. Although staining tended to be diffuse and difficult to ascribe to discrete cell populations, there were a small number of cells in the liver that were strongly stained (Fig. 3). By using RT-PCR, TNF transcripts were detected in all embryonic tissues tested and especially strongly in liver, spleen, and thymus (Fig. 4).
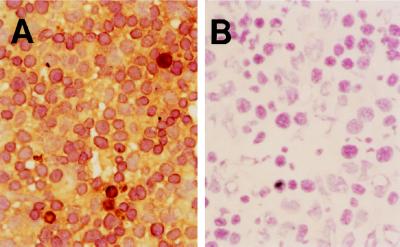
Figure 3.
Immunostaining of TNF in the liver of 14.5 dpc embryo with hamster anti-mouse TNF antibody. Shown are TNF+/+ (A) and TNF−/− (B) embryos. (×400)
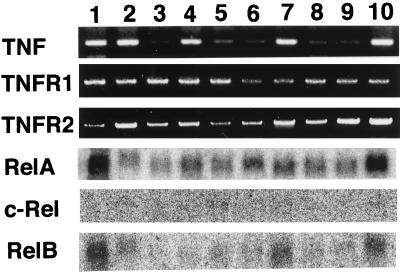
Figure 4.
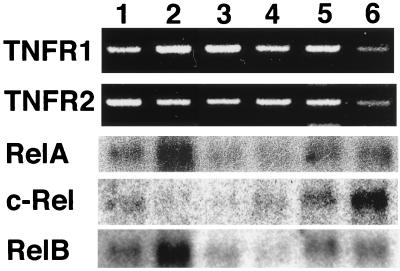
Expression of TNF, TNFR1, TNFR2, relA, c-rel, and relB. RNA was isolated from various tissues and cultured fibroblasts of TNF+/+RelA+/+ 14.5 dpc embryos. Lanes: (1) fibroblasts, (2) liver, (3) kidney, (4) spleen, (5) heart, (6) brain, (7) thymus, (8) lung, (9) intestine, and (10) placenta. The expression of TNF, TNFR1, and TNFR2 was analyzed by using RT-PCR. The expression of relA, c-rel, and relB was examined by using Northern blot analysis.
Expression of TNF Receptors and NF-κB Subunits in Tissues of Embryonic Mice.
Although TNF appears to be present ubiquitously in the embryo, tissue destruction was restricted to the liver at the time of death of TNF+/+ relA−/− mice. To explore the reason for this tissue specific destruction by TNF, the expression of TNF receptors and NF-κB family members was examined. By using RT-PCR, transcripts of TNFR1 and TNFR2 were detected in all embryonic tissues tested (Fig. 4). Among NF-κB family members, relA mRNA was ubiquitously expressed at high levels. relB mRNA was also widely expressed, but showed particularly high expression in placenta, thymus, and liver. Transcripts of the c-rel gene were below the level of detection in these embryonic tissues except for trace amounts in spleen. This was also the case in RelA-deficient mice, where no compensatory increase in expression of other NF-κB members was found (13).
TNF Sensitivity of Cultured Cells from RelA-Deficient Mice.
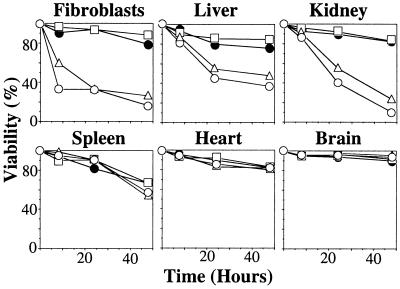
To analyze cell type differences in TNF sensitivity, cultured cells derived from various tissues of TNF−/−relA−/− or TNF−/−relA+/+ mice were examined for their sensitivity to TNF. As shown Fig. 5, cells from relA−/− liver and kidney, as well as embryonic fibroblasts, were sensitive to TNF, whereas cells from relA−/− spleen, heart, and brain were not. All cell types from relA+/+ mice were resistant to TNF treatment. Transcripts of TNFR1 and 2 were detectable in all these cultured cells (Fig. 6). The expression of NF-κB family members in cultured cells from relA wild-type mice was examined by using Northern blot analysis. Transcripts of relA and relB were present in all cells tested. Transcripts of c-rel were detected in cells derived from spleen, heart, and brain, but not in cells from kidney or embryonic fibroblasts. Low levels of c-rel expression were detected in cultured cells from the liver, but this may be accounted for by residual hematopoietic cells derived from the embryonic liver. The presence of c-rel expression generally correlates well with resistance of cells to TNF. Thus, c-rel, when it is expressed, may also mediate antiapoptotic signals that protect cells from TNF cytotoxicity.
Figure 5.
TNF cytotoxicity in vitro. Cultured cells were prepared as described in Materials and Methods and were treated with TNF for 8, 24, or 48 hours. TNF−/−relA−/− cells with no TNF (□); TNF−/−relA−/− cells with 10 ng/ml TNF (▵); TNF−/−relA−/− cells with 100 ng/ml TNF (○) and TNF−/−relA+/+ cells with 100 ng/ml TNF (●). Viability was examined by using trypan blue exclusion.
Figure 6.
The expression of TNFR1, TNFR 2, relA, c-rel, and relB in cultured cells. RNA was isolated from cultured cells of TNF −/−relA+/+ embryos or postnatal mice (see Fig. 5). Lanes: (1) hepatocytes (14.5 dpc) (2) fibroblasts (14.5 dpc), (3) kidney cells (3-day-postnatal), (4) heart cells (3-day-postnatal), (5) spleen cells (3-day-postnatal), and (6) brain cells (14.5 dpc). The expression of TNFR1 and TNFR2 was analyzed by using RT-PCR. The expression of relA, c-rel, and relB was examined by using Northern blot analysis.
Sensitivity of RelA-Deficient Mice to Exogenous TNF.
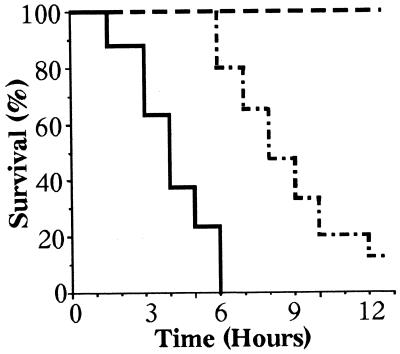
TNF was injected intravenously into newborn mice from intercrosses between TNF−/−relA+/− mice. With 1 μg of TNF, all TNF−/−relA−/− mice died within 6 hours, whereas TNF−/−relA+/− and TNF−/−relA+/+ mice survived longer than 12 hours (Fig. 7). Influence of relA gene dosage on TNF sensitivity was observed, with longer survival in relA+/+ than relA+/− mice. Similar results were obtained with 0.2 μg of TNF. In moribund double-deficient mice, no parenchymal damage was observed in liver or other organs. However, capillary damage was seen in liver, kidney, and lung, suggesting that endothelial cells are the main target of TNF injury in these mice. More extensive capillary damage was observed in TNF-treated TNF−/−relA−/− mice than in TNF−/−relA+/− and TNF−/−relA+/+ mice.
Figure 7.
Survival of newborn mice after injection of TNF. TNF−/− relA−/− (solid line; n = 8), TNF−/−relA+/− (dashed line; n = 15), TNF −/−relA+/+ (broken line; n = 5). Mice were injected with 1 μg of TNF intravenously.
DISCUSSION
Liver degeneration with absence of hepatocytes is the major pathology found in RelA-deficient 14.5-day embryos. TNF mediates this abnormality and the associated lethality, as viable RelA-deficient progeny without liver damage result from matings of TNF-deficient RelA-heterozygous mice with RelA-deficient mice. The hepatocytes derived from double-deficient mice are sensitive to the cytotoxic action of TNF in vitro, suggesting that loss of hepatocytes in RelA-deficient mice is caused by a direct action of TNF on hepatocytes. Another possibility is that the death of hepatocytes is secondary to endothelial cell apoptosis, and the capillary damage observed in the liver and other organs of TNF−/−relA−/− mice injected with TNF would support an indirect mechanism leading to hepatocyte loss. Whether caused by a direct or an indirect action of TNF on hepatocytes, the organ specificity (liver) and timing of TNF action in RelA deficient mice need explanation, especially because (i) TNF is widely expressed in embryonic tissues and is detectable before 14.5 days, (ii) organs other than liver (e.g., kidney) contain cells that are sensitive to TNF cytotoxicity, and (iii) capillary damage induced by TNF in double-deficient mice is not restricted to liver. Possibly, a surge of TNF production occurs around 14–15 days of embryonic life, and liver is the target of damage because a particularly high concentration of TNF is produced by Kuppfer cells in the developing liver.
In view of the cytotoxic action of TNF, we and others have long speculated that TNF is involved in the cell death that accompanies normal morphogenesis and organogenesis. However, no developmental abnormalities have been detected in TNF−/− mice, indicating that TNF does not have an irreplaceable role in this process (14, 23). Where TNF may be essential is in the elimination of cells damaged by chemical or physical agents or by infectious agents and their products. If this were the case, there would be a critical need for a mechanism, e.g., the NF-κB antiapoptotic signal, to protect normal cells. The increased TNF sensitivity of cells treated in vitro with protein or RNA synthesis inhibitors (3) may well have its counterpart in mice treated with these agents in vivo, and there is a speculation that TNF-mediated apoptosis is involved in the antitumor effects of irradiation and chemotherapeutic agents used in clinical practice. As a model for this surveillance role of TNF in eliminating damaged cells, mice injected with galactosamine (an agent that specifically inhibits transcriptional activity in hepatocytes) become exquisitely sensitive to bacterial lipopolysaccharide (25), with the liver of galactosamine/lipopolysaccharide-treated mice showing massive necrosis. This action of lipopolysaccharide and galactosamine is mediated by TNF (14, 23, 26, 27). In addition, cells infected with bacteria (28) or with a number of different viruses, including herpesviruses (29) and HIV (30), become sensitive to TNF-mediated cytotoxicity. Exotoxins, a product of Gram-positive organisms, also greatly enhance TNF cytotoxicity (31). In fact, the lethality of bacterial sepsis may well be due to the sensitization of cells in key organs by bacterial products to the apoptotic damage of TNF rather than to the currently held view that TNF-induced inflammatory damage and circulatory collapse are the primary pathogenic factors in septic shock, and recent evidence for TNF-mediated apoptosis of endothelial cells in lipopolysaccharide-treated mice (32) is consistent with this idea. It will be important to determine whether regulation of the NF-κB antiapoptotic signal is the unifying element in these examples of selective cytotoxic effects of TNF on damaged or infected cells. With the rapid expansion of the TNF ligand/TNFR superfamily, a number of new cytotoxic factors and new mechanisms for resistance to cytotoxicity have been identified, including expression of the decoy receptors, DcR1 and DcR2, in the TRAIL (Apo2)/DR5 and DR4 pathways (33–35), and tissue-restricted receptor expression in the Apo3L/DR3 (Apo3/Wsl1) system (36). However, recent evidence that NF-κB may have an important antiapoptotic role in the Apo3L/DR3 pathways (36, 37) suggests that activation of NF-κB may be a more general protective mechanism, and further studies of NF-κB-mediated antiapoptotic signals in other TNF ligand/TNFR superfamily are needed. TNF also is known to induce reactive oxygen species, and reactive oxygen species can mediate cytotoxicity and NF-κB activation (38). Defining the precise role of NF-κB in TNF-induced reactive oxygen species production, action, and protection of cells against reactive oxygen species cytotoxicity will be facilitated by availability of NF-κB-deficient mice. Similarly, the role of NF-κB in ceramide generation and apoptosis induction through the sphingomyelin pathway initiated by TNF can also be clarified in this way (39).
Cultured cells derived from several tissues of TNF/RelA double-deficient mice showed differential sensitivity to TNF; hepatocytes, kidney cells, and fibroblasts are sensitive to TNF cytotoxicity in vitro whereas those from heart, brain, and spleen are resistant. Given the capacity of high level c-rel expression to overcome the TNF sensitivity of cell lines expressing the IκB mutant (9), the expression of c-rel is a possible reason for the resistance of these cells to TNF. However, the contribution of c-rel to protection against TNF-mediated apoptosis or other cytotoxic agents has been difficult to determine because of the ubiquitous expression of RelA. TNF/RelA/c-rel triple-deficient and TNF/c-rel double-deficient offspring produced by a cross between TNF/RelA double-deficient mice and c-rel-deficient mice (40) should facilitate definition of the role of c-rel in NF-κB-dependent resistance to TNF.
The search for TNF-mediated/NF-κB-dependent “survival” genes has become a major objective in recent years. Several candidate “survival” genes have been identified, including IEX-1L (41), TRAF1 (TNFR-associated factor 1), TRAF2, c-IAP (cellular inhibitor of apoptosis)-1 and c-IAP-2 (42), and A20 (43). Transcription of IEX-1L is induced by TNF, and there is decreased IEX-1L transcription in NF-κB-compromised cells. Furthermore, IEX-1L expression in transfected relA−/− cells or in mutant IκBα cells renders these cells resistant to TNF treatment (41). The simultaneous expression of TRAF1, TRAF2, c-IAP-1, and c-IAP-2, but not the individual expression of these genes, was reported to protect relA−/− cells and mutant IκBα-transfected cells from TNF-mediated cytotoxicity (42). Although previous reports have established that TRAF2, c-IAP-1, and c-IAP-2 mediate NF-κB activation by TNF, recent evidence indicates that, like IEX-1L, all four of these genes appear to be rapidly up-regulated by NF-κB activation, (9, 44). Collectively, it seems likely that upon TNF receptor activation, the NF-κB/TRAF1, TRAF2, c-IAP-1, and c-IAP-2 pathways operate as a positive feedback system to amplify the survival signal to protect cells from TNF injury. With regard to the A20 zinc finger protein in TNF-mediated cytotoxicity, some studies show a protective role in TNF cytotoxicity (43), whereas others indicate that A20 inhibits NF-κB activation (45). Such contradictions might be resolved by the discovery of NF-κB-“independent” pathways, suggested already by studies on TRAF2-deficient mice and on transgenic mice with a dominant negative form of TRAF2 (46, 47). TRAF2-compromised cells showed increased sensitivity to TNF cytotoxicity despite normal activation of NF-κB by TNF. In TRAF2-compromised cells, Jun N-terminal kinase activation induced by TNF treatment was severely reduced, and genes downstream of Jun N-terminal kinase may also be involved in antiapoptosis. TNF/TRAF2 double- and TNF/TRAF2/RelA triple-deficient mice should facilitate testing of this hypothesis. How these candidate survival genes exert their antiapoptotic action is not resolved at present. A recent report indicated that the expression of TRAF1, TRAF2, c-IAP-1, and c-IAP-2 suppresses apoptosis by blocking activation of the initiator caspase, caspase-8 (42). Other studies, however, suggested that IAPs inhibit apoptosis by blocking activities of effector caspases (48). Furthermore, the NF-κB-dependent survival pathway has been thought to operate independently of Bcl-2, the other major antiapoptotic pathway. However, a recent study showed that Bcl-2 regulates NF-κB activation through induction of IκB degradation (49).
Clearly, a comprehensive picture of TNF-mediated/NF-κB-dependent antiapoptotic signaling has not yet emerged. Defining the nature of the NF-κB dependent antiapoptotic signal and identifying its site of action in the apoptotic cascade initiated by TNF are critical to understanding why normal cells are usually resistant to the cytotoxic actions of TNF, and why this resistance is sometimes compromised in injured cells. Furthermore, it will be important to know whether abnormalities in NF-κB signaling are central to the high sensitivity of certain transformed cells of mouse and human origin to TNF cytotoxicity that occurs in the absence of any added protein or RNA synthesis inhibitors.
ABBREVIATIONS
- TNF
tumor necrosis factor
- TNFR
TNF receptor
- RT-PCR
reverse transcription–PCR
- dpc
days postcoitum
References
- 1.Carswell E A, Old L J, Kassel R L, Green S, Fiore N, Williamson B. Proc Natl Acad Sci USA. 1975;72:3666–3670. doi: 10.1073/pnas.72.9.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williamson B D, Carswell E A, Rubin B Y, Prendergast J S, Old L J. Proc Natl Acad Sci USA. 1983;80:5397–5401. doi: 10.1073/pnas.80.17.5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruff M R, Gifford G E. Lymphokines. 1981;2:235–272. [Google Scholar]
- 4.Dealtry G B, Naylor M S, Fiers W, Balkwill F R. Eur J Immunol. 1987;17:689–693. doi: 10.1002/eji.1830170517. [DOI] [PubMed] [Google Scholar]
- 5.Rubin B Y, Smith L J, Hellermann G R, Lunn R M, Richardson N K, Anderson S L. Cancer Res. 1988;48:6006– 6010. [PubMed] [Google Scholar]
- 6.Ashkenazi A, Dixit V M. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 7.Baeuerle P A, Baichwal V R. Adv Immunol. 1997;65:111– 137. [PubMed] [Google Scholar]
- 8.Beg A A, Baltimore D. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 9.Liu Z, Hsu H, Goeddel D V, Karin M. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 10.Wang C-Y, Mayo M W, Baldwin A S., Jr Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 11.Van Antwerp D J, Martin S J, Kafri T, Green D R, Verma I M. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 12.Beg A A, Sha W C, Bronson R T, Ghosh S, Baltimore D. Nature (London) 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 13.Doi T S, Takahashi T, Taguchi O, Azuma T, Obata Y. J Exp Med. 1997;185:953–961. doi: 10.1084/jem.185.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marino M W, Dunn A, Grail D, Inglese M, Noguchi Y, Richards E, Jungbluth A, Wada H, Moore M, Williamson B, Basu S, Old L J. Proc Natl Acad Sci USA. 1997;94:8093–8098. doi: 10.1073/pnas.94.15.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blin N, Stafford D W. Nucleic Acids Res. 1976;3:2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 17.Nolan G P, Ghosh S, Liou H-C, Tempst P, Baltimore D. Cell. 1991;64:961–969. doi: 10.1016/0092-8674(91)90320-x. [DOI] [PubMed] [Google Scholar]
- 18.Bull P, Morley K L, Hoekstra M F, Hunter T, Verma I M. Mol Cell Biol. 1990;10:5473–5485. doi: 10.1128/mcb.10.10.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryseck R-P, Bull P, Takamiya M, Bours V, Siebenlist U, Dobrzanski P, Bravo R. Mol Cell Biol. 1992;12:674–684. doi: 10.1128/mcb.12.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheehan K C F, Pinckard J K, Arthur C D, Dehner L P, Goeddel D V, Schreiber R D. J Exp Med. 1995;181:607– 617. doi: 10.1084/jem.181.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohchi C, Noguchi K, Tanabe Y, Mizuno D, Soma G. Int J Biochem. 1994;26:111–119. doi: 10.1016/0020-711x(94)90203-8. [DOI] [PubMed] [Google Scholar]
- 22.Pollard J W, Walker J M, editors. Methods in Molecular Biology. Vol. 5. Clifton, NJ: Humana; 1990. [Google Scholar]
- 23.Pasparakis M, Alexopoulou L, Episkopou V, Kollias G. J Exp Med. 1996;184:1397–1411. doi: 10.1084/jem.184.4.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horwitz B H, Scott M L, Cherry S R, Bronson R T, Baltimore D. Immunity. 1997;6:765–772. doi: 10.1016/s1074-7613(00)80451-3. [DOI] [PubMed] [Google Scholar]
- 25.Galanos C, Freudenberg M A, Reutter W. Proc Natl Acad Sci USA. 1979;76:5939–5943. doi: 10.1073/pnas.76.11.5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfeffer K, Matsuyama T, Kundig T M, Wakeham A, Kishihara K, Shahinian A, Wiegmann K, Ohashi P S, Kronke M, Mak T W. Cell. 1993;73:457–467. doi: 10.1016/0092-8674(93)90134-c. [DOI] [PubMed] [Google Scholar]
- 27.Rothe J, Lesslauer W, Lotscher H, Lang Y, Koebel P, Kontgen F, Althage A, Zinkernagel R, Steinmetz M, Bluethmann H. Nature (London) 1993;364:798–802. doi: 10.1038/364798a0. [DOI] [PubMed] [Google Scholar]
- 28.Klimpel G R, Shaban R, Niesel D W. J Immunol. 1990;145:711–717. [PubMed] [Google Scholar]
- 29.Koff W C, Fann A V. Lymphokine Res. 1986;5:215–221. [PubMed] [Google Scholar]
- 30.Wong G H W, Krowka J F, Stites D P, Goeddel D V. J Immunol. 1988;140:120–124. [PubMed] [Google Scholar]
- 31.Morimoto H, Safrit J T, Bonavida B. J Immunol. 1991;147:2609–2616. [PubMed] [Google Scholar]
- 32.Haimovitz-Friedman A, Cordon-Cardo C, Bayoumy S, Garzotto M, McLoughlin M, Gallily R, Edwards C K, III, Schuchman E H, Fuks Z, Kolesnick R. J Exp Med. 1997;186:1831–1841. doi: 10.1084/jem.186.11.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan G, Ni J, Wei Y-F, Yu G-L, Gentz R, Dixit V M. Science. 1997;277:815–818. doi: 10.1126/science.277.5327.815. [DOI] [PubMed] [Google Scholar]
- 34.Sheridan J P, Marsters S A, Pitti R M, Gurney A, Skubatch M, Baldwin D, Ramakrishnan L, Gray C L, Baker K, Wood W I, et al. Science. 1997;277:818–821. doi: 10.1126/science.277.5327.818. [DOI] [PubMed] [Google Scholar]
- 35.Mongkolsapaya J, Cowper A E, Xu X-N, Morris G, McMichael A J, Bell J I, Screaton G R. J Immunol. 1998;160:3–6. [PubMed] [Google Scholar]
- 36.Chinnaiyan A M, O’Rourke K, Yu G-L, Lyons R H, Garg M, Duan D R, Xing L, Gentz R, Ni J, Dixit V M. Science. 1996;274:990–992. doi: 10.1126/science.274.5289.990. [DOI] [PubMed] [Google Scholar]
- 37.Marsters S A, Sheridan J P, Donahue C J, Pitti R M, Gray C L, Goddard A D, Bauer K D, Ashkenazi A. Curr Biol. 1996;6:1669–1676. doi: 10.1016/s0960-9822(02)70791-4. [DOI] [PubMed] [Google Scholar]
- 38.McGowan A J, Bowie A G, O’Neill L A J, Cotter T G. Exp Cell Res. 1998;238:248–256. doi: 10.1006/excr.1997.3853. [DOI] [PubMed] [Google Scholar]
- 39.Hannun Y A. Science. 1996;274:1855–1859. doi: 10.1126/science.274.5294.1855. [DOI] [PubMed] [Google Scholar]
- 40.Kontgen F, Grumont R J, Strasser A, Metcalf D, Li R, Tarlinton D, Gerondakis S. Genes Dev. 1995;9:1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- 41.Wu M X, Ao Z, Prasad K V S, Wu R, Schlossman S F. Science. 1998;281:998–1001. doi: 10.1126/science.281.5379.998. [DOI] [PubMed] [Google Scholar]
- 42.Wang C-Y, Mayo M W, Korneluk R G, Goeddel D V, Baldwin A S., Jr Science. 1998;281:1680–1683. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- 43.Opipari A W, Jr, Hu H M, Yabkowitz R, Dixit V M. J Biol Chem. 1992;267:12424–12427. [PubMed] [Google Scholar]
- 44.Chu Z-L, McKinsey T A, Liu L, Gentry J J, Malim M H, Ballard D W. Proc Natl Acad Sci USA. 1997;94:10057–10062. doi: 10.1073/pnas.94.19.10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song H Y, Rothe M, Goeddel D V. Proc Natl Acad Sci USA. 1996;93:6721–6725. doi: 10.1073/pnas.93.13.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee S Y, Reichlin A, Santana A, Sokol K A, Nussenzweig M C, Choi Y. Immunity. 1997;7:703–713. doi: 10.1016/s1074-7613(00)80390-8. [DOI] [PubMed] [Google Scholar]
- 47.Yeh W-C, Shahinian A, Speiser D, Kraunus J, Billia F, Wakeham A, de la Pompa J L, Ferrick D, Hum B, Iscove N, et al. Immunity. 1997;7:715–725. doi: 10.1016/s1074-7613(00)80391-x. [DOI] [PubMed] [Google Scholar]
- 48.Roy N, Deveraux Q L, Takahashi R, Salvesen G S, Reed J C. EMBO J. 1997;16:6914–6925. doi: 10.1093/emboj/16.23.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Moissac D, Mustapha S, Greenberg A H, Kirshenbaum L A. J Biol Chem. 1998;273:23946–23951. doi: 10.1074/jbc.273.37.23946. [DOI] [PubMed] [Google Scholar]